Abstract
Qatar is largely characterized by a hyper-arid climate and low soil fertility which create a stressful soil environment for arbuscular mycorrhizal (AM) fungi. In a study of AM fungal communities and their relationship with soil chemical characteristics, we used a high-throughput sequencing technique to explore AM fungal diversity and community composition in different habitats across Qatar. We identified a total of 79 AM fungal taxa, over 77% of which were species from the Glomeraceae family. The lowest AM fungal diversity was observed in saltmarsh and in one rawdha site, while the highest richness, effective number of species, and diversity were observed in rawdha and sabkha communities. NMDS and multiple regression analyses showed that AM fungi were negatively correlated with soil pH and TC, but positively correlated with K and NO3−. AM fungi also were positively correlated with Cd, with the latter suggesting that very low levels of heavy metals may not always be harmful to AM fungi. These findings provide baseline information on AM fungal assemblages and the chemical drivers of diversity across communities in Qatar. This work partly compensates for the current lack of broad-scale studies in the Arabian Peninsula by providing understanding of overall patterns of AM fungi and their drivers in the region.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Global drylands, including those in Qatar, cover approximately one-third of the Earth’s surface and therefore play a significant role in the global carbon cycle (Fitter 2005; Delgado-Baquerizo et al. 2015; Soudzilovskaia et al. 2015b; Neilson et al. 2017; Lüneberg et al. 2018). Because of high variability in rainfall and extended periods of drought in dryland regions, dryland ecosystems are highly susceptible to soil erosion and land degradation caused by climate variability, and often are described as unstable habitats for many microorganisms (Delgado-Baquerizo et al. 2015; Neilson et al. 2017; Laban et al. 2018). However, despite the severe environmental conditions they experience, research has shown that dryland ecosystems are extremely adaptable to environmental changes. For instance, re-establishment of natural grasslands in Africa since 1980 has led to resilience of many dryland microbes by natural selection through their evolutionary history (Reynolds et al. 2007; United Nations 2011). The diversity and colonization success of most underground dryland microorganisms depends on the fertility and overall condition of the soil environment, including adequate soil nutrients and interaction between biotic communities to sustain the integrity of dryland ecosystems (Jeffries et al. 2003; Neilson et al. 2017; Laban et al. 2018). Due to the adverse conditions in dryland habitats, the occurrence of mycorrhizal associations and the diversity of arbuscular mycorrhizal (AM) fungi driven by such habitats may be important. Drylands are known to host a large variety of endemic species, including AM fungi, which exhibit unique adaptations to the harsh conditions prevailing in drylands (Kamalvanshi et al. 2012; Neilson et al. 2017; Laban et al. 2018; Xu et al. 2018).
Among their many functions, AM fungi play an integral part in nutrient cycling and ecosystem functioning. Therefore, research pertaining to AM fungal communities is essential as a basis for understanding complex ecological relations. AM fungi primarily belong to the phylum Glomeromycota and form an obligatory symbiosis with plant roots (Smith and Read 2008; van der Heijden et al. 2015; Tedersoo et al. 2018). This symbiosis is usually mutualistic, so AM fungi play an important role in plant productivity and stress tolerance (Smith and Read 2008). The composition of AM fungal communities is highly influenced by edaphic factors (Dumbrell et al. 2010; Davison et al. 2015). Mycorrhizal associations are especially crucial in fragile ecosystems where access to soil resources is limited, and thus are of particular importance in semi-arid and hyper-arid ecosystems (Majid et al. 2016; Villalobos et al. 2016; Zhao et al. 2017). In this regard, the diversity of AM fungi in drylands may be vital for establishing symbiotic associations to maintain ecosystem function (Zhao et al. 2017; Mahmoudi et al. 2019). For instance, AM fungi support biologic carbon sequestration by improving soil aggregation (Xu et al. 2018), which in turn may increase net primary production (NPP) in AM-dominated habitats (Jeffries et al. 2003; Soudzilovskaia et al. 2015a). However, the extent of mycorrhization depends on the host plant and the AM fungal species. Diversity in AM fungi also may enhance ecosystem function (Delgado and Gómez 2016; Gerz et al. 2018). The positive effect on ecosystems is particularly important in climate-driven habitats, as potential changes in aridity due to climate change may influence the ability of dryland microbiomes to sustain geochemical cycles and ecosystem functions (Gustafson and Casper 2006; Delgado-Baquerizo et al. 2015; Majid et al. 2016; Neilson et al. 2017; Zhao et al. 2017). Soil disturbances influenced by agricultural management could have a positive impact on AM fungal diversity if agrosystem management is associated with increasing soil organic matter (Oehl et al. 2010). On the other hand, a disturbance associated with decreasing organic matter will likely reduce fungal diversity (Toljander et al. 2008). The diversity of AM fungal communities can be dependent on the chemical nature of the soil environment.
Differences in habitat also affect AM fungal associations. In China, the occurrence of mycorrhizal host plants is more frequent and diverse in grasslands which have high AM fungal diversity, whereas mycorrhizal plants in desert areas have no significant effect on AM fungal community composition, although both habitat types exist in drylands (Wang et al. 2018). Overall, that may reflect that AM fungi are biotrophs, receiving energy from host plants (Hammer et al. 2011). Addition of organic matter is a common agricultural practice to improve soil fertility in drylands, particularly saline drylands, in order to stimulate plant growth. Soil organic matter supports AM fungal growth by improving the water-holding capacity of the soil and increasing the supply of nutrients (Hammer et al. 2011). This in turn stabilizes the soil structure, facilitating formation of AM fungal aggregates. However, little is known about whether AM fungal growth is stimulated solely by the availability of nutrients or by a combination of these factors. Low phosphorus (P) concentrations have been shown to support a specific set of AM fungal taxa, leading to an increase in the distribution of these taxa relative to those at high P concentrations which suppress AM fungal diversity (Van Geel et al. 2017; Grant et al. 2005). In contrast, soil enrichment with nitrogen (N) through addition of fertilizer has positive correlations with AM fungal diversity. The direct benefits of soil N on AM fungi are not well known, but it has been suggested that N enrichment acidifies the soil environment (Jansa et al. 2014). However, other studies have found no observable effect of soil N on AM fungi, but the influence of soil pH on AM fungal community composition (Van Geel et al. 2017). In general, many studies have identified soil acidity as an important driver of AM fungal communities (Jansa et al. 2014; Van Geel et al. 2017).
Despite many studies of AM fungal populations in drylands, little is known about how these fungi survive in polluted environments, particularly in areas with heavy metal–contaminated soils. It has been suggested that soil pollution with cadmium (Cd) induces toxicity in mycorrhiza-associated plants, but that these plants later evolve to develop tolerance mechanisms (Shahabivand et al. 2012). The most common change in soil chemistry with heavy metal pollution is acidification, which in turn increases metal availability in the soil to toxic levels that could potentially disrupt mycorrhizal associations (Del Val et al. 1999). Heavy metal pollution in soils in Qatar is introduced through anthropogenic activities, with Cd and lead (Pb) pollution caused by years of exploitation of oil and gas resources/industrialisation in the region. Therefore, we examined the effects of these common heavy metal pollutants on AM fungal diversity and occurrence in soils in Qatar.
The overall aim of this study is to identify AM fungal communities along a stress-induced environmental gradient in Qatar. To achieve this aim, chemical drivers in the soil were compared with AM fungal richness and distribution across Qatar. Taxa of AM fungi over a range of habitat types were sequenced to determine the distribution and occurrence of AM fungi with respect to soil chemical parameters, specifically soil pH, salinity, total nitrogen (TN), nitrite (NO2−), nitrate (NO3−), and additional plant nutrients, and two heavy metals, Cd and Pb. The general expectation was that differences in the chemical properties of soils and in vegetation cover among locations would influence the diversity and composition of AM fungi. Specific hypotheses were that (1) the presence and diversity of AM fungi are positively associated with concentrations of plant nutrients in the soil; (2) increasing soil salinity and pH reduces the diversity of AM fungal species, as soil conditions become unfavorable for their growth; and (3) the presence of Cd and Pb in the soil environment has significant negative effects on the presence and abundance of AM fungal species, due to the toxicity effects of these heavy metals.
Materials and methods
Study area and sample collection
Soil samples were collected from 19 different locations across Qatar (Table S1, Fig. S1). Qatar is located between 24°–27°N and 50°–52°E on the north-eastern coast of the Arabian Peninsula and occupies a total land area of 11,571 km2 (Zahlan 2016). Most of the territory of Qatar is covered by drylands (with sparse/patchy and seasonal vegetation), with a highly arid climate and minimal annual rainfall (< 100 mm per year) that typically falls during the winter months (i.e., December–February). Mean monthly temperature ranges from 18.5 °C in January to 37 °C in July. However, maximum daytime temperatures during summer can rise above 40 °C, while other periods have humid days with highs of around 38 °C (Mamoon and Rahman 2016). The topography consists of mainly flat land surfaces, but with hills and undulating sand formations mostly found in the south-east of the country (Abulfatih et al. 2002; Norton et al. 2009). The soils are abundant in calcareous rocks, sand, and gravel. Most of Qatar’s soil surface is categorized as one of two habitat types, rawdha and sabkha. Rawdha refers to areas with relatively good soil quality and good water availability formed by a collapse below the surface or evaporation from the surface. Sand and minerals from higher ground often are washed into these pits during the rainy season (Al-Thani and Yasseen 2018). The terrain in rawdha is low-lying, with natural depressions and surrounding small hills with altitude of around 100 m (Mahasneh et al. 2008; Norton et al. 2009). The natural depressions can be up to 20 m deep and are generally richer in minerals than the surrounding higher soil (Norton et al. 2009). (2) Sabkha refers to salt pans that can be either barren or sparsely vegetated, have high salinity, and are inhabited by halophytic plants and extremophiles that are well-adapted to these highly saline and arid areas. However, most plant taxa lack the ability to grow in sabkha, while the few AM fungi found there are highly stressed by both aridity and salinity (Al-Thani and Yasseen 2018).
The 19 sampling locations in the present study differed in terms of topography, vegetation, and soil conditions and were classified into one of four habitat types: nine locations had calcareous soil depressions (i.e., rawdha), there was one salt marsh and one mangrove stand, and eight saline dryland areas had no or sparse vegetation (i.e., sabkha) (Table S1). Some of the locations were close to commercial districts and urban areas. Twenty subsamples were collected at each site (approximately 50 m × 50 m), with a roughly even distribution at opposite sides of plants when present, to about 5 cm beneath the soil surface. For each site, a composite sample was prepared by combining approximately 1–1.5 g of soil from each of the 20 subsamples. The composite soil samples were air-dried at room temperature in the laboratory for 4 days, and thereafter stored in zip-lock plastic bags with silica gel to absorb moisture.
Molecular analyses and bioinformatics
Environmental DNA was extracted from 0.25 g of homogenized soil sample using the DNeasy PowerSoil Kit (Qiagen GmbH, Hilden, Germany), following the manufacturer’s protocol. Soil samples were homogenized by first removing all roots, root fragments, and rock debris from the soil, and then finely grinding each sample manually by rubbing the bag of soil between the hands. Total genomic DNA was quantified and tested for extraction quality using 1% agarose gel electrophoresis. Environmental DNA of the composite soil samples was used as template in subsequent polymerase chain reaction (PCR) amplification. The small sub-unit (SSU) ribosomal RNA gene of AM fungal sequences was amplified using AM fungal-specific primers WANDA (Dumbrell et al. 2011) and AML2 (Lee et al. 2008). Each primer was tagged with 12 base multiplex identifier (MID) tags, as described in Tedersoo et al. (2014). PCR was carried out in a total reaction volume of 25 µL, comprising 1 µL of template DNA, 0.5 µL each of forward and reverse primer (20 µM), 18 µL of nuclease-free water, and 5 µL of 5 × Hot FIREPol Blend Master Mix (Solis Biodyne, Tartu, Estonia). Optimal PCR conditions used for amplifying fungal communities were as follows: 15 min at 95 °C, followed by 35 cycles of 30s at 95 °C, 30s at 55 °C, 1 min at 72 °C, and a final extension temperature of 72 °C for 10 min. PCR was performed on a GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA, USA).
The PCR products were pooled into two libraries, at approximately equimolar ratios as determined by the strength of the gel band (1.5% gel electrophoresis). They then were purified using the FavorPrep gel/PCR purification kit (Favorgen Biotech Corp, Ping Tung, Taiwan), following the manufacturer’s protocol. Each library was linked with Illumina MiSeq adapters using the TruSeq DNA PCR-free HT Library Prep kit (Illumina Inc., San Diego, CA, USA) and processed by the Illumina MiSeq 2X300 sequencing platform at the Estonian Genome Centre (Tartu, Estonia). A negative control (nuclease-free water instead of template DNA in the PCR mixture) and positive control (DNA extracted from mycorrhizal Plantago ovata roots) were used throughout the experiment, including sequencing runs.
Illumina paired-end raw sequences were demultiplexed and cleaned using a series of bioinformatics steps, following Vasar et al. (2017). Sequences were cleaned by selecting reverse and forward reads with average quality of at least 25, and both reads could have one match in forward and reverse primer sequences. Quality-filtered paired-end reads were combined with FLASH (v1.2.10) (Magoč and Salzberg 2011), using the default thresholds (overlap between 10 and 300 bp; overlap identity at least 75%). Sequences were pre-clustered with 98% identity using VSEARCH (v2.14.1) (Rognes et al. 2016) to reduce the computational complexity and time requirement. Cluster information was stored to allow clusters to be mapped back to individual reads. Putative chimeric sequences were removed using VSEARCH with the MaarjAM reference database, February 2020 (Öpik et al. 2010), and de novo chimera filtering algorithms. Sequences were clustered into operational taxonomic units (OTUs) using 97% similarity and were used as proxies for species. The cluster centroids obtained were used to gain a rough estimate of the taxonomy, by conducting a BLAST + search against the MaarjAM database (Öpik et al. 2010). All OTUs represented by a single sequence (singletons) were removed.
Chemical analyses
Prior to chemical analysis, 19 composite soil samples (one from each site) were oven-dried at 60–62 °C for 48h to prevent decomposition of organic material and to enhance the extractability of minerals (Erich and Hoskins 2011). The dried soil was then milled to a fine powder in a rotary ball mill (Retsch Mill, Haan, Germany) at a speed of 250 rpm for 40 min and passed through a standard sieve of 2 mm mesh, in preparation for downstream chemical analysis. Portions of processed soil used for chemical characterization were analyzed for the parameters: pH, salinity, total carbon (TC), TN, NO3−, NO2−, and concentrations of key chemical elements, including calcium (Ca), potassium (K), magnesium (Mg), P, Cd, and Pb, following methods validated by the International Organization for Standardization (ISO/IEC 17,025).
Soil pH and salinity were measured using a standard probe meter calibrated with 7.0 and 4.0 buffer solutions and 0.01 M potassium chloride (KCl) solution, respectively. Electrical conductivity (EC) was measured by an EC electrode (EC300 YSI EcoSense conductivity meter), and TC and TN contents in all soil samples and in glycine standard compound were determined by dry combustion in a CHN elemental analyzer (Skalar Analytical, Breda, Netherlands) (Jing et al. 2015). To ensure reliability of the values, the recoveries for theoretical TC and TN in glycine were compared with calculated values for unknown samples. Soil NO3− and NO2− were determined by extraction with KCl, followed by UV–Vis spectrophotometry (Fisher Scientific, Waltham, USA), after pretreatment with ethylenediaminetetraacetate solution and passing through a column containing copperized cadmium filing (Wood et al. 1967).
For quantification of element concentrations in soil, the samples were digested in a 54-well digestion hot block (Thomas Scientific, Swedesboro, USA) using a mixture of nitric acid (HNO3) and hydrofluoric acid (HF) (ratio 3:2) (Tighe et al. 2004). About 0.25 g of soil was weighed accurately and placed inside a PTFE Teflon digestion tube, followed by addition of 9.0 mL concentrated HNO3 (69% w/w) and 3.0 mL concentrated HF. The tubes were heated gradually to 135 °C for approximately 1h. Following acid digestion, evaporation was initiated by removing the reflux caps and increasing the temperature to 155 °C for an additional hour. Following near complete evaporation, 3.0 mL of HNO3 was added to the remaining residue and diluted to a total volume of 50 mL. The samples were then heated to boiling point, transferred to a 100-mL volumetric flask, and diluted with Milli-Q water. Sample solutions and quality controls (i.e., blanks, replicate analyses, and certified reference materials (CRM)) were analyzed by ICP-OES (Perkin Elmer Optima 7300DV System fitted with an S10 autosampler) for Mg, K, P, Ca, Cd, and Pb. The data obtained were processed using WinLab32 software. To ensure reliability of the data, internal standards were included during ICP-OES analysis to avoid potential interferences (Masson et al. 2010). The precision and accuracy of the chemical analyses were assessed using certified reference material (PACS-3, marine sediment).
Data analyses
Species richness was obtained from the bioinformatic analyses (OTUs). Hill diversity indices were calculated for species richness (q = 0), frequent (q = 1), and dominant (q = 2) species, using the iNEXT-PD package (Chao et al. 2010; Hsieh et al. 2016). Multiple regression analysis was conducted to evaluate the relationship between soil characteristics and AM fungal diversity indices using MiniTab (version 17).
The relative abundance of AM fungi at each location was determined and illustrated, based on the OTUs of each genus identified (Fig. S2). The indicator index was determined following the method in Dufrêne and Legendre (1997), which gives evidence of an environmental change. An indicator species that reflected the biotic and abiotic state of the habitat was selected for all environments.
Shapiro–Wilk (S-W) normality tests showed that the datasets for salinity, NO3 − , and TC were normally distributed. Soil chemical data that did not follow a normal distribution were normalized by logarithmic transformation prior to ANOVA. To test for differences in chemical properties among the 19 locations (with three replications selected among the site subsamples), we used one-way ANOVA, while we tested for differences between pairs of locations using post hoc tests (Tukey test). All these analyses were performed in MiniTab (version 17). We used principal component analysis (PCA) to evaluate combinations of chemical properties that could explain a common pattern of variation among the locations sampled. We used non-metric multidimensional scaling (NMDS) to visualize the separation of AM fungal communities and chemical properties among habitat types (i.e., mangrove, saltmarsh, rawdha, and sabkha), using PRIMER statistical package (PRIMER — E, Plymouth Marine Laboratory, Version 6.1.13) with the PERMANOVA add-on (Version 1.0.3).
Results
Community composition and occurrence of AM fungi
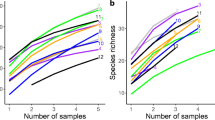
The Illumina 2 × 300 bp MiSeq platform generated 9,861,189 paired-end reads targeting the SSU region of the rRNA gene. These reads were quality-filtered, chimeras removed, and combined, which resulted in 2,474,899 sequences. Following removal of the chimeric reads, the positive control sample, and singletons (n = 6), the final dataset comprised 79 OTUs and 1,369,477 sequences from the 19 composite soil samples. Each sample contained 2–62,040 sequences (median 6588 sequences) and 1–28 OTUs (median 7 OTUs). The total of 79 AM fungal OTUs belonged to the families: Glomeraceae (61 OTUs), Diversisporaceae (4 OTUs), Claroideoglomeraceae (3 OTUs), Paraglomeraceae (2 OTUs), Archaeosporaceae (4 OTUs), Acaulosporaceae (2 OTUs), Ambisporaceae (2 OTUs), and Gigasporaceae (1 OTU) (Table S2). The dataset showed that 77% of the identified AM fungal OTUs were from the Glomeraceae, which was the most abundant AM fungal family, while 5.06% of the AM fungi in our study were from the Archaeosporaceae and Diversisporaceae, the second most abundant families (Fig. 1).
(a) Relative abundance of arbuscular mycorrhizal (AM) fungal genera, presented for the entire dataset and (b) AM fungal richness at each location in Qatar. The 19 locations are categorized as one of four habitat types: (1) rawdha: calcareous soil depressions with relatively good soil quality and water accessibility due to influxes of water and minerals from surrounding higher ground; (2) mangrove: mass of shrubs growing in coastal saline water; (3) saltmarsh: wet muddy soil with open access to coastal saltwater; and (4) sabkha: salt pans (either with or without vegetation) that tend to accumulate minerals due to calcareous surface evaporation
The AM fungal indicators (indicator index > 0.25) originated from two main taxa, Glomeraceae and Paraglomeraceae, with the indicator index value being higher for Glomeraceae (3.647) than Paraglomeraceae (0.306) (Table S2). Variation in the composition of AM fungal genera was observed among all soil samples. Glomus and Paraglomus were recorded at 89% of all sites and were therefore the dominant AM fungal genera in our study, with Glomus as the most diverse and species-rich AM taxon. These were followed by Claroideoglomus (68% of sites), Diversisporaceae (47% of sites), and Archaeospora (37% of sites) (Table S2), while the percentage frequency of occurrence of the remaining AM fungal taxa ranged from 5 to 11% (Table S2). Glomus species (61 OTUs) showed the highest species richness across all locations and Scutellospora species (Gigasporaceae) the lowest (1 OTU). Glomus was the most frequently detected AM fungus genus in the locations sampled, recorded at 17 sites (all except saltmarsh-3 and rawdha-9) (Fig. S2).
The AM fungal diversity indices and richness (Shannon diversity, Simpson diversity, richness, and effective number of species) were highest at the rawdha and sabkha sites, and lowest in the saltmarsh and exceptionally in one of the rawdha sites (Table 1). Species richness (Hill number q = 0) ranged from 49 OTUs at Site 15 (sabkha) to one at Site 3 (saltmarsh) and Site 9 (rawdha) (Table 1; Fig. 1). Effective number of species (q = 1) was highest at Site 6 (sabkha) and q = 2 was highest at Site 11 (rawdha), but both diversity indexes were the lowest at sites 3 and 9 (Table 1).
Soil Chemical Traits
The concentrations of Ca and Mg in the soil differed among sites, but were higher in soils in undisturbed areas than in areas close to a city, urban structure, or road (Table S3). Almost all measured TN levels were negligible, with values well below the limit of detection. The only measurable TN concentration was determined in samples taken from the mangrove stand (Site 2), but it was still close to zero (0.014%). Therefore, the TN measurements were excluded from further analysis. Spectrophotometric measurements of NO2− and NO3− showed various concentrations for different habitat types, although there were no statistically significant differences among sites (Table S4). Soil NO3− concentration displayed a wider range of concentrations than soil NO2− concentrations (Table 2), for which the values remained relatively constant among sites.
Slight alkalinity was detected in soil from sites 2 and 3, which was likely attributable to the large extent of inland saltmarsh, mangrove, and surrounding coastal areas in these locations. The pH levels were rather consistent across the whole dataset, with soil samples from sites 8 and 16 having the highest pH levels (Table S4). In contrast, the salinity levels fluctuated across the dataset and differed significantly at 12 of the 19 locations studied (Table S4). High salinity levels were found in soil samples from mangrove and saltmarsh (sites 2 and 3, respectively), as a result of the highly calcareous soil depressions at these locations, which were predominantly colonized by halophytic plant species, including Avicennia marina. A spike in salinity also was observed at sites 5, 11, and 13 (rawdha), most likely due to influx of minerals from surrounding higher ground into these lowland areas, creating inland salt pans that consist of fine silt and high salinity (Abulfatih et al. 2002; Majid et al. 2016).
In terms of heavy metals, sites 1, 4, and 5 were most strongly affected by soil Pb, while site 8 showed the least influence of the heavy metals measured in this study (Table S3). Our analyses also indicated that the environmental conditions at Site 8 were not directly defined by its basic soil chemistry but rather were most likely affected by the interaction of different environmental factors.
Among the chemical elements measured, Ca showed the highest concentrations in the soil samples, with values well over 90,000 mg/kg at all 19 locations studied (Table S3). The lowest measured concentrations were for the heavy metals Cd and Pb, with Cd concentrations ranging from 0.02 to 0.40 mg/kg and Pb concentrations ranging from 0.12 to 15.29 mg/kg (Table S3). In quality control of ICP-OES measurements using a certified reference material (PACS3), percentage recovery of all metals fell within 15% of the EPA criteria (accuracy 90–106%, n = 6). The PCA results showed that Mg and P concentrations were significantly positively correlated (r value close to 1), whereas Ca showed almost no correlation with the other elements (r value close to 0) (Fig. 2).
Effect of chemical properties and habitat on AM fungi
The NMDS and multiple regression analyses revealed that the AM fungal community was influenced by soil chemical parameters and habitat (Fig. 3; Table S5). TC had a significant negative effect on Simpon’s diversity and Shannon’s diversity (p = 0.036 and p = 0.013, Fig. S3). NO3− (p = 0.025) had a significant positive effect on Shannon’s diversity (Fig. S4), K had a significant positive effect on species richness (p = 0.047) (Table S5, Fig. S5). There also was a nearly significant negative effect of pH (p = 0.054) (Table S5; Fig. S6) and a positive effect of Cd (p = 0.052) on species richness (Table S5; Fig. S7). NO3− had a nearly significant positive effect (p = 0.054 and p = 0.076) on Simpson’s diversity and species richness (Table S5; Fig. S8). Overall, these factors had the greatest influence on Glomus followed by Claroideoglomus and Paraglomus. While saltmarsh and mangrove were only represented by one site each, they were most strongly associated with soil Ca, TC, and salinity (Fig. 2). The NMDS indicated that the effect of these chemical parameters was much lower on Scutellospora, Acaulospora and Ambiospora than on other AM fungal genera, which supports our finding that Glomus was the most frequently occurring AM fungal genus across our dataset. The AM fungal communities in rawdha and sabkha were intermixed in the NMDS plot, while the mangrove and saltmarsh communities, which represented single locations, were apart (Fig. 3).
Non-metric multidimensional scaling (NMDS) plots of arbuscular mycorrhizal (AM) fungal communities versus (a)chemical parameters and (b) AM fungal genera at 19 sites in Qatar, representing four type of habitat: mangrove, rawdha, sabkha, and saltmarsh. The circles represent correlation of 1 of vectors to variables
Discussion
Previous studies have highlighted the importance of soil type and chemical components for mycorrhizal communities (Tian et al. 2017; Xue et al. 2018). In line with this, we found that soil chemical parameters were the main drivers of variation in AM fungal communities in Qatar. The highest richness and greatest effective number of species (q = 1) of AM fungi were identified at sites 15 (49 OTUs), 11 (48 OTUs), and 6 (39 OTUs), representing sabkha and rawdha, communities. Simpson diversity was higher at site 11 (rawdha), while Shannon diversity was highest at site 6 (sabkha). This was supported by the results from the NMDS in which rawdha and sabkha communities were intermixed without clear separation. Glomeraceae was the dominant family in Qatar’s extreme environment. Fungi of the genus Glomus were most abundant and widely distributed among locations, most likely due to their capacity to inhabit a broad range of niches. However, AM fungi were not evenly distributed among the study sites and at some locations, such as sites 3 and 9, only a single species (Paraglomeraceae) was found, suggesting that specific AM fungi respond differently to prevailing environmental conditions which allow them to successfully occupy an area. However, it should be noted that Paraglomeraceae was found in all sites except sites 13 and 19. The infrequent occurrence of Acaulospora, Ambisporaceae, Gigasporaceae, and Paraglomeraceae could indicate scarcity of these taxa in highly arid regions which often are restricted in resources (Kamalvanshi et al. 2012; Zerihun et al. 2013).
Our first hypothesis, that AM fungal richness is influenced by availability of macronutrients in soil, was partly supported by the results, with K and NO3− being positively associated with AM fungal richness. Studies on soil fungi in semi-arid China have found that the richness and abundance of fungal communities are mainly driven by soil P content (Tian et al. 2017; Zhao et al. 2017; Xu et al. 2018). This suggests that the diversity and occurrence of AM fungi are generally positively correlated with the concentration of extractable soil P, which agrees with findings in some studies (Timmer and Leyden 1980; Bhat et al. 2014; Wu et al. 2018), but contradicts findings of a negative correlation between AM fungus abundance and P concentration in other studies (Khanam et al. 2006). However, the effect of soil P on AM fungi may depend on the level of available soil P, with both low and high levels of soil P having a negative effect. Furthermore, P fertilization is known to suppress mycorrhizas and reduce AM fungus occurrence in fertile soils (Treseder and Allen 2002; Grant et al. 2005; Van Geel et al. 2017). Addition of P to low-nutrient soils has been found to support AM fungus growth by supplying mycorrhizal plants with this essential nutrient (Treseder and Allen 2002). While we found no significant effect of P on AM fungal communities, the NMDS suggested a positive association between P and AM fungi. The importance of soil chemistry and macronutrients for microbial communities has been demonstrated in AM fungal niche studies with similar climate and environmental conditions to Qatar (e.g., Mendoza et al. 2011; Wang et al. 2018; Zhang et al. 2017; Xu et al. 2018). In our study, we found elevated concentrations of soil NO2− and NO3− in sabkhas and the saltmarsh. In terms of the elements present in soil, we also found that locations with abundant shrubs, such as sites 5 and 10, had higher concentrations of Mg, K, and P than locations with relatively barren soils. Study sites with areas of barren land showed lower concentrations of NO3−, NO2−, and soil elements. These results suggest that areas with high soil nutrient concentrations are likely associated with plant productivity, which in turn supports mycorrhiza formation and AM fungus occurrence.
As stated in our second hypothesis, pH was negatively associated with AM fungi. While we found no significant effect of salinity, the NMDS suggests that salinity is negatively associated with AM fungi. Highly saline soil conditions are known to have adverse effects on root colonization, preventing mycorrhizal associations from developing (Juniper and Abbott 2006; Becerra et al. 2014). Much of Qatar’s central peninsula comprises a plateau of limestone and sandstone outcrops, contributing to the saline nature of the soil environment (Babikir 1990; Abulfatih et al. 2002). Overall, our results strongly suggest that soil habitats in Qatar are highly saline. Based on the data obtained, soils in lowland saltmarsh and mangrove areas near Qatar’s coasts (e.g., sites 2 and 3) were the most saline of those studied here. While mangrove and saltmarsh were only represented by one site each, the diversity of AM fungi present in these locations was low, and they were mostly inhabited by species from Glomeraceae and Paraglomeraceae. Due to their proximity to the coast and low topography, the soils at these sites exhibited high pH and salinity. The pH values measured across Qatar were within the range 7.59–9.20, which is similar to that reported in other dryland studies (Delgado-Baquerizo et al. 2015; Wang et al. 2016; Tian et al. 2017). The highest pH was recorded at the coast (Site 16), but most locations had relatively high pH levels, most likely as a result of the calcareous soils. The majority of the Arabian Peninsula, including Qatar, is characterized by low rainfall and high atmospheric temperature, contributing to high evapotranspiration rates and minimal leaching capacity. This causes ions such as carbonates and bicarbonates of Ca and Mg to accumulate in the soil, especially in areas at low altitude due to their proximity to seawater (Khanam et al. 2006). This may be why TC could be high without TN being measurable. Moreover, highly alkaline soils are strongly associated with high, toxic quantities of iron (Fe), carbonate and bicarbonate (CO32−, HCO3−) nutrient deficiency, and elevated sodium (Na) levels (Wu et al. 2018). Accumulation of exchangeable Na ions in arid soils can potentially reduce the physical stability and overall fertility of the soil, resulting in an inadequate habitat for AM fungal root colonization.
We found that Cd had a positive association with AM fungi, contradicting our third hypothesis. This suggests that very low concentrations of Cd in the soil may not have adverse effects on the AM fungal communities, although the diversity of AM fungal families is usually strongly influenced by heavy metals, in addition to other well-known chemical drivers (pH, salinity, K, P, and NO2−) (Lauber et al. 2008; Kasel et al. 2008; Pan et al. 2020). Heavy metals, particularly Cd, not only alter the soil chemistry, but also have been found to have detrimental effects on soil microbial communities when present even at low concentrations, which could lead to losses of diversity and soil fertility (Bååth and Anderson 2003; Lv et al. 2019; Kasemodel et al. 2019; Lin et al. 2019). For example, Cd toxicity to hyphae in species within the genus Glomus has been reported at Cd concentrations above 0.38 mg/kg (Jiang et al. 2016). AM fungi are generally sensitive to changes in soil conditions, including pH, heavy metals, and micronutrients and are considered to be useful bioindicators of soil contamination (Oehl et al. 2010; Vyas and Gupta 2014; Dietterich et al. 2017; Kasemodel et al. 2019; Lin et al. 2019; Pan et al. 2020). Our results in this regard were inconsistent with those presented in other fungal diversity studies (Gaudino et al. 2007; Zarei et al. 2010; Rouphael et al. 2015; Song et al. 2018). At all our study locations in Qatar, Pb in soil did not affect AM fungi. However, Pb was present at very low concentrations (average 4.92 mg/kg soil), and the effect of this heavy metal on belowground fungal communities is generally reported to be largely dependent upon the concentration. It is likely that Pb had no negative effect on fungi because the concentrations were well below a threshold for effect.
Conclusions
Using the Illumina MiSeq sequencing platform, we identified a total of 79 AM fungal OTUs, eight families, and four orders from the phylum Glomeromycota in soils from across Qatar. The main conclusions of this work are:
-
1.
Among the eight AM fungal families identified, Glomeraceae was the dominant fungus family at sampling locations in Qatar. The genus Glomus had the highest species richness (61 OTUs), while the genus Scutellospora had the lowest species richness (1 OTU).
-
2.
Variations in the diversity, richness, and abundance of AM fungal OTUs between locations were mainly attributable to K and NO3− availability, while pH and TC had negative associations with AM fungi. The direct effect of soil parameters on AM fungi requires further assessment in studies that examine the effect of interrelations among factors and bioavailability of minerals on mycorrhiza formation. Overall, AM fungi inhabiting Qatar’s desert habitat appear to have relatively broad environmental niches, with their diversity remaining unaffected by changes in chemical conditions in their soil environment.
Availability of data and material
The data have been deposited with links to BioProject accession number PRJNA675322 in the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/sra/PRJNA675322).
Code availability
Not applicable.
References
Abulfatih HA, Abdel Bari EM, Alsubaey A, Ibrahim YM (2002) Halophytes and soil salinity in Qatar. Qatar Univ Sci J 22:119–135
Al-Thani RF, Yasseen BT (2018) Biological soil crusts and extremophiles adjacent to native plants at sabkhas and rawdahs Qatar: the possible roles. Front Environ Microbiol 4:55. https://doi.org/10.11648/j.fem.20180402.13
Bååth E, Anderson TH (2003) Comparison of soil fungal/bacterial ratios in a pH gradient using physiological and PLFA-based techniques. Soil Biol Biochem 35:955–963. https://doi.org/10.1016/S0038-0717(03)00154-8
Babikir AAA (1990) The vegetation of the State of Qatar as related to landform and soil. J Doc Humanit Res Cent 462–478. http://hdl.handle.net/10576/8409
Becerra A, Bartoloni N, Cofré N et al (2014) Arbuscular mycorrhizal fungi in saline soils: Vertical distribution at different soil depth. Braz J Microbiol 45:585–594
Bhat BA, Sheikh MA, Tiwari A (2014) Impact of various edaphic factors on AMF spore population and diversity in Catharanthus roseus at Gwalior. Int J Plant Sci 9:1–6
Chao A, Chiu C-H, Jost L (2010) Phylogenetic diversity measures based on Hill numbers. Philos Trans R Soc B Biol Sci 365:3599–3609. https://doi.org/10.1098/rstb.2010.0272
Davison J, Moora M, Öpik M et al (2015) Global assessment of arbuscular mycorrhizal fungus diversity reveals very low endemism. Fungal Symbionts 351:826. https://doi.org/10.1126/science.aad4228
Del Val C, Barea JM, Azcon-Aguilar C (1999) Diversity of arbuscular mycorrhizal fungus populations in heavy-metal-contaminated soils. Appl Environ Microbiol 65:718–723. https://doi.org/10.1128/AEM.65.2.718-723.1999
Delgado A, Gómez JA (2016) The soil Physical chemical and biological properties. In: Villalobos F, Fereres E (eds) Principles of Agronomy for Sustainable Agriculture. Springer, Cham, pp 15–27. https://doi.org/10.1007/978-3-319-46116-8_2
Delgado-Baquerizo M, Florentino A, Gozalo B et al (2015) Increasing aridity reduces soil microbial diversity and abundance in global drylands. Proc Natl Acad Sci USA 112:201516684. https://doi.org/10.1073/pnas.1516684112
Dietterich LH, Gonneau C, Casper BB (2017) Arbuscular mycorrhizal colonization has little consequence for plant heavy metal uptake in contaminated field soils. Ecol Appl 27:1862–1875
Dufrêne M, Legendre P (1997) Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol Monogr 67:345–366. https://doi.org/10.2307/2963459
Dumbrell AJ, Ashton PD, Aziz N et al (2011) Distinct seasonal assemblages of arbuscular mycorrhizal fungi revealed by massively parallel pyrosequencing. New Phytol 190:794–804. https://doi.org/10.1111/j.1469-8137.2010.03636.x
Dumbrell AJ, Nelson M, Helgason T et al (2010) Relative roles of niche and neutral processes in structuring a soil microbial community. ISME J 4:337–345. https://doi.org/10.1038/ismej.2009.122
Erich MS, Hoskins BR (2011) Effects of soil drying on soil ph and nutrient extractability. Commun Soil Sci Plant Anal 42:1167–1176. https://doi.org/10.1080/00103624.2011.566961
Fitter AH (2005) Darkness visible: reflections on underground ecology. J Ecol 93:231–243. https://doi.org/10.1111/j.1365-2745.2005.00990.x
Gaudino S, Galas C, Belli M et al (2007) The role of different soil sample digestion methods on trace elements analysis: a comparison of ICP-MS and INAA measurement results. Accreditation Qual Assur 12:84–93. https://doi.org/10.1007/s00769-006-0238-1
Gerz M, Guillermo Bueno C, Ozinga WA et al (2018) Niche differentiation and expansion of plant species are associated with mycorrhizal symbiosis. J Ecol 106:254–264. https://doi.org/10.1111/1365-2745.12873
Grant C, Bittman S, Montreal M, Plenchette C, Morel C (2005) Soil and fertilizer phosphorus: Effects on plant P supply and mycorrhizal development. Can J Plant Sci 85:3–14. https://doi.org/10.4141/P03-182
Gustafson DJ, Casper BB (2006) Differential host plant performance as a function of soil arbuscular mycorrhizal fungal communities: experimentally manipulating co-occurring Glomus species. Plant Ecol 183:257–263. https://doi.org/10.1007/s11258-005-9037-8
Hammer EC, Nasr H, Pallon J, Olsson PA, Wallander H (2011) Elemental composition of arbuscular mycorrhizal fungi at high salinity. Mycorrhiza 21:117–129
Hsieh TC, Ma KH, Chao A (2016) iNEXT: an R package for rarefaction and extrapolation of species diversity (Hill numbers). Methods Ecol Evol 7:1451–1456. https://doi.org/10.1111/2041-210X.12613
Jansa J, Erb A, Oberholzer HR, Šmilauer P, Egli S (2014) Soil and geography are more important determinants of indigenous arbuscular mycorrhizal communities than management practices in Swiss agricultural soils. Mol Ecology 23:2118–2135. https://doi.org/10.1111/mec.12706
Jeffries P, Gianinazzi S, Perotto S et al (2003) The contribution of arbuscular mycorrhizal fungi in sustainable maintenance of plant health and soil fertility. Biol Fertil Soils 37:1–16
Jiang QY, Zhuo F, Long SH et al (2016) Can arbuscular mycorrhizal fungi reduce Cd uptake and alleviate Cd toxicity of Lonicera japonica grown in Cd-added soils? Sci Rep 6:2–10. https://doi.org/10.1038/srep21805
Jing X, Sanders NJ, Shi Y et al (2015) The links between ecosystem multifunctionality and above-and belowground biodiversity are mediated by climate. Nat Commun 6:1–8. https://doi.org/10.1038/ncomms9159
Juniper S, Abbott LK (2006) Soil salinity delays germination and limits growth of hyphae from propagules of arbuscular mycorrhizal fungi. Mycorrhiza 16:371–379
Kamalvanshi M, Kumar A, Jha A, Dhyani SK (2012) Occurrence of arbuscular mycorrhizal fungi in rhizosphere of Jatropha curcas L. in arid and semi arid regions of India. Indian J Microbiol 52:492–494. https://doi.org/10.1007/s12088-011-0224-0
Kasel S, Bennett LT, Tibbits J (2008) Land use influences soil fungal community composition across central Victoria, south-eastern Australia. Soil Biol Biochem 40:1724–1732. https://doi.org/10.1016/j.soilbio.2008.02.011
Kasemodel MC, Sakamoto IK, Varesche MBA, Rodrigues VGS (2019) Potentially toxic metal contamination and microbial community analysis in an abandoned Pb and Zn mining waste deposit. Sci Total Environ 675:367–379. https://doi.org/10.1016/j.scitotenv.2019.04.223
Khanam D, Mridha MAU, Solaiman ARM, Hossain T (2006) Effect of edaphic factors on root colonization and spore population of arbuscular mycorrhizal fungi. Bull Inst Trop Agr Kyushu Univ 29:97–104
Laban P, Metternicht G, Davies J (2018) Soil biodiversity and soil organic carbon: keeping drylands alive. IUCN, Gland
Lauber CL, Strickland MS, Bradford MA, Fierer N (2008) The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol Biochem 40:2407–2415. https://doi.org/10.1016/j.soilbio.2008.05.021
Lee J, Lee S, Young JPW (2008) Improved PCR primers for the detection and identification of arbuscular mycorrhizal fungi. FEMS Microbiol Ecol 65:339–349. https://doi.org/10.1111/j.1574-6941.2008.00531.x
Lin Y, Ye Y, Hu Y, Shi H (2019) The variation in microbial community structure under different heavy metal contamination levels in paddy soils. Ecotoxicol Environ Saf 180:557–564. https://doi.org/10.1016/j.ecoenv.2019.05.057
Lüneberg K, Schneider D, Siebe C, Daniel R (2018) Drylands soil bacterial community is affected by land use change and different irrigation practices in the Mezquital Valley Mexico. Sci Rep 8:1–15. https://doi.org/10.1038/s41598-018-19743-x
Lv W, Qiu Y, Xie Z et al (2019) Gravel mulching effects on soil physicochemical properties and microbial community composition in the Loess Plateau, northwestern China. Eur J Soil Biol 94:103115. https://doi.org/10.1016/j.ejsobi.2019.103115
Magoč T, Salzberg SL (2011) FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. https://doi.org/10.1093/bioinformatics/btr507
Mahasneh IA, Al-Thani RF, Brown G (2008) The micro-organisms of sabkhat in Qatar. Sabkha Ecosystems. Springer, Netherlands, pp 89–97
Mahmoudi N, Cruz C, Mahdhi M et al (2019) Arbuscular mycorrhizal fungi in soil roots and rhizosphere of Medicago truncatula: diversity and heterogeneity under semi-arid conditions. PeerJ 7:e6401. https://doi.org/10.7717/peerj.6401
Majid SA, Graw MF, Chatziefthimiou AD et al (2016) Microbial characterization of Qatari Barchans and Dunes. PLoS ONE 11:1–22. https://doi.org/10.1371/journal.pone.0161836
Mamoon A, Rahman A (2016) Rainfall in Qatar: is it changing? Nat Hazards 85:453–470. https://doi.org/10.1007/s11069-016-2576-6
Masson P, Dalix T, Bussière S (2010) Determination of major and trace elements in plant samples by inductively coupled plasma-mass spectrometry. Commun Soil Sci Plant Anal 41:231–243. https://doi.org/10.1080/00103620903460757
Mendoza R, Cabello M, Anchorena J et al (2011) Soil parameters and host plants associated with arbuscular mycorrhizae in the grazed Magellanic steppe of Tierra del Fuego. Agric Ecosyst Environ 140:411–418. https://doi.org/10.1016/j.agee.2011.01.004
Neilson JW, Califf K, Cardona C et al (2017) Significant impacts of increasing aridity on the arid soil microbiome. mSystems 2:1–15. https://doi.org/10.1128/msystems.00195-16
Norton J, Majid SA, Debbie A et al (2009) An illustrated checklist of the flora of Qatar. Browndown Publications, Gosport
Oehl F, Laczko E, Bogenrieder A et al (2010) Soil type and land use intensity determine the composition of arbuscular mycorrhizal fungal communities. Soil Biol Biochem 42:724–738. https://doi.org/10.1016/j.soilbio.2010.01.006
Öpik M, Vanatoa A, Vanatoa E et al (2010) The online database MaarjAM reveals global and ecosystemic distribution patterns in arbuscular mycorrhizal fungi (Glomeromycota). New Phytol 188:223–241. https://doi.org/10.1111/j.1469-8137.2010.03334.x
Pan X, Zhang S, Zhong Q et al (2020) Effects of soil chemical properties and fractions of Pb Cd and Zn on bacterial and fungal communities. Sci Total Environ 715:136904. https://doi.org/10.1016/j.scitotenv.2020.136904
Reynolds JF, Stafford Smith DM, Lambin EF et al (2007) Ecology: global desertification: building a science for dryland development. Science 316:847–851. https://doi.org/10.1126/science.1131634
Rognes T, Flouri T, Nichols B et al (2016) VSEARCH: a versatile open source tool for metagenomics. PeerJ 2016:1–22. https://doi.org/10.7717/peerj.2584
Rouphael Y, Franken P, Schneider C et al (2015) Arbuscular mycorrhizal fungi act as biostimulants in horticultural crops. Sci Hortic 196:91–108. https://doi.org/10.1016/j.scienta.2015.09.002
Shahabivand S, Maivan HZ, Goltapeh EM, Sharifi M, Aliloo AA (2012) The effects of root endophyte and arbuscular mycorrhizal fungi on growth and cadmium accumulation in wheat under cadmium toxicity. Plant Physiol Biochem 60:53–58. https://doi.org/10.1016/j.plaphy.2012.07.018
Smith SE, Read D (2008) Mycorrhizal symbiosis, 3rd edn. Elsevier, Academic Press, Amsterdam
Song J, Shen Q, Wang L et al (2018) Effects of Cd, Cu, Zn and their combined action on microbial biomass and bacterial community structure. Environ Pollut 243:510–518. https://doi.org/10.1016/j.envpol.2018.09.011
Soudzilovskaia NA, Douma JC, Akhmetzhanova AA et al (2015a) Global patterns of plant root colonization intensity by mycorrhizal fungi explained by climate and soil chemistry. Glob Ecol Biogeogr 24:371–382. https://doi.org/10.1111/geb.12272
Soudzilovskaia NA, van der Heijden MGA, Cornelissen JHC et al (2015b) Quantitative assessment of the differential impacts of arbuscular and ectomycorrhiza on soil carbon cycling. New Phytol 208:280–293. https://doi.org/10.1111/nph.13447
Tedersoo L, Bahram M, Põlme S et al (2014) Global diversity and geography of soil fungi. Science 346:1052–1053. https://doi.org/10.1126/science.aaa1185
Tedersoo L, Tooming-Klunderud A, Anslan S (2018) PacBio metabarcoding of fungi and other eukaryotes: errors biases and perspectives. New Phytol 217:1370–1385. https://doi.org/10.1111/nph.14776
Tian Q, Taniguchi T, Shi WY et al (2017) Land-use types and soil chemical properties influence soil microbial communities in the semiarid Loess Plateau region in China. Sci Rep 7:1–9. https://doi.org/10.1038/srep45289
Tighe M, Lockwood P, Wilson S, Lisle L (2004) Comparison of digestion methods for ICP-OES analysis of a wide range of analytes in heavy metal contaminated soil samples with specific reference to arsenic and antimony. Commun Soil Sci Plant Anal 35:1369–1385. https://doi.org/10.1081/CSS-120037552
Timmer LW, Leyden RF (1980) The relationship of mycorrhizal infection to phosphorous-induced copper deficiency in sour orange seedlings. New Phytol 85:15–23. https://doi.org/10.1111/j.1469-8137.1980.tb04443.x
Toljander JF, Santos-González JC, Tehler A, Finlay RD (2008) Community analysis of arbuscular mycorrhizal fungi and bacteria in the maize mycorrhizosphere in a long-term fertilization trial. FEMS Microbiol Ecol 65:323–338. https://doi.org/10.1111/j.1574-6941.2008.00512.x
Treseder KK, Allen MF (2002) Direct nitrogen and phosphorus limitation of arbuscular mycorrhizal fungi: a model and field test. New phytologist 155:507–515. https://doi.org/10.1046/j.1469-8137.2002.00470.x
United Nations Environment Management Group (2011) Global Drylands: A UN system wide response. Geneva, Switzerland
van der Heijden MGA, Martin FM, Selosse M-A, Sanders IR (2015) Mycorrhizal ecology and evolution: the past the present and the future. New Phytol 205:1406–1423. https://doi.org/10.1111/nph.13288
Van Geel M, Verbruggen E, De Beenhouwer M, van Rennes G, Lievens B, Honnay O (2017) High soil phosphorus levels overrule the potential benefits of organic farming on arbuscular mycorrhizal diversity in northern vineyards. Agric Ecosyst Environ 248:144–152. https://doi.org/10.1016/j.agee.2017.07.017
Vasar M, Andreson R, Davison J et al (2017) Increased sequencing depth does not increase captured diversity of arbuscular mycorrhizal fungi. Mycorrhiza 27:761–773. https://doi.org/10.1007/s00572-017-0791-y
Villalobos FJ, Mateos L, Quemada M et al (2016) Control of salinity. In: Villalobos FJ, Fereres E (eds) Principles of Agronomy for Sustainable Agriculture. Springer International Publishing, Cham, pp 295–320
Vyas D, Gupta RK (2014) Effect of edaphic factors on the diversity of VAM fungi. Trop Plant Res 1:14–25
Wang J, Chen C, Ye Z, Li J, Feng Y, Lu Q (2018) Relationships between fungal and plant communities differ between desert and grassland in a typical dryland region of northwest China. Front Microbiol 9:2327. https://doi.org/10.3389/fmicb.2018.02327
Wang Z, Liu L, Chen Q et al (2016) Conservation tillage increases soil bacterial diversity in the dryland of northern China. Agron Sustain Dev 36:1–9. https://doi.org/10.1007/s13593-016-0366-x
Wood ED, Armstrong FAJ, Richards FA (1967) Determination of nitrate in sea water by cadmium-copper reduction to nitrite. J Mar Biol Assoc UK 47:23–31. https://doi.org/10.1017/S002531540003352X
Wu Z, Liu Q, Li Z et al (2018) Environmental factors shaping the diversity of bacterial communities that promote rice production. BMC Microbiol 18:1–11. https://doi.org/10.1186/s12866-018-1174-z
Xu Z, Yu G, Zhang X et al (2018) Biogeographical patterns of soil microbial community as influenced by soil characteristics and climate across Chinese forest biomes. Appl Soil Ecol 124:298–305. https://doi.org/10.1016/j.apsoil.2017.11.019
Xue PP, Carrillo Y, Pino V et al (2018) Soil properties drive microbial community structure in a large scale transect in South Eastern Australia. Sci Rep 8:1–11. https://doi.org/10.1038/s41598-018-30005-8
Zahlan RS (2016) The Creation of Qatar, 3rd edn. Routledge, Doha
Zarei M, Hempel S, Wubet T et al (2010) Molecular diversity of arbuscular mycorrhizal fungi in relation to soil chemical properties and heavy metal contamination. Environ Pollut 158:2757–2765. https://doi.org/10.1016/j.envpol.2010.04.017
Zerihun B, Mauritz V, Fassil A (2013) Diversity and abundance of arbuscular mycorrhizal fungi associated with acacia trees from different land use systems in Ethiopia. Afr J Microbiol Res 7:5503–5515. https://doi.org/10.5897/ajmr2013.6115
Zhang Y, Dong S, Gao Q et al (2017) Soil bacterial and fungal diversity differently correlated with soil biochemistry in alpine grassland ecosystems in response to environmental changes. Sci Rep 7:1–10. https://doi.org/10.1038/srep43077
Zhao H, Li X, Zhang Z et al (2017) Species diversity and drivers of arbuscular mycorrhizal fungal communities in a semi-arid mountain in China. PeerJ 5:e4155. https://doi.org/10.7717/peerj.4155
Acknowledgements
The authors thank Muhammad Sulaiman, Mohamed Faisal, Monzer Al Mandili, and Maktom Abdulrahman for assistance with the fieldwork and Pedro Range for help with the NMDS. We also thank the editor and reviewers for the constructive comments that improved the paper.
Funding
This work was supported by a grant from Qatar Petroleum to JMA (QUEX-CAS-QP-RD-18/19).
Author information
Authors and Affiliations
Contributions
The study was designed by JMA. JMA, JO, and TA conducted the fieldwork. SA, JO, AMS, and TA conducted the laboratory work. SA and JO analyzed the data. SA was the main author of the manuscript, JMA and TA revised the manuscript. All the authors (SA, JO, JMA, AMS, MA, LT, MZ, TA) discussed and contributed to the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Adenan, S., Oja, J., Alatalo, J.M. et al. Diversity of arbuscular mycorrhizal fungi and its chemical drivers across dryland habitats. Mycorrhiza 31, 685–697 (2021). https://doi.org/10.1007/s00572-021-01052-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00572-021-01052-3