Abstract
Background
Intrahepatic cholangiocarcinoma (ICC) is the second most common tumor in primary liver cancer, but the prognostic factors associated with long-term outcomes after surgical resection remain poorly defined. This study aimed to develop a novel prognostic classifier for patients with ICC after surgery.
Methods
Using a proteomics approach, we screened tumor markers that up-regulated in ICC tissues, and narrowed down by bioinformatics analysis, western blot and immunohistochemistry. Prognostic markers were identified using Cox regression analyses in primary training cohort and the predictive models for time to recurrence (TTR) were established. The predictive accuracy of predictive model was validated in external validation cohort and prospective validation cohort. MTT assay, clonal formation assay and trans-well assays were used to verify the effect on the proliferation and migration in ICC cell line.
Results
Triosephosphate isomerise (TPI1) was significantly up-regulated in ICC tissues and Kaplan–Meier analysis reveals that higher TPI1 expression was strongly correlated with higher recurrence rate of ICC patients. In the primary training cohort, mean TTR was significantly longer (p < 0.0001) than in the low-risk group (26.9 months for TTR, 95% CI 22.4–31.5) than in the high-risk group (14.5 months for TTR, 95% CI 10.6–18.4). Similar results were observed in two validation cohorts. In addition, a nomogram to predict recurrence was developed. Moreover, Knockdown of TPI1 by shRNA inhibited ICC cell growth, colony information, migration, invasion in vitro.
Conclusions
Current prognostic models were accurate in predicting recurrence for ICC patients after surgical resection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Intrahepatic cholangiocarcinoma (ICC) is a poorly understood biliary malignancy that originates from bile duct epithelium or hepatic progenitor cells within the liver [1]. ICC is the second-most-common primary liver cancer, accounting for an estimated 10–15% of all primary liver cancers [2]. The incidence and mortality of ICC has been increasing over the past 3 decades and only a low proportion of patients survive 5 years after diagnosis [3]. To date, surgery is the only potentially curative treatment for patients with resectable ICC [4], although a high risk of recurrence and poor prognosis are observed even after surgery due to the high grade of malignancy and ill-defined resection margin associated with ICC.
The 7th and 8th edition of the tumor–node–metastasis (TNM) staging system for ICC, proposed by the American Joint Committee on Cancer (AJCC), attempts to stratify patients according to overall survival (OS), while factors and predictors of time to recurrence (TTR) are not described [5]. The identification of new molecular markers for ICC to develop a novel prognostic scoring system might be beneficial to stratify patients for selecting an optimal treatment, and thus direct adjuvant therapy and the delivery of optimally targeted and timed interventions tailored to an individual’s molecular drivers of disease [6].
Proteomics analysis delivers on the promise of cancer biomarkers and remains an important technique for the screening of potential biomarkers [7, 8]. Proteomics techniques combined with bioinformatics analysis in the field of cancer research has an obvious advantage aim to identifying differentially expressed proteins [9], and successfully used for discovery of diagnostic biomarkers [10, 11], prognostic biomarkers [12, 13], associated with the development and progression of cancer and providing new opportunities to discover biomarkers and therapeutic targets as well as opportunities to reveal the molecular mechanism underlying the disease [14, 15].
In this study, we used an isobaric tag for relative and absolute quantitation (iTRAQ) technique combined with bioinformatics analysis to identify proteins for the prediction of recurrence in ICC. Proteomics analysis revealed that triosephosphate isomerise (TPI1) was significantly over-expressed in ICC tissue compared with intrahepatic bile duct (IHBD) tissue samples and it was further confirmed by western blotting and immunohistochemistry using tissue micro-array (TMA). We further evaluated the expression level of TPI1 by immunohistochemistry using TMA and by investigating its association with clinicopathological characteristics and prognosis especially time to recurrence and developed classifier and nomogram for predicting recurrence of ICC patients after surgical resection. The results may provide useful tool for predicting recurrence of ICC patients after surgery and will facilitate the identification of the most appropriate treatment strategy to improve clinical outcomes.
Patients and methods
Study design and patients
Study design of present study was indicated in Supplementary Figure 1. We used 1047 cases of ICC formalin-fixed paraffin-embedded (FFPE) tissue samples. Clinicopathological characteristics of 35 pairs of IHBD and ICC samples (32 pairs are FFPE and 3 pairs are fresh frozen tissues) were used in the screening and narrow down process was showed in Supplementary Table 1. iTRAQ/2DLC/ESI/MS/MS proteomics analysis used for screen of differentially expressed proteins, and bioinformatics analyses, western blotting, TMA-based immunohistochemistry were used for screen and narrow down of candidate biomarkers. OS and TTR data were obtained from 519 patients at the Eastern Hepatobiliary Surgery Hospital (EHBH), Second Military Medical University, Shanghai, China, between July 2000 and Dec 2008 as the primary training cohort. For the external validation cohort, these data were obtained from 318 patients from the Liver Cancer Institute, Zhongshan Hospital (ZSH), Fudan University, Shanghai, China, between March 2005 and March 2011. Data for the prospective cohort were obtained from 178 patients at the EHBH, Second Military Medical University, Shanghai, China, between December 2011 and December 2012. Patients in the training cohort were followed up until September 2014 [16], and patients in the external validation cohort were followed-up until April 2015 [17]; those patients in the prospective validation cohort were followed-up until Jan 2017. Patients had no signs of distant metastasis and had not received anti-cancer treatments before surgery. OS was defined as the length of time between surgery and death, or the last follow-up examination. TTR was defined from the date of tumor resection until the detection of tumor recurrence, death or last observation. Recurrence data were adopted for the patients who were able to determine recurrence or non-recurrence during the follow-up period, while those patients who were uncertain for recurrence were censored.
Clinicopathological characteristics of three cohort of ICC patients were used in the prognostic evaluation was shown in Supplementary Table 2. The detailed follow-up procedure was shown in Supplementary Methods Follow-up Procedure.
This study was approved by the institutional ethical review boards of both participating hospitals, and written informed consent was obtained from all patients.
Proteomics analysis and selection of proteins
We extracted protein from 10 cases of IHBD and 10 cases of ICC FFPE specimens and performed iTRAQ labelling, 2DLC-ESI–MS/MS analysis, database searching, and criteria as reported previously [18]. Proteins were labelled with iTRAQ isobaric tags of 118 for IHBD and 121 for ICC, respectively. For enhancement of protein coverage, three replicates were performed on RPLC-ESI–MS/MS, and quantified proteins were subsequently filtered with manually selected filter exclusion parameters (at least 2 peptides, p < 0.05 and expression level differed at least 1.25- or 0.8-fold in ICC compared to IHBD). We performed bioinformatics analysis focused on these up-regulated proteins in ICC.
Evaluation of protein expression level
Protein extraction from fresh frozen tissue was performed and TMA were utilized in primary training cohort and external validation cohort were constructed using two representative cores from each specimen as described previously [18]. In the prospective validation cohort, the regular large sections were used for immunohistochemistry.
Primary antibodies used for western blotting and immunohistochemistry (IHC) were as noted in Supplementary Table 3. The omission of primary antibody served as an additional negative control. For evaluating the expression levels of three proteins in IHBD and ICC tissue samples using IHC, the protein expression levels (IOD/um × um) were determined using an imaging system comprising a CCD camera (DFC420) connected to a DM IRE2 microscope (Leica Microsystems Imaging Solutions, Cambridge, UK) and Leica QW in Plus v3.0 software. The mean expression levels from two images for each sample were used to represent expression level using Image-Pro Plus v6.0 software (Media Cybernetics, Bethesda, MD, USA), as reported previously (Supplementary Figure 2) [18]. According to our experience of thousands of cases, the maximum ratio (diffuse strong positive) IOD / um × um is 150, and the minimum ratio (negative) is 0.
Construction and validation of prognostic classifier for ICC
For construction of prognostic classifiers in the training cohort, protein expression levels and clinicopathologic characteristics with continuous variables were subjected to X-tile software analysis (v3.6.1, Yale University, New Haven, CT, USA), and the optimum cut-off points based on outcome were obtained [19]. Cut-off points for each continuous variable of IHC tumor markers, were obtained using X-tile analyses from the training cohort, and this cut-off points were applied to external validation cohort and prospective validation cohort.
Using the primary training cohort, prognostic factors (p < 0.05) were selected by univariate Cox analysis, and these factors were included in multivariate Cox regression analyses to construct a prognostic classifier for OS and TTR, respectively. The predictive accuracy of the prognostic classifier was further validated in the external validation cohort and prospective validation cohort of ICC patients using Kaplan–Meier analysis and AUC analysis.
Cell culture, short hairpin RNA and real-time PCR
The ICC cell line RBE was purchased from the Cell Center of the Chinese Academy of Sciences. The cells were maintained in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum (Invitrogen) and cultured at 37 °C in 5% CO2. TPI1 short hairpin RNA (shRNA) and unspecific scrambled shRNA plasmids were purchased from Sunbio Medical Biotechnology (Shanghai, China) (Supplementary Figure 3 A). RBE cells were digested, and 1 × 105 of the cells were seeded in 6-well plates. The cells were transfected with TPI-shRNA or scrambled shRNA using Lipofectamine 2000 reagent (Invitrogen) and 5 ng of shRNA plasmid per well according to the reagent manufacturer’s instructions. The lentivirus vectors were transfected into 293 T packaging cells to obtain control (pMagic 1.1) and shRNA-lentivirus (PLVE2089, PLVE2090, PLVE2091). After transfecting shRNA-lentivirus, interference efficiency was confirmed by real-time PCR and Actin as internal control gene. Science, shRNA-lentivirus (PLVE2091) was shows better interference efficiency by real-time PCR (Supplementary Figure 3 B), we further confirmed interference efficiency at protein level by western blot (Supplementary Figure 3 C).
Cell proliferation and Colony formation assay
A number of transfected cells in exponential growth were seeded into 96-well plates with 2 × 104 cells per well. Then the plates were added with 10 μl CCK-8 (Beyo Biotechnology, China) after incubated for 0, 24, 48 and 72 h. The cell activity was detected by Microplate System using 450 nm and 620 nm spectrum. 500 cells were inoculated into 6-well plates and hatched in 37 °C, 5% CO2 for 12 days. Afterwards the 6-well plates were fixed by methyl alcohol and dyed with Giemsa (Invitrogen Life Technologies) and counted cells colonies and then figured up the cloning efficiency.
Migration and invasion assay
1 × 105 RBE cells transfected with TPI1-shRNA-lentivirus and control lentivirus in 100 μl serum-free media were planted into the upper chamber of the transwell chambers (Corning, USA). While the following wells were filled with 600 μl 10% FBS DMEM as a function of chemoattractant which can induce the cells in the upper chamber to the lower one. After incubated for 48 h, cells non-invaded were rubbed away using cotton swab carefully, while cells that invaded to the lower chamber were fixed with 4% formaldehyde and stained using HE reagent. We observed at least six separate fields of cells by fluorescence microscope. The invasion assay was similar to the migration assay but the upper chamber was coated with Matrigel (Sigma-Aldrich, USA).
Statistical analysis
For comparing quantitative variables, the Mann–Whitney test was performed using GraphPad Prism (v5.01). To calculate the optimum cut-off point for OS and TTR, the X-tile statistical package (v3.6.1) was used. Other statistical analyses were performed using SPSS v22.0 software (SPSS, Chicago, IL, USA). Chi-square tests were used to compare qualitative variables, while Kaplan–Meier analysis and a log-rank test were used to compare the time of categorization of patients. Univariate and multivariate Cox regression analyses were performed to determine the predictors of ICC. To evaluate the prediction efficacy, ROC analysis was performed. Nomogram and calibration plots were done with the rms package of R software (version 3.3.1). All statistical tests were two-sided, and a p value of < 0.05 was considered statistically significant.
Results
Screening and identification of differentially expressed proteins in ICC
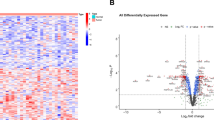
To identify proteins that were differentially expressed in ICC, an iTRAQ technique was first used ICC tissues and normal IHBD tissues (Fig. 1a). According to the criteria for protein quantification described previously [18]. 1066, 665, and 537 proteins were quantified by each technical replicate, respectively (data not shown). From three replicate, 1283 proteins (data not shown) were quantified and subsequently filtered with manually selected filter exclusion parameters (at least 2 peptides, p < 0.05, and at least 1.25- or 0.8-fold change in expression level in ICC compared with IHBD). Finally, 107 proteins were screened out, including 47 proteins up-regulated in ICC and 60 proteins down-regulated in ICC (Supplementary Table 4). In turn, 47 up-regulated proteins in ICC were selected for bioinformatics analysis. Gene ontology (GO) classification showed that a significant role of glycometabolism (glycolysis, glucose metabolic process, gluconeogenesis) was involved in ICC (Fig. 1b and Supplementary Table 5). Pathway analysis was also revealed that glycometabolism (glycolysis/gluconeogenesis, pentose phosphate pathway, fructose and mannose metabolism) is mainly involved in ICC (Supplementary Table 6 and Supplementary Figure 4). Taken together, Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH), Pyruvate Kinase M1/2 (PKM2), Enolase 1 (ENO1), tansketolase (TKT), fructose-bisphosphate aldolase A (ALDOA), TPI1 were selected as preliminary candidate biomarkers, which up-regulated in ICC and participate in glycometabolism. After literature review, GAPDH was abandoned for further analysis, because it is the internal reference, ENO1 and PKM2 were also abandoned for further analysis, because these two proteins were have been reported as prognostic markers in cholangiocarcinoma and ICC [20, 21]. While, TKT, ALDOA, and TPI1 proteins have never been reported as prognostic markers for ICC, thus, we chose these three proteins as candidate proteins for further validation using western blotting and IHC for find new prognostic factors of ICC. Western blotting analysis indicates TKT and ALDOA expression was not differed between IHBD and IHC, while, TPI1 was significantly up-regulated in ICC (Fig. 1c). IHC result indicates TKT was significantly down-regulated in ICC, while ALDOA and TPI1 was significantly up-regulated in ICC (Fig. 1d). Finally, we selected ALDOA and TPI1 as tumor markers for IHC, and further evaluated the prognosis value of these two proteins.
Screening process for proteins up-regulated in ICC. a The reporter ion assignments are as follows: 118-IHBD, 121-ICC. The intensity of reporter ions from precursor peptides indicates protein expression levels. Representative MS/MS spectrum showing the peptide of TPI1 protein and the MS/MS spectra show identified peptide sequences with QSLGELIGTLNAAK leading to the identification of TPI1. b Gene ontology (GO) classification showed that a significant role of glycometabolism (glycolysis, glucose metabolic process, gluconeogenesis) was involved in ICC. c Expression level of TKT, ALDOA, and TPI proteins in 3 pairs of IHBD and ICC tissues quantified by western blot analysis. d Representative expression of TKT, ALDOA, TPI1 in IHBD and ICC tissues by immunohistochemistry (left figure) and quantitative analyses these 3 proteins by immunohistochemistry in 22 pairs of IHBD and ICC tissue samples (right figure). Two tailed Mann–Whitney tests, error bars represent mean ± s.e.m
Determination of optimum cut-off point by X-tile
The optimum cut-off point was determined by X-tile analysis for quantitative variables, including clinicopathologic characteristics (CA199, ALB, tumor size), ALDOA, TPI1 expression. The representative X-tile plot for tumor size is shown in Supplementary Figure 5 (Table 1).
Construction and validation of the prognostic classifier
To build a prognostic classifier, factors that were significant on univariate Cox regression analysis in primary training cohort was used. As results, the prognostic classifier for OS was constructed from serum CA199, serum ALB, tumor size, tumor number, direct invasion; and the prognostic classifier for TTR was constructed from tumor size, tumor number, direct invasion, and TPI1. Because serum CA199, serum ALB, tumor size, tumor number, direct invasion has been reported as the prognostic factor for OS, while, TPI1 was identified as novel prognostic factor for TTR, thus we only focus on the prognostic classifier for TTR (Table 2).
The formula for recurrence was below:
In this formula, low status equal 0; high status equal 1 for tumor size, TPI1 and single tumor equal 1; multiple tumor equal 2; and no direct invasion equal 0, and direct invasion equal 1, respectively. From the equation, a risk score could be calculated which was a linear predictor variable for each patient, and was able to estimate recurrence risk for each case. In general, among patients with risk score of 0 or lower the patient at low risk of recurrence (low-risk group) would be low; those patients with high risk score that is greater than 0 had a high risk of recurrence (high-risk group) and this was also confirmed using X-tile plots (Supplementary Figure. 6).
Patients with lower risk scores generally had a better recurrence status versus patients with higher risk scores in the three cohorts (Fig. 2). Kaplan–Meier analyses in the primary training cohort demonstrate that mean TTR in training cohort among the high risk group was 13.2 months (95% CI 9.5–17.0) versus 26.9 (95% CI 22.5–31.4) months in the low risk group (p < 0.0001) (Fig. 2a). In the external validation cohorts, mean TTR was 29.5 months (95% CI 22.4–36.5) in the high risk group versus 48.2 months (95% CI 40.6–55.7) in low risk group (p < 0.0001) (Fig. 2b), and 11.6 months (95% CI 8.2–15.1) versus 26.4 (95% CI 21.0–31.8) months in the prospective validation cohort (p < 0.0001) (Fig. 2c).
Risk sore by the classifier and Kaplan–Meier curves for recurrence in the primary training cohort, external validation cohort, and prospective validation cohort. a Risk score distribution for TTR and Kaplan–Meier plot for primary training cohort. b Risk score distribution for TTR and Kaplan–Meier plot for external validation cohort. c Risk score distribution for TTR and Kaplan–Meier plot for prospective validation cohort. TTR time to recurrence
Evaluation of predictive efficacy
Using ROC analysis, we compared sensitivity and specificity for recurrence prediction. Area under the curve (AUC) analyses demonstrated that the classifier for TTR (AUC = 0.644 for primary training cohort; AUC = 0.639 for external validation cohort; AUC = 0.700 for prospective validation cohort) was better than tumor size (AUC = 0.591; AUC = 0.552; AUC = 0.607), tumor number (AUC = 0.546; AUC = 0.566; AUC = 0.582), direct invasion (AUC = 0.523; AUC = 0.560; AUC = 0.581), and TPI1 (AUC = 0.599; AUC = 0.579; AUC = 0.611) in all three cohorts (Fig. 3). Furthermore, we calculated AUCs using ROC analyses at 1, 3, and 5 years of recurrence to assess time dependent prognostic accuracy. Specifically, the classifier had a better predictive performance for 1-, 3-, and 5-year recurrence than individual prognostic factors (Supplementary Figure. 7, Supplementary Figure. 8, and Supplementary Figure. 9). Collectively, the results suggested that the classifier could better prognostically stratify ICC patients into low-risk or high-risk for recurrence following surgery.
ROC curves compare the prognostic accuracy of the classifier with clinicopathological risk factors in the primary training cohort, external validation cohort, and prospective validation cohort. ROC curves and AUC for TTR in a primary training cohort, b external validation cohort, c prospective validation cohort. TTR time to recurrence, ROC receiver operator characteristic. AUC area under curve. TPI1 triosephosphate isomerase 1
Nomogram for recurrence
To provide the clinician with a quantitative method to predict probability of recurrence, nomogram was constructed using the primary training cohort data (Fig. 4). Calibration plots showed that the nomograms performed well compared with an ideal model in the primary training cohort (Supplementary Figure. 10), external validation cohort (Supplementary Figure. 11) and prospective validation cohort (Supplementary Figure. 12) at 1-, 3-, and 5-year recurrence.
Time to recurrence nomogram for patient with intrahepatic cholangiocarcinoma after surgery. a Nomogram for predicting proportion of patient recurrence. (To use the nomogram, an individual patient’s value is located on each variable axis, and a line is drawn upward to determine the number of points received for each variable value. The sum of these numbers is located on the Total points axis, and a line is drawn downward to the survival axes to determine the likelihood of 1-, 3-, 5-year recurrence). TPI1 triosephosphate isomerase 1
TPI1-shRNA suppressed growth, colony number, migration and invasion of RBE cells
Based on the clinical data, we speculated that TPI1 might influence aggressiveness of RBE cells. As expect, knockdown of TPI1 by shRNA significantly decreased growth of RBE cells, as revealed by cell proliferation assay (p < 0.001) (Fig. 5a) and colony formation assay (p < 0.001) (Fig. 5b). Furthermore, knockdown of TPI1 significantly inhibited migration (p < 0.001) and invasion (p < 0.001) of RBE cells (Fig. 5c, d). These observations were consistent with increased TPI1 expression in clinical ICC samples.
Discussion
In this study, tumor markers for IHC were screened using an iTRAQ technique, bioinformatics analyses, western blotting, and TMA-based IHC. After screening for up-regulated proteins in ICC, a novel prognostic classifier based on a combination of widely used clinicopathological features and tumor markers was developed and validated. The novel prognostic classifiers were able to improve the prediction of TTR among ICC patients after surgery compared with other traditional, single prognostic markers. In turn, a risk score calculated by the prognostic classifier could classify patients into high-risk recurrence and low-risk recurrence of populations that more accurately stratified patients than any single prognostic factor, such as tumor size, tumor number, direct invasion, and TPI1. Importantly, while these findings were initially developed in a primary training cohort, the data were subsequently confirmed in both an external validation cohort and a prospective cohort. Furthermore, ROC analyses demonstrated that 1-, 3-, and 5-year recurrence were better stratified by the novel prognostic classifier than any single prognostic factor when trying to predict TTR in three cohorts (Supplementary Figure. 7, Supplementary Figure. 8, and Supplementary Figure. 9). In addition, calibration curve also showed good agreement between prediction and observation in the probability of 1-, 3-, and 5-year TTR n three cohorts (Supplementary Figure. 10, Supplementary Figure. 11, and Supplementary Figure. 12.
The TNM staging system is currently the most commonly used schema for prognostic purposes among patients with ICC. However, large variations in the clinical outcomes of patients within the same cancer stage have been reported [22] and the current ICC TNM staging system (8th edition) addresses only OS. Therefore, the TNM staging system lacks the ability to predict recurrence [5]. Similarly, ICC staging systems proposed by Okabatashi [23], Nathan [24], and Wang [25] had significant predictive value, but did not include IHC tumor markers and/or TTR predictors. Importantly, the prognostic classifier developed in this study includes comprehensive features such as TNM staging, tumor characteristics (size and number), IHC tumor markers (TPI1). Importantly, comparing the present predictors with Wang’s predictors [25] and without IHC tumor markers, the present predictor had superior predictive accuracy for TTR (Supplementary Figure. 13).
TPI1 catalyses the stereospecific 1,2-proton shift at dihydroxyacetone phosphate (DHAP) to give (R)-glyceraldehyde 3-phosphate through a pair of isomeric enzyme-bound cis-enediolate phosphate intermediates [26]. TPI1 expression levels are higher in metastatic tumors than in primary tumors of ovarian cancers [27]. Results from the current study demonstrated for the first time that higher TPI1 expression might indicate higher recurrence rate of ICC patients, TPI1 expression levels are higher in no recurrence tumors than in recurrence tumors of ICC (Supplementary Figure. 14), and TPI1-shRNA suppressed growth, colony number, migration and invasion of ICC cells. However, further studies are required on the transcriptional regulation mechanism of TPI1, its effect on glucose metabolism and its relationship with tumor recurrence and metastasis need to be clarified.
In summary, using up-regulated proteins in ICC, a prognostic classifier was developed based on clinicopathological features and IHC tumor markers. The classifiers for TTR were validated in an independent external cohort and a prospective cohort. The prognostic and predictive accuracy of the classifier was noted to be superior to other independent prognostic factors, including tumor size, tumor number, and direct invasion. These data provide a crucial first framework for the development of prospective risk-stratified clinical trials for TTR in patients with ICC after surgery. The proposed prognostic classifiers facilitate prognostic differentiation of ICC patients after surgery and will facilitate the identification of the most appropriate treatment strategy to improve clinical outcomes.
Conclusion
TPI1 was identified as novel prognostic factors for ICC, and proposed prognostic classifiers may classify postoperative ICC patients in high or low risk, and the present classifiers may be a useful tool for the most appropriate treatment strategy to improve clinical outcomes.
Abbreviations
- ICC:
-
Intrahepatic cholangiocarcinoma
- AUC:
-
Area under the curve
- OS:
-
Overall survival
- TTR:
-
Time to recurrence
- TNM:
-
Tumor–node–metastasis
- AJCC:
-
American Joint Committee on Cancer
- iTRAQ:
-
Isobaric tag for relative and absolute quantitation
- PKM2:
-
Pyruvate kinase isozymes M1/M2
- TPI1:
-
Triosephosphate isomerise
- FFPE:
-
Formalin-fixed paraffin-embedded
- IHBD:
-
Intrahepatic bile duct
- TMA:
-
Tissue microarray
- EHBH:
-
Eastern Hepatobiliary Surgery Hospital
- ZSH:
-
Zhongshan Hospital
- IOD:
-
Integrated optical density
- TKT:
-
Tansketolase
- ALDOA:
-
Fructose-bisphosphate aldolase A
References
Patel T. Cholangiocarcinoma–controversies and challenges. Nat Rev Gastroenterol Hepatol. 2011;8:189–200.
Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90.
Rizvi S, Gores GJ. Pathogenesis, diagnosis, and management of cholangiocarcinoma. Gastroenterology. 2013;145:1215–29.
Khan SA, Thomas HC, Davidson BR, et al. Cholangiocarcinoma Lancet. 2005;366:1303–14.
Edge SB, Byrd DR, Compton CC. et al. AJCC Cancer Staging Manual. 7th ed. New York: Springer; 2010.
Amin MB, Greene FL, Edge SB, et al. The eighth edition AJCC cancer staging manual: continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J Clin. 2017;67:93–9.
Powell K. Proteomics delivers on promise of cancer biomarkers. Nat Med. 2003;9:980.
Zhang B, Wang J, Wang X, et al. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513:382–7.
Djomehri SI, Gonzalez ME, da Veiga LF, et al. Quantitative proteomic landscape of metaplastic breast carcinoma pathological subtypes and their relationship to triple-negative tumors. Nat Commun. 2020;11:1723.
Park JH, Lee C, Han D, et al. Moesin (MSN) as a novel proteome-based diagnostic marker for early detection of invasive bladder urothelial carcinoma in liquid-based cytology. Cancers (Basel). 2020;12:4.
Sakaguchi Y, Yamamichi N, Tomida S, et al. Identification of marker genes and pathways specific to precancerous duodenal adenomas and early stage adenocarcinomas. J Gastroenterol. 2019;54:131–40.
Ribeiro IP, Esteves L, Anjo SI, et al. Proteomics-based predictive model for the early detection of metastasis and recurrence in head and neck cancer. Cancer Genom Proteom. 2020;17:259–69.
Zhou Q, Bauden M, Andersson R, et al. YAP1 is an independent prognostic marker in pancreatic cancer and associated with extracellular matrix remodeling. J Transl Med. 2020;18:77.
Fotouhi O, Kjellin H, Juhlin CC, et al. Proteomics identifies neddylation as a potential therapy target in small intestinal neuroendocrine tumors. Oncogene. 2019;38:6881–97.
Singh AN, Sharma N. Quantitative SWATH-based proteomic profiling for identification of mechanism-driven diagnostic biomarkers conferring in the progression of metastatic prostate cancer. Front Oncol. 2020;10:493.
Jin GZ, Dong W, Dong H, et al. The diagnostic and prognostic value of MRP8/MRP14 in intrahepatic cholangiocarcinoma. Oncotarget. 2015;6:39357–64.
Gao Q, Zhao YJ, Wang XY, et al. Activating mutations in PTPN3 promote cholangiocarcinoma cell proliferation and migration and are associated with tumor recurrence in patients. Gastroenterology. 2014;146:1397–407.
Jin GZ, Li Y, Cong WM, et al. iTRAQ-2DLC-ESI-MS/MS based identification of a new set of immunohistochemical biomarkers for classification of dysplastic nodules and small hepatocellular carcinoma. J Proteome Res. 2011;10:3418–28.
Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252–9.
Rucksaken R, Pairojkul C, Pinlaor P, et al. Plasma autoantibodies against heat shock protein 70, enolase 1 and ribonuclease/angiogenin inhibitor 1 as potential biomarkers for cholangiocarcinoma. PLoS ONE. 2014;9:e103259.
Qian Z, Hu W, Lv Z, et al. PKM2 upregulation promotes malignancy and indicates poor prognosis for intrahepatic cholangiocarcinoma. Clin Res Hepatol Gastroenterol. 2020;44:162–73.
Liu N, Chen NY, Cui RX, et al. Prognostic value of a microRNA signature in nasopharyngeal carcinoma: a microRNA expression analysis. Lancet Oncol. 2012;13:633–41.
Okabayashi T, Yamamoto J, Kosuge T, et al. A new staging system for mass-forming intrahepatic cholangiocarcinoma: analysis of preoperative and postoperative variables. Cancer. 2001;92:2374–83.
Nathan H, Aloia TA, Vauthey JN, et al. A proposed staging system for intrahepatic cholangiocarcinoma. Ann Surg Oncol. 2009;16:14–22.
Wang Y, Li J, Xia Y, et al. Prognostic nomogram for intrahepatic cholangiocarcinoma after partial hepatectomy. J Clin Oncol. 2013;31:1188–95.
Richard JP. A paradigm for enzyme-catalyzed proton transfer at carbon: triosephosphate isomerase. Biochemistry. 2012;51:2652–61.
Yoshida A, Okamoto N, Tozawa-Ono A, et al. Proteomic analysis of differential protein expression by brain metastases of gynecological malignancies. Hum Cell. 2013;26:56–66.
Acknowledgements
We thank Lu Xin Yuan (Department of Pathology, Eastern Hepatobiliary Surgery Hospital, Second Military Medical University) for review of HE stained sections in the initial stage of the study.
Funding
This work was supported by grants from the National Natural Science Foundation of China; 81972574, 81472769 (Guang-Zhi Jin), 81572856 (Guanzhen Yu), 81472278 (Wen-Ming Cong). National S&T Major Project of China, 2017ZX10203205 (Guang-Shun Yang). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Study concept and design: G-ZJ, W-LY. Drafting of the manuscript: G-ZJ, W-LY. Acquisition of data, analysis and interpretation of data: G-ZJ, W-LY, GY, HD, KC, JX, HY. Critical revision of the manuscript: W-MC, A-JL. Statistical analysis: G-ZJ, JX. Study supervision: G-SY. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Ethics approval and consent to participate
Each patient or their guardians provided informed consent and the Ethics Committee of Eastern Hepatobiliary Surgery Hospital and Zhongshan Hospital Research Ethics Committee approved the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yu, WL., Yu, G., Dong, H. et al. Proteomics analysis identified TPI1 as a novel biomarker for predicting recurrence of intrahepatic cholangiocarcinoma. J Gastroenterol 55, 1171–1182 (2020). https://doi.org/10.1007/s00535-020-01729-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00535-020-01729-0