Abstract
Background
Pancreatic cancer (PC) is categorized as a neoplasm associated with Lynch syndrome; however, the precise proportion of PC patients harboring DNA mismatch repair genes (MMR genes) remains unclear, especially in the Asian population.
Methods
Among 304 Japanese patients with pathologically proven pancreatic ductal adenocarcinoma, we selected 20 (6.6%) patients with a personal or family history involving first- or second-degree relatives fulfilling the revised Bethesda guidelines (RBG), defined as RBG-compatible cases. We analyzed germline variants in 21 genes related to a hereditary predisposition for cancer as well as clinical features in all 20 cases.
Results
The RBG-compatible cases did not show any unique clinicopathological features. Targeted sequencing data revealed three patients carrying deleterious or likely deleterious variants. Specifically, these three patients harbored a nonsense variant in ATM, a frameshift variant in ATM, and a concurrent nonsense variant in PMS2 and missense variant in CHEK2 (double-mutation carrier), respectively. Although an MMR gene mutation was identified in only one of the 20 patients, up to 15% of the RBG-compatible PC cases were associated with germline deleterious or likely deleterious variants.
Conclusions
These findings showed that these guidelines could be useful for identifying PC patients with DNA damage repair genes as well as MMR genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Overcoming the poor prognosis of pancreatic cancer (PC) remains one of the most challenging problems for oncological researchers and clinicians. PC was ranked as the seventh leading cause of cancer-related mortality worldwide in 2012, contributing to approximately 331,000 deaths per year [1]. Although new diagnosis and treatment strategies have shown steady clinical impacts on patients with PC, the prognosis remains dismal with a 5-year survival rate of 8% in the USA and Japan [http://seer.cancer.gov/statfacts/html/pancreas.html; http://ganjoho.jp/data/reg_stat/statistics/brochure/2016/cancer_statistics_2016_date_J.pdf].
Research in the last few decades has provided a deeper understanding of hereditary cancer risks, led to the identification of hereditary cancer susceptibility genes, and contributed significant advancements in genetic testing technology. Several hereditary syndromes with potential germline mutations, such as hereditary breast and ovarian cancer syndrome, are considered to be high-risk factors for the development of PC [2]. According to the revised Bethesda guidelines (RBG) published in 2004, PC is categorized as a neoplasm associated with a form of hereditary non-polyposis colorectal cancer termed Lynch syndrome (LS), which is genetically characterized by the existence of germline mutations in DNA mismatch repair genes (MMR genes), including MLH1, MSH2, MSH6, PMS1, and PMS2 [3]. A recent prospective observational study based on the LS database demonstrated that the relative cumulative incidence of PC at 75 years was 7.8% among MLH1 mutation carriers [4]. LS represents a social concern since this disorder is genetically inherited from parents to offspring; thus, gene screening and genetic counseling for relatives are typically included upon diagnosis, which is expected to help achieve early detection or prevention in healthy relatives. In addition, an immune checkpoint blockade treatment strategy has shown striking anti-tumor activity for an increasing number of neoplasms. The dramatic success of immune checkpoint inhibitors, especially for the treatment of MMR-deficient tumors, is currently one of the most exciting topics in cancer treatment [5, 6]. Although there have been a few studies related to MMR-deficient PC, these have been mainly conducted in Caucasian populations, and the prevalence of MMR gene mutations in the Asian population has not yet been fully revealed [7,8,9,10]. To fill this knowledge gap, in the present study, we screened Japanese PC patients meeting the RBG, which is currently the most sensitive criteria for the identification of MMR gene mutation carriers. The RBG was originally established for testing colorectal cancers for microsatellite instability (MSI), and the compatibility of each case is comprehensively judged following the age of onset, the presence of synchronous/metachronous colorectal or other LS-associated tumors, histology, and a family history [3]. Here, we selected PC patients with a personal or family history of first- or second-degree relatives fulfilling the RBG. We defined these patients as RBG-compatible cases, and analyzed their clinical features and germline variants among genes known to be related to a hereditary predisposition for cancer. These results and further investigation of patients with deficient MMR proteins will have great clinical relevance with promise to improve treatment success and options, despite the generally low prevalence of these mutations.
Methods
Study design
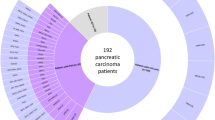
We reviewed the National Cancer Center Hospital database of patients with pathologically proven pancreatic ductal adenocarcinoma between 2007 and 2013. Clinical data reviewed included gender, age, tumor location, histology of PC, UICC stage at diagnosis, smoking history, history of any cancer, and family history of any cancer within first- and second-degree relatives. After excluding cases with insufficient data, a total of 304 patients were further analyzed. Among these, we ultimately selected 20 patients with a personal or family history of first- or second-degree relatives fulfilling the RBG criteria and compared their clinical features with those of 284 other patients. This study was reviewed and approved by the Institutional Review Board of the National Cancer Center and was conducted in accordance with the precepts established by the Helsinki Declaration. A flow diagram of the patient selection process is shown in Fig. 1.
Flow diagram of the patient selection process. Among 304 patients with pathologically proven pancreatic ductal adenocarcinoma, 20 patients with a personal or family history of first- or second-degree relatives fulfilling the revised Bethesda guidelines were selected, and germline variants for 21 genes related to a hereditary predisposition for cancer were analyzed for these cases. NCCH National Cancer Center Hospital, PDAC pancreatic ductal adenocarcinoma, RBG the revised Bethesda guidelines
Next-generation sequencing of 21 genes associated with hereditary predispositions for cancer
Germline DNA samples were available for all 20 PC patients compatible with RBG from the National Cancer Center Biobank, Japan. A custom targeted-capture kit was designed using NimbleDesign (NimbleGen, Madison, WI, USA) targeting the exons and splice sites of 21 genes known to be associated with hereditary predispositions for pancreatic, breast, and ovarian cancers (ATM, BARD1, BRCA1, BRCA2, BRIP1, CDH1, CHEK2, MLH1, MRE11, MSH2, MSH6, MUTYH, NBN, PALB2, PMS1, PMS2, PTEN, RAD50, RAD51C, STK11, and TP53). Manual library preparation was conducted using the SeqCap EZ Library (NimbleGen) and KAPA Library Preparation Kits (Kapa Biosysytems, Wilmington, MA, USA). Targeted-capture sequencing was performed on Illumina HiSeq2500 platforms (Illumina, San Diego, CA, USA). With the intent to maximize sensitivity of detecting variants, no variant quality filters were applied. Bases were called using Illumina BCLFAST2 (Illumina). Paired-end reads were aligned to the human reference genome (GRCh37) using the Burrows–Wheeler Aligner (BWA), and Genome Analysis Toolkit (GATK) was used to detect single-nucleotide substitutions and small insertions and deletions (https://www.broadinstitute.org/gatk/) [11, 12].
Classification of detected variants
Variants in 21 genes were considered for variant characterization if they were: (1) called as non-reference by GATK; (2) predicted to affect the protein sequence or the splice site (i.e., ± 5 base pairs); and (3) had an allele frequency of less than 1% in the 1000 Genomes Project [13, 14], dbSNP [15], or Japanese Genetic Variation database (Human Genetic Variation Browser, http://www.genome.med.kyoto-u.ac.jp/SnpDB/ and Integrative Japanese Genome Variation Database, https://ijgvd.megabank.tohoku.ac.jp/).
All variants were classified according to ClinVar [16]. Moreover, variants in MLH1, MSH2, MSH6, MUTYH, PMS2, and STK11, and those in BRCA1, BRCA2, BRIP1, PALB2, and RAD51C were analyzed based on the InSiGHT consortium database (http://insight-group.org/variants/database/) [17] and the Leiden Open Variation Database (http://www.lovd.nl/3.0/home) [18], respectively.
Rare non-synonymous variants not found in these databases were classified based on the predicted effect on the protein product. Nonsense variants and variants changing the canonical splice sites (i.e., ± 2 base pairs), and frameshift insertions and deletions were judged as deleterious unless they occurred in the last exon. As for the identification of functional missense mutations, SIFT (http://sift.jcvi.org) [19], PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/) [20], MutationTaster (http://www.mutationtaster.org) [21], and Functional Analysis through Hidden Markov Models (FATHMM) (http://fathmm.biocompute.org.uk) [22,23,24] were employed. According to the above algorithm and a literature review, each variant was comprehensively classified as deleterious, benign, or VUS.
Sanger sequencing for validation of variants
Variants classified as deleterious or likely deleterious in targeted-capture sequencing were validated by Sanger sequencing. Polymerase chain reaction (PCR) amplification was performed using 20 ng of gDNA with intronic primers flanking the targeted exons, and the products were sequenced with the M13F primer (5′-GTAAAACGACGGCCAGT-3′) or M13R primer (5′-CAGGAAACAGCTATGACC-3′). These results were analyzed with Sequencher 5.0.1 software (Gene Codes, Ann Arbor, MI, USA).
Statistical analysis
Differences in categorical variables between RBG-compatible patients and other patients were analyzed using Fisher’s exact test, and two-sided P values below 0.05 were considered statistically significant. Statistical analysis was performed using EZR version 1.32 (Saitama Medical Center, Jichi Medical University), which is a graphical user interface for R (The R Foundation for Statistical Computing, Vienna, Austria, 2007. http://www.R-project.org/, version 3.2.2).
Results
Clinical data in patients with pathologically proven pancreatic ductal adenocarcinoma
Clinical data for the 304 patients included in this study are summarized in Table 1. Among the 20 RBG-compatible patients, the age at diagnosis ranged from 46 to 79, the percentage of smokers was 45%, and UICC stage at diagnosis was IA–IIB in 8 (40%), III in 6 (30%), and IV in 6 (30%) patients, respectively. There were no significant differences between RBG-compatible patients and the other 284 cases in terms of age, gender, smoking history, tumor location, and UICC stage. The clinicopathological features in the RBG-compatible cases are summarized in Table 2. All cases were histologically typical adenocarcinoma, and no unique subtypes were observed. Nine patients had a history of any cancer, including seven with colorectal cancer and one with cecal cancer. Most of above eight cases received treatments for colorectal or cecal cancer in another hospital, and tumor tissues for immunohistochemistry of MMR proteins or MSI testing were unavailable. In addition, 8 patients met the RBG criteria themselves, whereas RBG compatibility was evident for first- or second-degree relatives in the 13 other patients; in one case, both the patient (ID-17) and his relatives concurrently met the criteria. No patients or their relatives compatible with RBG criteria fulfilled the Amsterdam II criteria, which is more specific for the identification of MMR gene mutation carriers.
Analysis of germline variants
The average unique coverage depth was 575 × (range 217–1129 ×). The patients with a family history of LS had 18 polymorphisms on average (range 13–22) in the above 21 targeted genes. Four variants in three patients were considered to be deleterious or likely deleterious, including two in ATM, one in PMS2, and one in CHEK2, which were verified successfully with Sanger sequencing (Table 3; Fig. 2a–c). Double germline mutations in PMS2 and CHEK2 were detected in Patient ID-5. In detail, the ATM variant p.Arg2486Ter detected in Patient ID-3 was a nonsense variant and was categorized as deleterious according to ClinVar. Another variant of ATM, p.Ala2391ValfsX10, detected in Patient ID-19 was a frameshift mutation and was categorized as deleterious on the basis of causing a structural change of the protein product. The PMS2 gene variant p.Arg211Ter was detected as a nonsense variant in Patient ID-5, which was classified as deleterious according to ClinVar, and is listed in the InSiGHT consortium database under an interpretation of unknown pathogenicity. The missense CHEK2 variant p.His371Tyr was also detected in Patient ID-5 and was classified as likely deleterious based on dbSNP data, in spite of a conflicting interpretation in ClinVar. Liu et al. [25] identified this variant in Chinese breast cancer patients and concluded it was significantly associated with increased breast cancer risk. The same variant was detected in a breast cancer patient by Baloch et al. [26] and Chen et al. [27], and in diffuse large B-cell lymphoma patients by de Miranda et al. [28]. This amino acid change (p.His371Tyr) occurs within the activation loop of the kinase domain of CHEK2 protein, and was comprehensively considered as likely deleterious. The other 21 variants detected in the 13 patients were classified as variants of unknown significance (VUS), which are listed in Supplementary Table S1.
Discussion
We retrospectively selected 20 RBG-compatible patients among 304 PC patients and analyzed their germline variants for genes known to be related to a hereditary predisposition for cancer. Among the PC patients, 6.6% were identified as RBG-compatible, 5% of whom (0.3% of all PC patients) harbored a germline mutation in an LS-related gene (PMS2). In addition, the PMS2 mutation carrier was also identified to harbor a CHEK2 mutation as double-mutation carrier, and two other patients were identified to harbor an ATM mutation. In terms of histopathology, previous studies have described poorly differentiated or medullary carcinoma as a characteristic for LS-related PC, whereas the histopathology of our patient (ID-5) harboring a PMS2 germline mutation was moderately differentiated adenocarcinoma [25, 26].
In regard to LS-related genes, Gargiulo et al. [9] reported 14% (19/135) of Italian PC patients as RBG-compatible. DNA material was available for 11 patients, and 36% (4/11) of the cases harbored a germline mutation in MLH1 or MSH2. In our study, the prevalence of RBG-compatible patients and that of patients harboring germline mutation of MMR genes were both lower compared with Gargiulo’s report. Although the precise reason for this difference is unclear, it could be related to the smaller sample size or difference in the ethnicity between the two cohorts, among other factors. Approximately 20–40% of colorectal cancer patients have been reported to fulfill the RBG criteria, and 0.7–3.6% of all colorectal cancer patients harbor germline mutations of MMR genes [29,30,31,32,33,34,35]. This prevalence was also much lower in our study, which could be mainly explained by the lower baseline incidence of LS among PC patients. As another possible reason, a previous study revealed that a considerable number of colorectal cancer patients with LS failed to meet the RBG criteria, which might also be true for PC patients [35]. Overall, our results suggest that LS can be identified by the RBG in only a small portion of PC patients. Recently, universal tumor screening, which entails routine MSI and/or immunohistochemistry testing for all colorectal and endometrial cancers, has been proposed as a highly sensitive screening option in Western countries [36, 37]. However, because of the overall low prevalence of LS patients, this exhaustive approach is not realistic in terms of cost-effectiveness in clinical practice for patients with PC, although it is an interesting research topic for achieving a more accurate estimation of the prevalence.
As mentioned above, two variants in ATM and one in CHEK2 were detected as deleterious or likely deleterious, indicating that up to 15% of the patients in our RBG-compatible cohort harbored germline mutations in some cancer-predisposition genes. This prevalence is comparable to that reported for familial PC, which is defined as at least one pair of first-degree relatives diagnosed with PC [38, 39]. Therefore, the RBG might be useful for selecting PC patients with any cancer-predisposition genes, including non-LS-related genes. ATM was originally considered to be related to the onset of ataxia telangiectasia, and a deleterious ATM variant was also shown to increase the risk of breast cancer [40, 41]. Roberts et al. [42] first identified a deleterious ATM variant in two relatives with hereditary PC, and additional analysis showed that the prevalence of this mutation was high among familial PC probands or in families with three or more affected members. Kim et al. [43] also reported that the loss of ATM expression (determined by immunohistochemistry) was conspicuous in patients with a family history of PC. ATM plays a central role in the repair of DNA double-strand breaks, and the activation of this gene results in the phosphorylation and the consequent activation of other substrates such as p53 and BRCA1. This mechanism suggests potential treatment strategies using synthetic lethal interactions worthy of clinical investigation [44, 45]. In our analysis, one patient (Patient ID-3) with a deleterious ATM variant had a family history of quadruple cancer, including PC, in her father. The CHK2 protein (encoded by the CHEK2 gene) is activated in response to DNA damage and is involved in cell-cycle arrest [45]. Deleterious CHEK2 variants have also been reported to increase the risk of breast cancer, and another report suggested a possible contribution of CHEK2 mutations to a small subset of familial PC cases [46, 47].
There are some limitations of this study that should be noted. First, tumor specimens were mainly biopsy samples, and immunohistochemistry for MMR proteins and MSI testing were not conducted because of the limited sample quantity. For the identification of MMR mutation carriers, the RBG proposes a two-step screening strategy with MSI testing as the first step and analysis of germline variants in MMR genes as the second. In this case, MSI-high or MMR protein deficiency is considered as the most important biomarker of immune checkpoint inhibitors, regardless of whether the patient harbors mutations in MMR genes [6]; thus, further investigation including MSI or MMR protein data would be warranted. Second, the number of analyzed patients was relatively small, which might not be a sufficient sample size to draw a definite conclusion. Moreover, we did not analyze germline variants in 284 cases not corresponding to the RBG criteria, which complicated statistical comparisons about mutation prevalence in DNA damage repair genes and MMR genes. Finally, our approach of classifying variants was conservative, in which all rare non-synonymous variants were classified as VUS. If some of these VUS are in fact pathogenic, we might have underestimated the overall prevalence of mutation carriers.
In conclusion, we retrospectively investigated Japanese PC patients based on the RBG and analyzed germline variants for genes related to a hereditary predisposition for cancer. Although an MMR gene mutation was identified in only one patient, 15% of the patients compatible with the RBG criteria were found to harbor some germline deleterious variants. Thus, while the RBG appears to be useful for identifying LS in only a small portion of PC patients, these guidelines could be useful for identifying PC patients with DNA damage repair genes as well as MMR genes.
References
Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86.
Matsubayashi H. Familial pancreatic cancer and hereditary syndromes: screening strategy for high-risk individuals. J Gastroenterol. 2011;46:1249–59.
Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–8.
Møller P, Seppälä TT, Bernstein I, et al. Cancer risk and survival in path_MMR carriers by gene and gender up to 75 years of age: a report from the Prospective Lynch Syndrome database. Gut. 2017. https://doi.org/10.1136/gutjnl-2017-314057.
Smyrk TC, Watson P, Kaul K, et al. Tumor-infiltrating lymphocytes are a marker for microsatellite instability in colorectal carcinoma. Cancer. 2001;91:2417–22.
Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–20.
Humphris JL, Patch AM, Nones K, et al. Hypermutation in pancreatic cancer. Gastroenterology. 2017;152:68–74.
Laitman Y, Herskovitz L, Golan T, et al. The founder Ashkenazi Jewish mutations in the MSH2 and MSH6 genes in Israeli patients with gastric and pancreatic cancer. Fam Cancer. 2012;11:243–7.
Gargiulo S, Torrini M, Ollila S, et al. Germline MLH1 and MSH2 mutations in Italian pancreatic cancer patients with suspected Lynch syndrome. Fam Cancer. 2009;8:547–53.
Grant RC, Selander I, Connor AA, et al. Prevalence of germline mutations in cancer predisposition genes in patients with pancreatic cancer. Gastroenterology. 2015;148:556–64.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60.
McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303.
1000 Genomes Project Consortium, Abecasis GR, Altshuler D, Auton A, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73.
1000 Genomes Project Consortium, Abecasis GR, Auton A, Brooks LD, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65.
Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–11.
Landrum MJ, Lee JM, Riley GR, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–5.
Thompson BA, Spurdle AB, Plazzer JP, et al. Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat Genet. 2014;46:107–15.
Vallée MP, Francy TC, Judkins MK, et al. Classification of missense substitutions in the BRCA genes: a database dedicated to Ex-UVs. Hum Mutat. 2012;33:22–8.
Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–81.
Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9.
Schwarz JM, Cooper DN, Schuelke M, et al. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361–2.
Shihab HA, Gough J, Cooper DN, et al. Predicting the functional, molecular and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat. 2013;34:57–65.
Shihab HA, Gough J, Cooper DN, et al. Predicting the functional consequences of cancer-associated amino acid substitutions. Bioinformatics. 2013;29:1504–10.
Shihab HA, Gough J, Mort M, et al. Ranking non-synonymous single nucleotide polymorphisms based on disease concepts. Hum Genom. 2014;8:11.
Liu Y, Liao J, Xu Y, et al. A recurrent CHEK2 p.H371Y mutation is associated with breast cancer risk in Chinese women. Hum Mutat. 2011;32:1000–3.
Baloch AH, Daud S, Raheem N, et al. Missense mutations (p. H371Y, p.D438Y) in gene CHEK2 are associated with breast cancer risk in women of Balochistan origin. Mol Biol Rep. 2014;41:1103–7.
Chen W, Yurong S, Liansheng N. Breast cancer low-penetrance allele 1100delC in the CHEK2 gene: not present in the Chinese familial breast cancer population. Adv Ther. 2008;25:496–501.
de Miranda NF, Peng R, Georgiou K, et al. DNA repair genes are selectively mutated in diffuse large B cell lymphomas. J Exp Med. 2013;210:1729–42.
Hampel H, Frankel WL, Martin E, et al. Feasibility of screening for Lynch syndrome among patients with colorectal cancer. J Clin Oncol. 2008;26:5783–8.
Julié C, Trésallet C, Brouquet A, et al. Identification in daily practice of patients with Lynch syndrome (hereditary nonpolyposis colorectal cancer): revised Bethesda guidelines-based approach versus molecular screening. Am J Gastroenterol. 2008;103:2825–35.
Rodríguez-Moranta F, Castells A, Andreu M, et al. Clinical performance of original and revised Bethesda guidelines for the identification of MSH2/MLH1 gene carriers in patients with newly diagnosed colorectal cancer: proposal of a new and simpler set of recommendations. Am J Gastroenterol. 2006;101:1104–11.
Piñol V, Castells A, Andreu M, et al. Gastrointestinal Oncology Group of the Spanish Gastroenterological Association. Accuracy of revised Bethesda guidelines, microsatellite instability, and immunohistochemistry for the identification of patients with hereditary nonpolyposis colorectal cancer. JAMA. 2005;293:1986–94.
Hampel H, Frankel WL, Martin E, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer). N Engl J Med. 2005;352:1851–60.
Yurgelun MB, Kulke MH, Fuchs CS, et al. Cancer susceptibility gene mutations in individuals with colorectal cancer. J Clin Oncol. 2017;35:1086–95.
Pérez-Carboneli L, Ruiz-Ponte C, Guarinos C, et al. Comparison between universal molecular screening for Lynch syndrome and revised Bethesda guidelines in a large population-based cohort of patients with colorectal cancer. Gut. 2012;61:865–72.
Giardiello FM, Allen JI, Axilbund JE, et al. Guidelines on genetic evaluation and management of Lynch syndrome: a consensus statement by the US Multi-Society Task Force on colorectal cancer. Am J Gastroenterol. 2014;109:1159–79.
Vasen HF, Blanco I, Aktan-Collan K, et al. Revised guidelines for the clinical management of Lynch syndrome (HNPCC): recommendations by a group of European experts. Gut. 2013;62:812–23.
Klein AP. Identifying people at a high risk of developing pancreatic cancer. Nat Rev Cancer. 2013;13:66–74.
Takai E, Yachida S, Shimizu K, et al. Germline mutations in Japanese familial pancreatic cancer patients. Oncotarget. 2016;7:74227–35.
Renwick A, Thompson D, Seal S, et al. Breast Cancer Susceptibility Collaboration (UK). ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat Genet. 2006;38:873–5.
Thompson D, Duedal S, Kirner J, et al. Cancer risks and mortality in heterozygous ATM mutation carriers. J Natl Cancer Inst. 2005;97:813–22.
Roberts NJ, Jiao Y, Yu J, et al. ATM mutations in patients with hereditary pancreatic cancer. Cancer Discov. 2012;2:41–6.
Kim H, Saka B, Knight S, et al. Having pancreatic cancer with tumoral loss of ATM and normal TP53 protein expression is associated with a poorer prognosis. Clin Cancer Res. 2014;20:1865–72.
Choi M, Kipps T, Kurzrock R. ATM mutations in cancer: therapeutic implications. Mol Cancer Ther. 2016;15:1781–91.
Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–21.
Cybulski C, Wokołorczyk D, Jakubowska A, et al. Risk of breast cancer in women with a CHEK2 mutation with and without a family history of breast cancer. J Clin Oncol. 2011;29:3747–52.
Lener MR, Kashyap A, Kluźniak W, et al. The prevalence of founder mutations among individuals from families with familial pancreatic cancer syndrome. Cancer Res Treat. 2017;49:430–6.
Acknowledgements
We wish to thank all of the patients and their families who contributed to this study. We also thank Dr. Kokichi Sugano and Dr. Teruhiko Yoshida (Department of Genetic Medicine and Services, National Cancer Center Hospital).
Funding
This work was supported by the National Cancer Center Research and Development Fund (28-A-1 to S.Y. and C.M.), the Takeda Science Foundation (to S.Y.), and the Pancreas Research Foundation of Japan (to A.O.). The National Cancer Center Biobank is supported by the National Cancer Center Research and Development Fund, Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare no potential conflicts of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ohmoto, A., Morizane, C., Kubo, E. et al. Germline variants in pancreatic cancer patients with a personal or family history of cancer fulfilling the revised Bethesda guidelines. J Gastroenterol 53, 1159–1167 (2018). https://doi.org/10.1007/s00535-018-1466-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00535-018-1466-y