Abstract
Cryptosporidium is one of the most common, worldwide diarrheal diseases caused by parasites. Due to absence of an effective treatment, determining the prevailing species of Cryptosporidium is a key in identifying its transmission dynamics and a necessary precursor required for the planning and implementation of effective preventive and control strategies. This PCR-RFLP study was done to determine the prevalence of Cryptosporidium species in the stool of a cohort of Egyptian children and evaluate/assess associated risk factors for susceptibility to cryptosporidiosis, due to the lack of existent studies addressing Cryptosporidium transmission dynamics in humans and assessed risk factors in Egypt. Stool samples were collected from 431 children; 331 diarrheic and 100 apparently healthy non-diarrheic children; their data were recorded. Samples were processed for Copro-nPCR targeting Hsp90 gene and PCR-RFLP analysis for species identification. Variables which showed statistical significance for Cryptosporidium were included in a logistic regression analysis to identify the estimated risk. Out of 84 (19.5%) Cryptosporidium-positive samples (78 diarrheic and 6 non-diarreic), 75 (89.3%) were Cryptosporidium hominis, 6 (7.1%) were Cryptosporidium parvum, and 3 (3.6%) were non-typed. There was a significant association between Cryptosporidium detection in stool and the estimated risk factors: diarrhea, soft stool, and drinking from tap water. Cryptosporidium is an indigenous, prevailing intestinal parasite among children in Cairo that physicians must consider, especially in diarrheic, preschool-aged children, who drink from tap water. The finding of a predominance of C. hominis indicates anthroponotic rather than zoonotic transmission.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cryptosporidium possesses 24 valid species and more than 44 genotypes infecting many vertebrates; including humans and animals, which differ significantly in their molecular signatures (Cama et al. 2008). Thirteen cases of intestinal and gastric Cryptosporidium species infecting immunocompetent and immunocompromised humans have been reported until now (C. parvum, C. hominis, C. meleagridis, C. felis, C. canis, C. suis, C. muris, C. andersoni, C. fayeri, C. cuniculus, C. ubiquitum, and C. viatorum) (Fayer et al. 2010; Elwin et al. 2012). Among them, C. parvum (which infects not only humans but also ruminants and perhaps a few other animals) and C. hominis (which is almost exclusively a human parasite) are the most common species involved in clinical infections (Sulaiman et al. 2005).
Cryptosporidium is listed as a neglected disease by the World Health Organization, largely due to a lack of studies in developing countries, but is now gaining increasing attention (Savioli et al. 2006).
Because of the limitations in the specific detection of Cryptosporidium using microscopic, immunological, and/or flowcytometric methods, a wide range of nucleic acid-based methods had been developed and evaluated for the identification of species, as well as the detection of genetic variation within and among species of Cryptosporidium with a high diagnostic yield (Smith et al. 2006).
The current study aimed to determine the prevalence of Cryptosporidium among a cohort of Egyptian children targeting the Hsp90 gene and RFLP analysis for species identification. Due to the lack of existent studies assessing risk factors in Egypt, this study was designed to investigate the role of collected data variables for susceptibility to Cryptosporidium infection among individuals.
Methods
Sampling and data collection
A cross-sectional study was designed involving 431 stool samples (331 diarrheic and 100 apparently healthy non-diarrheic children) collected from children of both sexes ranging in age from 1 to 12 years, presenting with diarrhea and/or other GIT symptoms recruited from the outpatient clinic in Abu El Rish pediatric hospital, Kasr Al-Ainy School of Medicine, and Cairo University from April 2013 to January 2014. Data included the recording of their sociodemographic and environmental data. Parents of young children provided consent forms and responded to the questionnaire.
Extraction of the genomic DNA
Genomic DNA was extracted using Favor Prep stool DNA isolation Mini Kit (Favorgen Biotech corporation ping-Tung 908, Taiwan, Cat. No. FASTI001) with modification in the form of prolongation of incubation to 95 °C for 1 h after thermal shock (cycling of deep freezing in liquid nitrogen for 5 min and immediately transferred into a water bath 95 °C for 5 min; repeated for 5 cycles) then the purified DNA was measured for concentration and purity.
Copro-nPCR/RFLP analysis
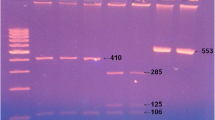
The nPCR was done by analysis of Hsp90 gene. Amplification of 835–844 for the primary reaction and a fragment of 676–685 for the secondary reaction, using two sets of oligonuclutide primers (Table 1) with the reaction components and the cycling conditions carried out according to Feng et al. (2009) with modification in the form of 12.5 μl master mix, 200 nM from each primer, and 3 μl of the template DNA for the primary reaction and 1 μl for the secondary one in a total volume of 25 μl and 50 °C annealing temperature for the primary and the secondary reactions. The amplified products were visualized with 1.5% agarose gel electrophoresis after ethidium bromide staining. The amplified products of nPCR of positive samples were digested using StyI and HphI endonuclease (Table 2) according to manufacturer’s instruction and resolved, using 3% metaphor electrophoresis after ethidium bromide staining.
Statistical analysis
Data were statistically analyzed using the statistical package SPSS version 17 (Chicago, IL, USA). Data were analyzed with Fisher᾿s exact test and multiple logistic regression. All variables that were significantly associated with the Cryptosporidium prevalence in the univariate model were included in a multivariate logistic regression.
Results
Out of 431 examined stool samples with nPCR, 84 (19.5%) stool samples were Cryptosporidium positive (78 diarrheic and 6 non-diarrheic); among them 75 (89.3%) were C. hominis, 6 (7.1%) were C. parvum, and 3 (3.6%) were non-typed. Their relative data were tabulated in Table 3.
Among the studied variables, only diarrhea, the type of water, and stool consistency were significantly associated (P˂0.05) with detection of Cryptosporidium. After these variables were subjected to a multivariate analysis using logistic regression, there was an estimated increase in the risk of Cryptosporidium among children suffering from diarrhea 4.8 times greater than the non-diarrheic children, with soft and liquid stools, 6.5 and 3.3 times, respectively, relative to formed stool, and 4.4 times for children drinking tap water relative to mineral water (Table 4).
In contrast, none of the studied variables, including gender distribution, age distribution, animal contact habits, and gross features of stool showed any significant association among genotypes (P > 0.05) (Table 4).
Discussion
Cryptosporidium is a highly prevailing parasite among studied diarrheic children for about a quarter of them (23.6%) using nPCR, with an overall prevalence of 19.5% in both groups. There is a difference in the reported prevalence of Cryptosporidium in Egypt in the last decades; most of the studies report a high molecular prevalence of up to a quarter of examined patients (El-Settawy and Fathy 2012; Fathy et al. 2014; Ghallab et al. 2016), however, much lower results (4.6%) were also reported in Egypt using PCR (Abd El-Kader et al. 2011) and these studies claimed that the presence of fecal inhibitors and contaminants such as bilirubin, bile salts, and others can inhibit DNA amplification, yielding less accurate detection results (Abd El-Kader et al. 2011).
Until now, few studies in Egypt have addressed the Cryptosporidium transmission dynamics in humans, despite it being pivotal in terms of establishing effective prevention and disease control policies. In the present study, C. hominis, an anthroponotic Cryptosporidium species, was significantly more prevalent (89.3%) than C. parvum (7.1%). Mixed infections were not detected, indicating that the main source of Cryptosporidium infection in the study group was of human rather than zoonotic source. Akiyoshi et al. (2003) reported that in mixed infections, C. parvum predominates and rapidly displaces C. hominis which adds more proof that in our study, it is a C. hominis predominance and may explain the non-typed species we obtained. Our results are in concordance with those of only one study (Abd El-Kader et al. 2011) and contradictory to those of other Egyptian studies and most studies from other Middle Eastern countries (Eida et al. 2009; Al-Brikan et al. 2008; Hijjawi et al. 2010; Iqbal et al. 2011). Our data and the data of Abd El-Kader et al. (2011) suggest a clear anthroponotic transmission; this discrepancy in the results may be explained by the fact that Abd El-Kader et al. (2011) and our study were conducted in Great Cairo, while the contrasting results were found in more rural parts of Egypt.
This predominance of C. hominis is in accordance with molecular studies in Australia, Canada, Japan, USA, and developing countries (Xiao and Fayer 2008). In contrast, studies in the UK have shown C. parvum predominance (McLauchlin et al. 1999, 2000).
Among studied variables, diarrhea, the type of water, and stool consistency showed statistically significant association with cryptosporidiosis. Our results were in agreement with those of other reports in Mexico and Iran, in which there were no reported significant differences between cases with cryptosporidial diarrhea and age, race, ethnicity, or sex (Nair et al. 2008; Saneian et al. 2010; Bushen et al. 2007). It was reported that C. hominis is contracted at an earlier age than C. parvum. Samie et al. (2006) reported that C. hominis was equally distributed between males and females. GIT symptoms, including diarrhea, abdominal pain, flatulence, itching, vomiting, and/or appetite loss (Chauret et al. 1999), animal contact (Hijjawi et al. 2010), and type of water supply (Goh et al. 2004; Beach 2008; Garvey and McKeown 2009; Al-Warid et al. 2012) are well-known risk factors for cryptosporidiosis.
The occurrence of clinical manifestations in Cryptosporidium can be related in part to the different Cryptosporidium species and subtypes of C. hominis. Cama et al. (2008) reported that C. hominis was associated with diarrhea, vomiting, nausea, and general malaise, while C. parvum, C. canis, C. felis, and C. meleagridis were associated with diarrhea only. Environmental, clinical, and host behavioral factors may act as important risk factors for Cryptosporidium infection, susceptibility, or disease prevalence, but do not affect the pathogenicity or the course of the disease (Mumtaz et al. 2010).
In the present study, besides the studied variables that may induce susceptibility of cryptosporidiosis in Egypt, there are added variables such as higher population densities with a greater chance of person-to-person transmission and variation in socioeconomic status within the same geographical areas. This might also explain the discrepancies in the reported results for the prevalence of the disease between past studies and our current research findings.
Conclusion
Cryptosporidium is an indigenous, prevailing intestinal parasite among children in Cairo with a predominance of C.hominis, suggesting anthroponotic transmission rather than zoonotic. The significant majority of Cryptosporidium infections were detected in diarrheic patients, who relied upon tap water sources; their stool was soft and yielded more parasites than the patients with liquid stool. Due to the absence of any effective curative treatments, determining the prevailing species of Cryptosporidium and identifying the significant etiological risk factors are essential in order to identify its transmission dynamics and plan preventive and control strategies.
Referenes
Abd El-Kader MN, Blanco M, Tammam MA, Abd El Ghaffar AB, Osman A, El Sheikh N, Rubio JM, de Fuentes I (2011) Detection of Cryptosporidium parvum and Cryptosporidium hominis in human patients in Cairo, Egypt. Parasitol Res 110(1):2465–2470. doi:10.1007/s00436-011-2465-6
Akiyoshi DE, Mor S, Tzipori S (2003) Rapid displacement of Cryptosporidium parvum type 1 by type 2 in mixed infections in piglets. Infect Immun 71(10):5765–5771
Al-Brikan FA, Salem HS, Beeching N, Hilal N (2008) Multilocus genetic analysis of Cryptosporidium isolates from Saudi Arabia. J Egypt Soc Parasitol 38:645–658
Al-Warid HS, AL-Saqur IM, Mahmood SH (2012) Occurrence of Cryptosporidium spp. among people live in north of Baghdad. Eur J Sci Res 78(4):539–545
Beach MJ (2008) Waterborne: recreational water in: Fayer, R. and Xiao, L. eds. 2008. Cryptosporidium and cryptosporidiosis. 2nd edition USA: Taylors & Francis Group 355–361.
Bushen OY, Kohli A, Pinkerton RC, Dupnik K, Newman RD, Sears CL, Fayer R, Lima AA, Guerrant RL (2007) Heavy cryptosporidial infections in children in northeast Brazil: comparison of Cryptosporidium hominis and Cryptosporidium parvum. Trans R Soc Trop Med Hyg 101(4):278–284
Cama VA, Bern C, Roberts J, Cabrera L, Sterling CR, Ortega Y, Gilman RH, Xiao L (2008) Cryptosporidium species and subtypes and clinical manifestation in children, Peru. Emerg Infect Dis 14(10):1567–1574
Chauret C, Springthorpe S, Sattar S (1999) Fate of Cryptosporidium oocysts, Giardia cysts, and microbial indicators during wastewater treatment and anaerobic sludge digestion. Can J Microbiol 45(3):257–262
Eida AM, Eida MM, El-Desoky A (2009) Pathological studies of different genotypes of human Cryptosporidium Egyptian isolates in experimentally mice. J Egypt Soc Parasitol 39(3):975–990
El-Settawy MA, Fathy GM (2012) Evaluation and comparison of PCR, coproantigen ELISA and microscopy for diagnosis of Cryptosporidium in human diarrhoeic specimens. J Am Sci 8(12):1378–1385
Elwin K, Hadfield SJ, Robinson G, Crouch ND, Chalmers RM (2012) Cryptosporidium viatorum sp. among travellers returning to Great Britain from the Indian subcontinent, 2007–2011. Int J Parasitol 42:675–682
Fathy MM, Abdelrazek NM, Hassan FA, El-Badry AA (2014) Molecular copro-prevalence of cryptosporidium in Egyptian children and evaluation of three diagnostic methods. Indian Pediatr 51(8):727–729
Fayer R, Santín M, Macarisin D (2010) Cryptosporidium ubiquitum sp. in animals and humans. Vet Parasitol 172:23–32
Feng Y, Dearen T, Cama V, Xiao L (2009) 90-kilodalton heat shock protein, Hsp90, as a target for genotyping Cryptosporidium spp. known to infect humans. Eukaryot Cell 8(4):478–482
Garvey P, McKeown P (2009) Epidemiology of human cryptosporidiosis in Ireland 2004-2006: analysis of national notification data. Eur Secur 14(8):1–5
Ghallab MMI, Abdel-Aziz IZ, Shoeib EY, El-Badry AA (2016) Laboratory utility of coproscopy, copro immunoassays and copro nPCR assay targeting Hsp90 gene for detection of Cryptosporidium in children, Cairo, Egypt. J Parasit Dis 40(3):901–905
Goh S, Reacher M, Casemore DP, Verlander NQ, Chalmers R, Knowles M, Williams J, Osborn K, Richards S (2004) Sporadic cryptosporidiosis, North Cumbria, England, 1996–2000. Emerg Infect Dis 10(6):1007–1015
Hijjawi N, Ng J, Yang R, Atoum MF, Ryan U (2010) Identification of rare and novel Cryptosporidium GP60 subtypes in human isolates from Jordan. Exp Parasitol 125:161–164
Iqbal J, Khalid N, Hira PR (2011) Cryptosporidiosis in Kuwaiti children: association of clinical characteristics with Cryptosporidium species and subtypes. J Med Microbiol 60(5):647–652
McLauchlin J, Pedraza-Diaz S, Amar-Hoetzeneder C, Nicholas GL (1999) Genetic characterization of Cryptosporidium strains from 218 patients with diarrhea diagnosed as having sporadic cryptosporidiosis. J Clin Microbiol 37:3153–3158
McLauchlin J, Amar C, Pedraza-Diaz S, Nichols GL (2000) Molecular epidemiological analysis of Cryptosporidium spp. in the United Kingdom: results of genotyping Crvptosporidium spp. in 1705 fecal samples from humans and 105 fecal samples from livestock animals. J Clin Microhiol 38:3984–3990
Mumtaz S, Ahmed J, Ali L (2010) Frequency of Cryptosporidium infection in children under five years of age having diarrhea in the north west of Pakistan. Afr J Biotechnol 9(8):1230–1235
Nair P, Jamal A, Mohamed HL, DuPont JF, Figueroa LG, Carlin ZJ, Jiang ZD, Belkind-Gerson J, Martinez-Sandoval FG, Okhuysen PC (2008) Epidemiology of cryptosporidiosis in North American travelers to Mexico. AmJTrop Med Hyg 79(2):210–214
Samie A, Bessong PO, Obi CL, Sevilleja JE, Stroup S, Houpt E, Guerrant RL (2006) Cryptosporidium species: preliminary descriptions of the prevalence and genotype distribution among school children and hospital patients in the Venda Region, Limpopo Province, South Africa. Exp Parasitol 114(4):314–322
Saneian H, Yaghini1 O, Yaghini A, Modarresi1 MA, Soroshnia M (2010) Infection rate of Cryptosporidium parvum among diarrheic children in Isfahan. Iran J Pediat 20(3):343–347
Savioli L, Smith H, Thompson A (2006) Giardia and Cryptosporidium join the ‘Neglected Diseases Initiative’. Trends Parasitol 22(5):203–208
Smith HV, Cacciò SM, Tait A, McLauchlin J, Thompson RC (2006) Tools for investigating the environmental transmission of Cryptosporidium and Giardia infections in humans. Trends Parasitol 22:160–167
Sulaiman IM, Hira PR, Zhou L, Al-Ali FM, Al-Shelahi FA, Shweiki HM, Iqbal J, Khalid N, Xiao L (2005) Unique endemicity of cryptosporidiosis in children in Kuwait. J Clin Microbiol 43(6):2805–2809
Xiao L, Fayer R (2008) Molecular characterisation of species and genotypes of Cryptosporidium and Giardia and assessment of zoonotic transmission. Int J Parasitol 38(11):1239–1255
Acknowledgments
We would like to thank Mrs. Sieglear for language editing and proofreading of this manuscript.
Author information
Authors and Affiliations
Contributions
All manuscript authors contributed to every aspect of the reported research: the idea for the research paper, the study design, the collection of materials, the methodology, the writing of the paper, and the reviewing/editing of the material presented in it.
Corresponding author
Ethics declarations
Funding
Self funded by authors.
Ethical approval
This study was ethically approved by the “Research Ethical Committee,” Deanship of postgraduate education & scientific research, Faculty of Medicine, Cairo University in compliance with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from patients or their relatives and parents of young children before they responded to the questionnaire.
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
El-Badry, A.A., Abdel Aziz, I.Z., Shoeib, E.Y. et al. Cryptosporidium genotypes and associated risk factors in a cohort of Egyptian children. Comp Clin Pathol 26, 1017–1021 (2017). https://doi.org/10.1007/s00580-017-2477-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00580-017-2477-4