Abstract
A linear polyacrylamide (LPA) coating and a sulfonate-silica hybrid strong cation exchange (SCX) monolith were prepared sequentially in a single fused silica capillary. The SCX-LPA capillary was used for sample preconcentration using solid-phase extraction (SPE) followed by capillary zone electrophoresis (CZE) separation and mass spectrometry detection. Samples were prepared in 1 M acetic acid, loaded by pressure onto the SCX SPE monolith, and eluted using 200 mM ammonium bicarbonate (pH 8). The background electrolyte for CZE was 1 M acetic acid. This combination of elution buffer and background electrolyte results in formation of a dynamic pH junction, which further improves the preconcentration performance. All experiments were performed using a CZE autosampler and an electrokinetically pumped nanospray interface. By loading 50 ng of Xenopus laevis eggs protein digest, 330 protein groups and 872 peptides were identified. The system was also applied to the analysis of 11 μL of a 10−4 mg⋅mL−1 (1.1 ng) bovine serum albumin tryptic digest; 12 peptides were identified and the protein coverage was 25%. Finally, by loading 5.5 μL of a 10−3 mg⋅mL−1 (5.5 ng) E. coli digest, 145 protein groups and 365 peptides were identified.

A linear polyacrylamide coating and a sulfonate-silica hybrid cation exchange monolith were prepared sequentially in a single fused silica capillary and coupled to the capillary zone electrophoresis-mass spectrometry for automated online sample preconcentration.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Capillary zone electrophoresis-mass spectrometry (CZE-MS) has been widely used for analysis of proteins and peptides [1–5]. This method typically employs small sample injection volumes that lead to low column capacity and poor concentration detection limits. To overcome this limitation, various preconcentration techniques have been developed, including sample stacking [6, 7], dynamic pH junction [8–10], and solid phase extraction (SPE) [11–17], which have been summarized in reviews [18–22].
We have reported the preparation of a strong cationic exchange (SCX) hybrid monolith at the inlet of the separation capillary and its use for sample preconcentration, processing, and subsequent separation using CZE with high concentration of acetic acid as background electrolyte [15, 16, 22]. Samples are loaded in an acidic solution and desorbed using a basic solution. This plug of sample forms a pH junction in the low-pH background electrolyte, leading to outstanding preconcentration of tryptic peptides.
In our first generation device, the monolith was prepared in an uncoated capillary [15]. After preconcentration and elution, analyte were separated by CZE in the uncoated capillary. The high electro-osmotic flow produced in the uncoated capillary results in a short separation window, which limits the number of tandem mass spectra that can be generated for peptide identifications. In our second generation device, the monolith was prepared independently and connected to a linear polyacrylamide-coated capillary with a zero-dead volume connector [16]. After preconcentration and elution, analyte were separated in the coated capillary, which provided low electroosmotic flow and a 30-min separation window. A union is required to couple the SPE column or microreactor to the coated separation capillary [13, 18, 19]. However, the connector can introduce dead volume that can degrade separation, and coupling of the monolith to the separation capillary requires a fair degree of training and manual dexterity. To overcome these problems and to take advantage of the well-developed methods for the preparation of SCX hybrid monoliths and LPA coated capillaries [23, 24], we now report the preparation of an LPA coating and a SCX hybrid monolith in a single capillary, and employ it for ultrasensitive bottom-up analysis of complex proteomes.
Experimental section
Reagents and chemicals
Bovine serum albumin (BSA), bovine pancreas TPCK-treated trypsin, poly(ethylene glycol) (PEG, MW = 10,000), urea, ammonium bicarbonate (NH4HCO3), dithiothreitol (DTT), iodoacetamide (IAA), γ-methacryloxypropyltrimethoxysilane (γ-MAPS), tetramethoxysilane (TMOS), vinyltrimethoxysilane (VTMS), 3-sulfopropyl methacrylate potassium salt, 2,2′-azobis(2-isobutyronitrile) (AIBN), and acetic acid were purchased from Sigma-Aldrich (St. Louis, MO, http://www.sigmaaldrich.com/united-states.html). Formic acid (FA) was purchased from Fisher Scientific (Pittsburgh, PA, https://www.fishersci.com/us/en/home.html). Methanol was purchased from Honeywell Burdick & Jackson (Wicklow, Ireland, https://labchemicals-honeywell.com/). Water was deionized by a Nano Pure system from Thermo Scientific (Marietta, OH, http://www.thermofisher.com/us/en/home.html). Fused silica capillaries were purchased from Polymicro Technologies (Phoenix, AZ, USA, http://www.molex.com/molex/home). The Xenopus laevis eggs were donated by Prof. Paul W. Huber from the University of Notre Dame and the E. coli cells (Dh5-Alpha) were cultured in our lab.
Preparation of the LPA coated capillary
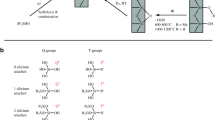
A diagram of the procedure is shown in Fig. 1. The fused silica capillary (365 μm OD × 50 μm ID, 95 cm total length) was pretreated by flushing with 0.1 M NaOH for 2 h, water until the outflow reached pH 7.0, 0.1 M HCl for 24 h, and water until the outflow reached pH 7.0. The capillary was dried with a nitrogen stream for 12 h at room temperature. Next, the γ-MAPS solution in methanol at a volume ratio of 1:1 was injected with a syringe to fill 90 cm (out of 95 cm total length) of the capillary, leaving ~5 cm untreated for the subsequent preparation of the SCX hybrid monolith. Then the capillary was sealed with rubber at both ends and submerged in a water bath at 45 °C overnight [25]. Finally, the capillary was rinsed with methanol and water to flush out residual reagents and dried with a nitrogen stream. The preparation of the LPA coated capillary followed the method reported previously [24]. Briefly, 40 mg of acrylamide was dissolved in 1 mL water. 2 μL of 5% (w/v) ammonium persulfate (APS) was added to the 500 μL of acrylamide solution. The mixture was vortexed for 30 s and degassed for 5 min using nitrogen. Then the mixture was used to fill the first 90-cm of the pretreated capillary. With both ends sealed, the capillary was incubated at 50 °C in a water bath for 30 min. Unreacted reagents were flushed by water and the coated capillary was stored at room temperature.
Preparation of the sulfonate-silica hybrid SCX-SPE monolith
The preparation of the sulfonate SCX hybrid monolithic column was similar to published methods [13, 14, 18, 20]. As shown in Fig. 1, the SCX monolith was prepared in the 5 cm uncoated section of the capillary. A prehydrolyzed mixture was prepared by mixing and stirring acetic acid (10 mM, 5.0 mL), PEG (MW = 10,000, 540 mg), urea (450 mg), TMOS (1.8 mL), and VTMS (0.6 mL) for 1 h at 0 °C to form a homogeneous solution. Then, 37 mg of 3-sulfopropyl methacrylate potassium salt and 2 mg of AIBN were added into 0.5 mL of the hydrolyzed mixture for 10 min with sonication. The polymerization mixture was then introduced into 5 cm long, uncoated section of the LPA coated capillary using a syringe. Both ends of the capillary were then sealed with pieces of rubber, and the capillary was incubated at 37 °C for 12 h for polycondensation and at 60 °C for 12 h for polymerization. The sulfonate SCX hybrid column was then flushed with methanol and water to remove the porogen and other residual reagents. Finally, the sulfonate SCX hybrid monolithic column was trimmed to ~1.5 cm.
Column capacity measurement
To determine the dynamic binding capacity of the SCX hybrid monolith, a Xenopus protein digest was used to saturate the sulfonate SCX hybrid monolithic SPE column. Briefly, different amounts of 0.5 mg⋅mL−1 Xenopus protein digest in 1 M acetic acid were loaded onto the sulfonate SCX hybrid monolithic SPE column. After flushed with 1 M acetic acid, the protein digest trapped on the SCX hybrid monolithic column was eluted by 100 nL 200 mM NH4HCO3 and analyzed by CZE-MS/MS. The numbers of peptide and protein identifications were used to evaluate the dynamic binding capacity of the SCX hybrid monolithic SPE column.
Sample preparation
Details of sample preparation are provided in the Supporting information.
Automated on-line SCX-SPE CZE-ESI-MS/MS analysis
The CZE system consists of a PrinCE Next 840 Series autosampler (from PrinCE Technologies), high-voltage power supplies (Spellman CZE 1000R), and an electrokinetically pumped nanospray interface that coupled the CZE separation capillary to a LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) [26–28]. The electrospray emitter was a borosilicate glass capillary (1.0 mm OD × 0.75 mm ID, 10 cm long) pulled with a Sutter instrument P-1000 flaming/brown micropipette puller; the emitter inner diameter was 15–20 μm. The electrospray sheath flow electrolyte was 10% (v/v) methanol with 0.5% aqueous FA. The separation background electrolyte was 1 M acetic acid in water. The injection end of the capillary was held in the auto sampler that allowed application of either pressure to pump fluids or voltage to drive electrophoresis. The separation capillary and the SCX hybrid monolith were first conditioned by washes with 1 M acetic acid at 1500 psi for 30 min. The samples prepared in 1 M acetic acid were hydrodynamically injected for varying lengths of time. Unretained components were removed between runs by rinsing with 1 M acetic acid. Peptide elution was performed by hydrodynamic injection of 200 mM NH4HCO3 (pH 8). After elution, the electrolyte was changed back to 1 M acetic acid and normal CZE-ESI-MS/MS analysis was carried out with a separation voltage of 19.8 kV and a spray voltage of 1.8 kV. Between runs, the SCX hybrid monolithic column was rinsed with 100 nL 200 mM NH4HCO3 (pH 8) to minimize carry-over.
Mass spectrometer operating parameters
An LTQ-Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) was used in this experiment. The ion transfer tube temperature was held at 280 ˙C. The S-Lens RF level was 50.00. The mass spectrometer was programmed in data-dependent mode. A top 10 method was used. Full MS scans were acquired with the Orbitrap mass analyzer over m/z 380–1800 range with resolution of 70,000 (m/z 200) and the number of microscans set to 1. The target value was 1.00E + 06, and maximum injection time was 250 ms. For MS/MS scans, the twenty most intense peaks with charge state ≥2 were sequentially isolated and further fragmented by Collision-Induced Dissociation (CID) following one full MS scan. The normalized collision energy was 33%.
Database searching
Database searching of the raw files was performed in Proteome Discoverer 1.4 with MASCOT 2.2.4. For BSA digest, ipi.bovin.v3.68.fasta was used for database searching. For the E. coli cell lysate, an NCBI database of E. coli DH1 (4160 sequences) was used. For the Xenopus digest, the UniProt Xenopus reference protein database (48,523 protein sequences) was used for database searching. Database searching using the reversed databases were also performed to evaluate the false discovery rates. The database searching parameters included full tryptic digestion and allowed up to two missed cleavages, precursor mass tolerance 10 ppm, and fragment mass tolerance 0.5 Da. Carbamidomethylation (C) was set as fixed modifications. Oxidation (M) and deamidated (NQ) were set as variable modifications. On the peptide level, peptide confidence value as high was used to filter the peptide identification, and the corresponding false discovery rate on peptide level was less than 1%. On the protein level, protein grouping was enabled.
Results and discussion
Choice of materials
To improve the CZE separation, capillaries with neutral coatings are usually used to minimize electroosmotic flow and to reduce the adsorption of solutes onto its inner surface [5, 16, 24]. Linear polyacrylamide (LPA) coated capillaries are most widely used for the analysis of proteins and peptides due to the good hydrophilic property of the coating and the simple coating protocol [16, 24]. As a result, we used LPA coating in our work.
Solid phase extraction (SPE) monoliths typically fall into two types: reversed phase (RP) [12–14], and ion exchange [15, 16, 29]. We have observed a number of problems when coupling a reversed phase SPE monolith to a LPA coated capillary. The low electroosmotic flow of the LPA coated capillary, coupled with low electrophoretic migration of the sample after elution with organic solvents from the reversed phase monolith, led to a very long migration time and significant band broadening. In contrast, the SCX phase generated much better results with the LPA coated capillary because no organic solvent is required for elution. Moreover, the formation of a dynamic pH junction after elution from the SCX-SPE monolith further improved the CZE separation performance. Therefore, the SCX phase was used as SPE column in this work.
Loading capacity of the SCX SPE column
A Xenopus protein digest was loaded onto the SCE-SPE. When the loading amount increased from 5 to 50 ng, the number of protein group IDs increased from 225 to 330 and the number of peptide IDs increased from 528 to 872, Fig. 2. The protein groups and sequences of the identified peptides are listed in supporting information.
Column capacity of the SCX hybrid monolith using a Xenopus laevis protein digest. Experimental conditions: 50 μm ID × 365 μm OD × 1.5 cm SCX microreactor, 50 μm ID × 365 μm OD × 90 cm long LPA coated capillary as separation capillary, 100 nL of 200 mM NH4HCO3 (pH 8.2) elution buffer, 1 M acetic acid separation background electrolyte, 19.5 kV separation voltage, and 1.5 kV spray voltage
Representative electropherograms are shown in Fig. 3. The larger sample loading resulted in a minor delay in the appearance of peptides during electrophoresis, presumably due to the presence of the large loading electrolyte.
Butterfly plots of base peak electropherograms of a Xenopus protein digests. Experimental conditions are same as Fig. 2
Large-volume proteomic analysis
To evaluate the performance of the SCX SPE-LPA system for the analysis of low concentration digests, 11 μL of a 10−4 mg⋅mL−1 BSA tryptic digest was loaded onto the SCX hybrid monolith and eluted with 100 nL of 200 mM NH4HCO3 (pH 8). 12 peptides were identified and the protein coverage was 25%, which is two times higher than that reported previously without LPA coating [18]. The sequences of the identified peptides are listed in the supporting information.
To further evaluate the performance of the system, E. coli digests were prepared in 1 M acetic acid. A 5.5 μL aliquot of a 10−3 mg⋅mL−1 tryptic digest was hydrodynamically loaded onto the SCX SPE monolith and eluted with 100 nL of 200 mM NH4HCO3 (pH 8), 145 protein groups and 365 peptides were identified, a ~ 33% improvement compared to previous results [18]. The protein groups and sequences of the identified peptides are listed in the supporting information. The results also confirmed the importance of LPA coating for the improvement of separation performance and the decrease of the sample adsorption on the inner surface of the capillary.
Repeatability and stability
The repeatability was evaluated by loading 10 ng of BSA digestion and eluting using 100 nL of 200 mM NH4HCO3 buffer. The RSD for protein coverage was 5% (n = 2) and for number of unique peptides was 4% (n = 2). For column-to-column, the RSD for protein coverage was 7% (N = 2) and for the number of unique peptides was 1% (n = 2). The sequences of the identified peptides are listed in the supporting information.
References
Robledo VR, Smyth WF (2014) Review of the CE-MS platform as a powerful alternative to conventional couplings in bio-omics and target-based applications. Electrophoresis 35(16):2292–2308. doi:10.1002/elps.201300561
Stepanova S, Kasicka V (2016) Recent developments and applications of capillary and microchip electrophoresis in proteomic and peptidomic analyses. J Sep Sci 39(1):198–211. doi:10.1002/jssc.201500973
Stepanova S, Kasicka V (2016) Recent applications of capillary electromigration methods to separation and analysis of proteins. Anal Chim Acta 933:23–42. doi:10.1016/j.aca.2016.06.006
Heemskerk AAM, Deelder AM, Mayboroda OA (2016) CE-ESI-MS for bottom-up proteomics: advances in separation, interfacing and applications. Mass Spectrom Rev 35(2):259–271. doi:10.1002/mas.21432
Mesbah K, Mai TD, Jensen TG, Sola L, Chiari M, Kutter JP, Taverna M (2016) A neutral polyacrylate copolymer coating for surface modification of thiol-ene microchannels for improved performance of protein separation by microchip electrophoresis. Microchim Acta 183(7):2111–2121. doi:10.1007/s00604-016-1825-4
Quirino JP, Terabe S (1999) Sample stacking of fast-moving anions in capillary zone electrophoresis with pH-suppressed electroosmotic flow. J Chromatogr A 850(1–2):339–344. doi:10.1016/S0021-9673(99)00204-6
Chien RL (2003) Sample stacking revisited: a personal perspective. Electrophoresis 24(3):486–497. doi:10.1002/elps.200390057
Britz-McKibbin P, Otsuka K, Terabe S (2002) On-line focusing of flavin derivatives using dynamic pH junction-sweeping capillary electrophoresis with laser-induced fluorescence detection. Anal Chem 74(15):3736–3743. doi:10.1021/ac025701o
Zhu GJ, Sun LL, Yan XJ, Dovichi NJ (2014) Bottom-up proteomics of Escherichia coli using dynamic pH junction preconcentration and capillary zone electrophoresis-electrospray ionization-tandem mass spectrometry. Anal Chem 86(13):6331–6336. doi:10.1021/ac5004486
Kazarian AA, Hilder EF, Breadmore MC (2011) Online sample pre-concentration via dynamic pH junction in capillary and microchip electrophoresis. J Sep Sci 34(20):2800–2821. doi:10.1002/jssc.201100414
Ye H, Xia S, Lin W, Yu L, Xu X, Zheng C, Liu X, Chen G (2010) CE-ESI-MS coupled with dynamic pH junction online concentration for analysis of peptides in human urine samples. Electrophoresis 31(20):3400–3406. doi:10.1002/elps.201000194
Tomlinson AJ, Benson LM, Guzman NA, Naylor S (1995) Preliminary investigations of preconcentration-capillary electrophoresis-mass spectrometry. J Chromatogr A 669(1):67–73. doi:10.1016/0378-4347(95)00127-5
Figeys D, Ducret A, Yates JR, Aebersold R (1996) Protein identification by solid phase microextraction-capillary zone electrophoresis-microelectrospray-tandem mass spectrometry. Nat Biotechnol 14(11):1579–1583. doi:10.1038/nbt1196-1579
Wang YJ, Fonslow BR, Wong CCL, Nakorchevsky A, Yates JR (2012) Improving the comprehensiveness and sensitivity of sheathless capillary electrophoresis-tandem mass spectrometry for proteomic analysis. Anal Chem 84(20):8505–8513. doi:10.1021/ac301091m
Zhang ZB, Sun LL, Zhu GJ, Yan XJ, Dovichi NJ (2015) Integrated strong cation-exchange hybrid monolith coupled with capillary zone electrophoresis and simultaneous dynamic pH junction for large-volume proteomic analysis by mass spectrometry. Talanta 138:117–122. doi:10.1016/j.talanta.2015.01.040
Zhang ZB, Yan XJ, Sun LL, Zhu GJ, Dovichi NJ (2015) Detachable strong cation exchange monolith, integrated with capillary zone electrophoresis and coupled with pH gradient elution, produces improved sensitivity and numbers of peptide identifications during bottom-up analysis of complex proteomes. Anal Chem 87(8):4572–4577. doi:10.1021/acs.analchem.5b00789
Benavente F, Vescina MC, Hernández E, Sanz-Nebot V, Barbosa J, Guzman NA (2007) Lowering the concentration limits of detection by on-line solid-phase extraction-capillary electrophoresis-electrospray mass spectrometry. J Chromatogr A 1140(1–2):205–212. doi:10.1016/j.chroma.2006.11.092
Breadmore MC, Thabano JRE, Dawod M, Kazarian AA, Quirino JP, Guijt RM (2009) Recent advances in enhancing the sensitivity of electrophoresis and electrochromatography in capillaries and microchips (2006-2008). Electrophoresis 30(1):230–248. doi:10.1002/elps.200800435
Ramautar R, Somsen GW, de Jong GJ (2016) Developments in coupled solid-phase extraction-capillary electrophoresis 2013-2015. Electrophoresis 37(1):35–44. doi:10.1002/elps.201500401
Breadmore MC, Tubaon RM, Shallan AI, Phung SC, Abdul Keyon AS, Gstoettenmayr D, Prapatpong P, Alhusban AA, Ranjbar L, See HH, Dawod M, Quirino JP (2015) Recent advances in enhancing the sensitivity of electrophoresis and electrochromatography in capillaries and microchips (2012-2014). Electrophoresis 36(1):36–61. doi:10.1002/elps.201400420
Puig P, Borrull F, Calull M, Aguilar C (2007) Recent advances in coupling solid-phase extraction and capillary electrophoresis (SPE-CE). TrAC Trends Anal Chem 26(7):664–678. doi:10.1016/j.trac.2007.05.010
Zhu GJ, Sun LL, Dovichi NJ (2016) Dynamic pH junction preconcentration in capillary electrophoresis-electrospray ionization-mass spectrometry for proteomics analysis. Analyst 141(18):5216–5220. doi:10.1039/c6an01140c
Zhang ZB, Wang FJ, Xu B, Qin HQ, Ye ML, Zou HF (2012) Preparation of capillary hybrid monolithic column with sulfonate strong cation exchanger for proteome analysis. J Chromatogr A 1256:136–143. doi:10.1016/j.chroma.2012.07.071
Zhu GJ, Sun LL, Dovichi NJ (2016) Thermally-initiated free radical polymerization for reproducible production of stable linear polyacrylamide coated capillaries, and their application to proteomic analysis using capillary zone electrophoresis-mass spectrometry. Talanta 146:839–843. doi:10.1016/j.talanta.2015.06.003
Wu RA, Zou HF, Fu HJ, Jin WH, Ye ML (2002) Separation of peptides on mixed mode of reversed-phase and ion-exchange capillary electrochromatography with a monolithic column. Electrophoresis 23(9):1239–1245. doi:10.1002/1522-2683(200205)23:9<1239::AID-ELPS1239>3.0.CO;2-X
Wojcik R, Dada OO, Sadilek M, Dovichi NJ (2010) Simplified capillary electrophoresis nanospray sheath-flow interface for high efficiency and sensitive peptide analysis. Rapid Commun Mass Spectrom 24:2554–2560. doi:10.1002/rcm.4672
Sun LL, Zhu GJ, Zhao YM, Yan XJ, Mou S, Dovichi NJ (2013) Ultrasensitive and fast bottom-up analysis of femtogram amounts of complex proteome digests. Angew Chem Int Ed Eng 52(51):13661–13664. doi:10.1002/anie.201308139
Sun LL, Zhu GJ, Zhang ZB, Mou S, Dovichi NJ (2015) Third-generation electrokinetically pumped sheath-flow nanospray interface with improved stability and sensitivity for automated capillary zone electrophoresis-mass spectrometry analysis of complex proteome digests. J Proteome Res 14(5):2312–2321. doi:10.1021/acs.jproteome.5b00100
Zhang ZB, Sun LL, Zhu GJ, Cox OF, Huber PW, Dovichi NJ (2016) Nearly 1000 protein identifications from 50 ng of Xenopus laevis zygote homogenate using online sample preparation on a strong cation exchange monolith based microreactor coupled with capillary zone electrophoresis. Anal Chem 88(1):877–882. doi:10.1021/acs.analchem.5b03496
Acknowledgements
We thank Dr. William Boggess in the Notre Dame Mass Spectrometry and Proteomics Facility for his help with this project. We also thank Professor Paul Huber of the University of Notre Dame for providing the Xenopus eggs used in this experiment.
This work was funded by the National Institutes of Health (Grants R01GM096767 and R01HD084399). Elizabeth H. Peuchen acknowledges support from a National Science Foundation Graduate Research Fellowship (Grant No. DGE-1313583).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no competing interests.
Additional information
Associated content
Additional information as noted in text. This material is available free of charge via the Internet.
Rights and permissions
About this article
Cite this article
Zhang, Z., Zhu, G., Peuchen, E.H. et al. Preparation of linear polyacrylamide coating and strong cationic exchange hybrid monolith in a single capillary, and its application as an automated platform for bottom-up proteomics by capillary electrophoresis-mass spectrometry. Microchim Acta 184, 921–925 (2017). https://doi.org/10.1007/s00604-017-2084-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00604-017-2084-8