Abstract
Komagataella phaffii, formerly Pichia pastoris (P. pastoris), is a promising methylotrophic yeast used in industry to produce recombinant protein and valuable metabolites. In this study, a genome-scale metabolic model (GEMs) was reconstructed and used to assess P. pastoris’ metabolic capabilities for the production of S-adenosyl-l-methionine (AdoMet or SAM or SAMe) from individual carbon sources along with the addition of l-methionine. In a model-driven P. pastoris strain, the well-established genome-scale metabolic model iAUKM can be implemented to predict high valuable metabolite production. The model, iAUKM, was created by merging the previously published iMT1026 model and the draught model generated using Raven toolbox from the KEGG database which covered 2309 enzymatic reactions associated with 1033 metabolic genes and 1750 metabolites. The highly curated model was successful in capturing P. pastoris growth on various carbon sources, as well as AdoMet production under various growth conditions. Many overexpression gene targets for increasing AdoMet accumulation in the cell have been predicted for various carbon sources. Inorganic phosphatase (IPP) was one of the predicted overexpression targets as revealed from simulations using iAUKM. When IPP gene was integrated into P. pastoris, we found that AdoMet accumulation increased by 16% and 14% using glucose and glycerol as carbon sources, respectively. Our in silico results shed light on the factors limiting AdoMet production, as well as key pathways for rationalized engineering to increase AdoMet yield.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Over the last century, Pichia pastoris has emerged as the preferred model for metabolite and heterologous protein production. P. pastoris can grow on a minimal medium at very high cell densities (over 100 g cell dry weight/L) to secrete the heterologous protein and exhibit post-translational processing capabilities [1]. The presence of a tightly controlled promoter from the gene of alcohol oxidase (pAOX1) and a strong constitutive Glyceraldehyde 3 phosphate dehydrogenase promoter (pGAP) is the most notable feature of P. pastoris. Consequently, glucose, glycerol, and methanol can be used as a carbon source as well as an inducer of heterologous protein expression and metabolite production. Repression/de-repression of AOX1 promoter in the presence of glucose or glycerol results in decreased utilization of methanol as a substrate [2, 3], preventing simultaneous utilization of methanol in the presence of other carbon sources, such as glucose or glycerol. As a result, the analysis of these carbon sources for product formation in P. pastoris is typically done in an exclusive manner.
AdoMet is a key methyl group donor in a variety of biochemical reactions. It has potential applications in the treatment of diseases such as Alzheimer’s, osteoarthritis, and liver disease [4,5,6]. Chemical, enzymatic, and fermentation processes might all be used to make AdoMet. Chemical approaches need strict conditions for chiral separation of the molecule, whereas enzymatic methods are not economically viable due to the high cost of Adenosine triphosphate (ATP). Chu et al. examined the progress of AdoMet manufacture in great detail (2013). Microbial cells store AdoMet in varying amounts, with Saccharomyces cerevisiae being able to accumulate huge amounts [7]. Shiozaki et al. [8] discovered that yeasts acquire huge amounts of AdoMet after screening diverse bacteria. In yeast, two AdoMet synthetase genes, sam1 and sam2, have been discovered of the two genes, sam2 is L-met concentration insensitive [9, 10]. The excess AdoMet generated in l-methionine (L-Met) supplemented media is sequestered into vacuoles, leading in hyper-accumulation of this molecule in yeast [11]. Overexpression of methionine adenosyltransferase (MAT or SAM synthetase, EC2.5.1.6), which catalyzes the formation of AdoMet from ATP and L-Met, has been the mainstay of metabolic engineering attempts to enhance AdoMet production [12,13,14]. The second strategy for promoting AdoMet synthesis is to restrict competing routes in order to increase precursor availability or impede AdoMet use. Synthetic siRNA was used to decrease the transcript levels of the gamma-glutamate kinase, glutamine synthetase, and acetylglutamate kinase genes, which increased ATP supply and AdoMet build-up in Escherichia coli [15]. Adenosine kinase or cytosolic malate synthase gene disruption has been shown to increase L-Met biosynthesis and, as a result, AdoMet production [16]. Furthermore, knocking down the S-adenosylmethionine decarboxylase gene [17] and the cystathionine-synthase (CBS) gene CYS4 [18] made it easier to block the AdoMet decarboxylation pathway and the elimination of homocysteine from the methionine and AdoMet cycle. CBS malfunction, on the other hand, resulted in the production of cysteine auxotroph’s [11], requiring cysteine or glutathione (GSH) supplementation for cell proliferation [18]. If the target genes in the competing pathways are required for cell development, downregulation will be preferable to deletion. For example, downregulation of the cystathionine-synthase gene (CYS4) using a weak promoter PG12 reduced homocysteine removal from the AdoMet cycle, resulting in a 48.8% increase in AdoMet titer (1.68 g/L) in the P. pastoris strain G12-CBS. Moreover, this strategy prevented cysteine auxotrophy caused by deletion of this essential gene [6]. Following that, employing the ideal l-methionine feeding method, the AdoMet titer of G12-CBS increased to 13.01 g/L. Moreover, in shake flask culture, further overexpression of glutamate dehydrogenase 2 (GDH2) resulted in a 52.3% rise in titer (2.71 g/L) [6].
Efforts to optimize the fermentation process have been done in tandem with strain creation. Since ATP is the precursor and driving force in AdoMet biosynthesis, a number of ATP-focused strategies were used, including metabolome-based culture medium optimization [19], alternate methanol and glycerol feeding [20], two-stage amino acid addition, and sodium citrate feeding as an auxiliary energy source [21]. Optimization of L-Met feeding in an engineered P. pastoris strain [13] and employing DL-Met (a blend of D-Met and L-Met) as a substrate for AdoMet production in a mutant Saccharomyces cerevisiae were two additional strategies used [22]. In reality, because physiological properties are strain/clone specific and not foreseeable a priori, optimizing culture conditions for a freshly generated strain is critical. Because of the increasing commercial demand for AdoMet, the low yield of AdoMet production should be addressed. Previous work in our lab improved AdoMet precursor accessibility by combining methionine permease (mup1) and adenylate kinase (adk1) gene overexpression strategies along with sam2, as well as improving L-met process conversion efficiency in P. pastoris. The expression of mup1 and sam2 did not improve AdoMet production when l-methionine and methanol were combined. On the other hand, co-expression of adk1 and sam2 increased AdoMet production (276.10 mg/L), indicating that ATP is the main limiting factor in AdoMet production. Furthermore, overexpression of all three genes together resulted in a synergistic 77% improvement in AdoMet (423.31 mg/L), when compared to overexpressing sam2 alone [23]. These findings support the idea of using metabolic engineering as a tool to improve bioprocesses.
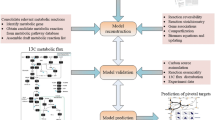
Flux balance analysis (FBA) is a mathematical method for analyzing metabolite flow through a metabolic network. Genome-scale metabolic models (GEMs) are in silico representations of the entire set of metabolic reactions that occur in a cell [24]. GEMs can be used to understand and predict how organisms respond to genetic and environmental changes via FBA. Recent research has shown that FBA can be used to discover novel metabolic engineering strategies for protein and metabolite production [24,25,26,27]. The RAVEN (Reconstruction, Analysis, and Visualization of Metabolic Networks) toolbox was created to help with GEM reconstruction, curation, and simulation in order to meet the growing demand for metabolic network modeling [27]. The use of the KEGG and MetaCyc databases in assisting draught model reconstruction is a notable enhancement of RAVEN 2.0. MetaCyc is a pathway database that only includes experimentally validated pathways with curated reversibility information and mass-balanced reactions [26]. The updated P. pastoris GEMs has been named as iAUKM, which can be used as an upgraded platform for future P. pastoris systems biology research.
In this paper, we extend the iAUKM model’s capabilities by accurately describing P. pastoris growth phenotype and AdoMet accumulation when using glucose, glycerol, or methanol as sole carbon sources, along with l-methionine supplementation. These carbon substrates have also been used to identify in silico gene overexpression targets for improving AdoMet production.
Materials and methods
In silico analysis of P. pastoris model
Flux balance analysis (FBA) analysis and Flux scanning based on enforced objective flux (FSEOF) analysis was performed using the genome-scale model constructed iAUKM with the help of (Reconstruction, Analysis and Visualization of Metabolic Networks) RAVEN toolbox in Matlab. The simulations were done for the prediction of preferable carbon source for biomass production and AdoMet production as a constraint with and without the supplement of l-methionine. FSEOF simulation were done to predict the overexpression target for enhance production of AdoMet.
Materials
P. pastoris X33 the vectors pPICZB and Zeocin were purchased from the Invitrogen Co. Ltd, USA. pGAP(modified) vector was designed and developed inhouse with 3’AOX site for insertion. PCR reagents, restriction endonucleases were purchased from the TaKaRa Biotech Co. Ltd, Japan. AdoMet standard samples were purchased from the Sigma, Co. Ltd. Multi-Copy Pichia Expression Kit was purchased from the Invitrogen Co. Ltd. Yeast nitrogen base was purchased from the Himedia, India. All other reagents and chemicals were purchased from Qualigens, Thermofisher. Primers used for the amplification of genes are listed in Table 1.
Construction of the plasmid pGAP(modified)-ADK-Sam2-MUP and integrated into P. pastoris X33
SAM 2, ADK1, MUP and IPP were amplified using the primer set listed in Table 1. PCR products were purified and digested by XhoI and XbaI for SAM2, ADK1 and MUP, and for IPP digested with EcoR1 and Xho1 and ligated into the XhoI-XbaI digested plasmid pGAP(modified) for SAM2, ADK1 and MUP, and ligated into EcoR1-XhoI digested plasmid pGAP(modified) for IPP. The plasmid constructs were named as pGAP(M)-Sam2, pGAP(M)-ADK, pGAP(M)-MUP and pGAP(M)-IPP, respectively. As per the protocol mentioned in Multi-Copy Pichia Expression Kit, the cassette of pGAP(M)-Sam2-TT was extracted out by digesting with BglII and BamHI. The plasmid of PGAP(M)-ADK was linearized with BamHI and the cassette of pGAP(M)-Sam2-TT was ligated as BglII and BamHI have compatible cohesive ends to form a plasmid of pGAP(M)-ADK-SAM2.
Similarly, the cassette of pGAP(M)-MUP-TT was extracted with amplifying the whole cassette with GAP forward and TT reverse primer and the plasmid pGAP(M)-ADK-SAM2 linearized with BamHI and ligated to form pGAP(M)-ADK-SAM2-MUP plasmid. The plasmids pGAP(M)-SAM2, pGAP(M)-ADK1, pGAP(M)-MUP, pGAP(M)-IPP, pGAP(M)-ADK-SAM2, and pGAP(M)-ADK-SAM2-MUP were linearized with EcoNI that was transformed into P. pastoris X33 by the electroporation method with parameters: 1.5 kV, 200 μF and 200 Ω. Transformants were screened by Yeast Extract Peptone Dextrose (YPD) agar plates and Zeocin as antibiotic. The genomic DNA of selected transformants was isolated according to the easy select manual provided by the manufacturer. PCRs were carried out to confirm whether all the genes were integrated into the genomic DNA of P. pastoris X33.
P. pastoris culture for the AdoMet production in flasks
P. pastoris X33-pGAP(M)-SAM2, X33-pGAP(M)-ADK, X33-pGAP(M)-MUP, X33-pGAP(M)-IPP, X33-pGAP(M)-ADK-SAM2 and X33-pGAP(M)-ADK-SAM2-MUP were inoculated, respectively, into a 3 mL of YPD medium and incubated at 28 °C for 24 h as seed cultures. A 1 mL aliquot of the seed culture was added to 100 mL BM(D/G)Y medium (yeast extract 1%, peptone 2%, 2% of glucose/1% of glycerol, 10 × YNB10%, 0.1 M potassium phosphate buffer, pH 6.0) supplemented with 1% L-Met in 500 mL flasks and cultured at 28 °C for 4 days. 2% (v/v) of glucose/glycerol used as an inducer and carbon source was added daily for three times after 1 day of growth. After 96 h of culture, 1 mL of sample was taken out for determining the yield of AdoMet.
Bioreactor setup
The pGAP(M)-ADK-SAM2-MUP strain was grown in a YPD medium at 28 °C for 24 h, and 100 mL of cultures was inoculated in a 3.7 L KLF Bioengineering fermentor containing 2 L of FM22 fermentation medium consisting of FM22, which was composed of KH2PO4 42.9 g/L, (NH4)2SO4 5 g/L, CaSO4·2H2O 1.0 g/L, K2SO4 14.3 g/L, MgSO4·7H2O 11.7 g/L and glycerol 40 g/L; Pichia trace minerals 4 (PTM4) salt solution was prepared with CuSO4·5H2O 2.0 g/L, NaI 0.08 g/L, MnSO4·H2O 3.0 g/L, Na2 MoO4·2H2O 0.2 g/L, H3BO3 0.02 g/L, CaSO4·2H2O 0.5 g/L, CoCl2 0.5 g/L, ZnCl2 7 g/L, FeSO4·7H2O 22 g/L, biotin, 0.2 g/L and 1 ml of concentrated H2SO4. Initial fermentation conditions were as follows: dissolved oxygen (DO) was set to 100% and maintained above 20% by the automatic control of agitation, pH was 4.8 (adjusted with liquid ammonium), and temperature was set at 28 °C. Cells were grown until glycerol depleted completely. This was indicated by a dramatic increase in the DO to above 80%. Glycerol feeding was then initiated to increase the cell biomass as well as to induce the GAP(M) Promoter under limited conditions: 1000 mL of 70% glycerol containing 12 mL of PTM4 trace salts was fed at 10 mL/L/h. 10 gm of L-Met was fed every 12 h. Samples were taken out at different times for determining the yield of AdoMet and biomass.
Determination of the yield of AdoMet
Biomass was calculated using both dry cell weight (DCW) and optical density at 600 nm (OD600). AdoMet was extracted from P. pastoris cultures using the perchloric acid method, but with 0.75 N perchloric acid instead of 0.4 N, as originally described by Wang et al. [28]. AdoMet was quantified using a reversed-phase YMCR ODS C-18 analytical column (4.6 mm 250 mm, 5 m particle size) equipped with a DAD detector in high-performance liquid chromatography (UHPLC, Agilent) (254 nm). The mobile phase was made up of two buffers: buffer-1: 20 mM citric acid and 10 mM sodium dihydrogen orthophosphate in HPLC grade water, and buffer-2: 100% HPLC grade acetonitrile. The HPLC buffer was made by combining 56 volumes of buffer-1 and 44 volumes of buffer-2 along with adding 0.4% (w/v) sodium lauryl sulfate. Ultrasonication was used to degas the buffer, which was then filtered through 0.2 μM membrane filters. The sample was injected after the HPLC system had been equilibrated with buffer, and the separation was obtained using isocratic flow. The flow rate was kept constant at 1.2 ml per minute, and the detection was measured at 254 nm.
Results and discussion
Reconstruction of the P. pastoris GEM iAUKM
The P. pastoris GEM iAUKM was updated using previously available iMT1026 models [29, 30], as well as a draught model created with RAVEN Toolbox. Gap filling was also done, and gaps were filled by manually incorporating essential reactions from KEGG and other databases. A few more reactions are also added to improve the model’s ability to cover 2309 enzymatic reactions associated with 1033 metabolic genes involving 1750 metabolites.
Model verification by growth capabilities on different carbon sources
P. pastoris iAUKM prediction capacity for growth was tested with 28 carbon sources and 18 nitrogen substrates from the data obtained from previous studies [29,30,31]. The growth potential of each carbon or nitrogen source was assessed using FBA. Only ammonia was employed as a nitrogen source for carbon utilization prediction, but other metabolites required for growth, such as oxygen, biotin, phosphorus, and sulfur, were kept at adequate levels. The other carbon source which may contribute the growth were set to zero at the same time. Similarly, sole carbon sources glucose, glycerol, and methanol were set individually to test the growth for nitrogen source. Except the specified nitrogen source other nitrogen sources were set to zero. If the expected specific growth rate was greater than zero, the target carbon or nitrogen source was considered growth supportive. iAUKM showed growth response to 28 carbon sources out of 31 and 18 out of 21 nitrogen sources which were listed in supplement 4.
Maximization of biomass in silico analysis
The FBA generated the exchange fluxes of the updated iAUKM model. Compounds that needed to be consumed in order to maximize biomass production have been indicated by exchange reactions with negative fluxes. It was crucial to include these components in the culture medium. The fluxes with positive value represent metabolites that are produced. The list of exchange reactions and their fluxes when equimolar flux of glucose, glycerol and methanol were selected as carbon sources is summarized in Table 2. When 1 mmol of glucose, glycerol and methanol was given as a sole carbon source, glucose gives the maximum growth rate. Moreover, glucose has higher carbon moles compared to glycerol and methanol. Consequently, simulation was done with matching the carbon moles. The simulation predicted to give higher yield in glycerol followed by glucose and greater than that of methanol.
The findings on biomass maximization suggest that glycerol is a preferred choice of carbon source followed by glucose and methanol for maximizing the yield of biomass. This could be attributed to the limitations associated with methanol metabolism which produces hydrogen peroxide (H2O2) and formaldehyde formed as byproducts in the peroxisomes. Though these products are further metabolized into non-toxic products, it sets a constrain on the content of peroxisomes for the survivability of P. pastoris. The findings in our simulation with growth in methanol produced 10.477 mmol/gDW/h of H2O, a product of methanol detoxification suggests the detoxification scenario as a potential limiting factor for biomass yield [32]. This is reinforced by the findings that peroxisomes may fill up to 90% of the cell volume during growth in the presence of methanol, while their existence is undetectable in the presence of other carbon sources such as glucose or glycerol [3]. Moreover, the elevated utilization of sulfate in the presence of glycerol observed in our simulation (Table 2) appears to be both beneficial towards endogenous methionine synthesis and hence the increase in biomass and AdoMet.
Maximization of AdoMet formation in silico analysis
Balancing the carbon moles simulation was done with an objective function of maximizing AdoMet formation on glycerol, glucose and methanol as a sole carbon source. The simulation with glycerol showed higher flux for AdoMet accumulation followed by glucose and methanol. It is observed that l-methionine is a limiting precursor for AdoMet synthesis and most of the flux is higher towards NADP/NADPH dependent reaction on all carbon sources used. Moreover, when glucose and methanol were used as a sole carbon source, the oxidative phase of pentose phosphate pathway (PPP) was prominent but it is lower in case of glycerol. The metabolic flux from all the three investigated carbon sources, viz., glucose, glycerol and methanol converge to generate fructose 1,6 bisphosphate (1,6FBP). 1,6FBP is then metabolized into fructose 6-phosphate which can have two fates, viz., (1) utilization towards biomass production or (2) pentose phosphate pathway. In this connection, the earlier analysis on maximizing biomass production suggested that growth in glycerol had higher phosphate utilization suggesting the prominence of PPP.
The major role of oxidative phase of PPP is to produce NADPH, which further helps in enhancing the l-methionine production in the cell via aspartate metabolism. Glucose 6 phosphate dehydrogenase (G6PDH2) and phosphogluconate dehydrogenase (GND) produces two moles of NADPH which is utilized in Aspartate-methionine pathway where it consumes two moles of NADPH. In a rate-limiting step, G6PDH2 catalyzes the irreversible oxidation of Glucose 6 Phosphate to 6-phosphogluconolactone; the first molecule of NADPH is produced during this reaction. G6PDH2 serves as the pathway’s “gatekeeper” and is therefore the rate-limiting enzyme in the PPP. Phosphogluconate dehydrogenase (GND), a NADP-dependent enzyme, catalyzes the formation of NADPH and Ribulose 5-phosphate from 6-Phospho-d-gluconate. These two enzymes would be a better gene candidate for overexpression which enhance the NADPH/NADP+ pool to improve the precursor l-methionine. NADP-dependent glycerol dehydrogenase upregulation was predicted when glycerol is used as the sole carbon source. On all carbon sources, genes correspond to L-serine and folate metabolism, i.e., formation of 5-methyltetrahydrofolate from L-serine is involved in NADP/NADPH dependent pathway. 5-Methyltetrahydrofolate is directly involved in the conversion of l-Homocysteine to l-Methionine and Tetrahydrofolate. All the above predictions show that methionine was the limiting substrate compared to ATP for AdoMet production. Biomass formation was zero on maximizing AdoMet production and AdoMet production was zero on maximum biomass formation (Table 3). By comparing the other intracellular fluxes, it clearly shows that the precursor for AdoMet is completely utilized for biomass formation.
Identification of overexpression targets
For further optimization, flux scanning based on enforced objective flux (FSEOF) was used to identify the overexpression target to produce AdoMet. In addition, l-methionine was supplemented as it was the limiting substrate along with carbon source. Table 4 represents the overexpression targets for AdoMet production. Methionine adenosyltransferase (MAT or METAT or SAM2) and adenylate kinase (ADK1) are predicted by the simulation to be overexpressed. This was already proved in our lab by overexpressing these two enzymes [23]. Glutamate synthase and glutamine synthetase were also predicted as overexpression targets for AdoMet production. Considering methionine metabolic flux, active site for methionine synthase is known to have binding sites for zinc and the substrates, viz., l-homocysteine and 5-methyltetrahydrofolate-glutamate [33]. Methionine synthase takes part in methionine cycle and folate cycle. The methionine cycle utilizes the methionine produced by methionine synthase, while the folate cycle produces tetrahydrofolate. Since ATP is a direct substrate for AdoMet synthesis, the essentiality of methionine synthase and glutamate are notable. Glutamate can be formed from glutamine by the action of glutaminase and glutamate can be converted back into glutamine by the action of glutamine synthase [34]. However, Glutamine synthetase is an ATP dependent enzyme while glutaminase is not. In addition to the formation of glutamate via glutaminase, glutamate synthase catalyzes the formation of glutamate using TCA cycle intermediate 2-oxoglutarate in the presence of NADPH [34]. This reaction could drive the export of 2-oxoglutarate from the mitochondria through the Malate-2-oxoglutarate antiporter and sustain the TCA cycle flux utilizing the malate derived from fumarase, an additional overexpression target predicted in our analysis (Fig. 1).
In addition, considering the requirement of 2-oxoglutarate towards glutamate synthase, which is prominently cytosolic [34], there arises a need for the export of 2-oxoglutarate (α-ketoglutarate) formed via the TCA cycle within mitochondria into the cytosolic compartment. In this regard, the export of 2-oxoglutarate into cytosol is possible through malate-α-ketoglutarate antiporter which is usually involved in malate-aspartate shuttle [35]. However, in this scenario, for the export of α-ketoglutarate from the mitochondrial matrix, two essential criteria should be met, viz., (1) elevated synthesis of malate and (2) elevated presence of synthesized malate outside the mitochondrial matrix so that the antiporting could occur. This possibility can be achieved by increased activity of fumarase, a TCA cycle enzyme involved in the formation of malate and the presence of malate/phosphate exchanger which exports malate from the mitochondrial matrix by simultaneously importing inorganic phosphate inside the matrix. Thus, through the action of fumarase and malate/phosphate exchanger a sufficient build-up of malate in the outside compartment of mitochondrial matrix can be achieved which can then export α-ketoglutarate outside of the matrix for further utilization towards synthesis of glutamate involved in methionine cycle and de novo purine synthesis. Interestingly, the predictions using FSEOF analysis showed that the enzymes involved in malate and inorganic phosphate synthesis, viz., fumarase and inorganic diphosphatase as potential targets for overexpression. Thus, the predictions from FSEOF are also in agreement with the mechanistic pathway towards increased metabolic flux yield of AdoMet (Fig. 1).
Thus, out of the 30 genes predicted by FSEOF analysis, the 5 genes, viz., (1) fumarase, (2) glutamate synthase, (3) glutamine synthetase, (4) inorganic diphosphatase and (5) methionine adenosyltransferase directly match with the mechanistic formation of AdoMet. The additional gene, adenylate kinase, predicted from the FSEOF analysis has been already experimentally confirmed by previous studies in our lab [23]. Moreover, considering the formation of glutamine from the metabolic pathway catalyzed by glutamine synthetase, the prediction is also supported by our previous work [11]. Specifically, addition of l-glutamine in the presence of AOX promoter increased AdoMet production [11]. The other overexpression gene candidates for all three different carbon sources with and without the supplement of l-methionine are listed in Supplementary file 3. Considering the prediction from the model that glutamine and glutamate synthase overexpression would increase the AdoMet production, we questioned whether the availability of the metabolic outcomes of these genes could have a positive effect over AdoMet production. In this regard, we supplemented the culture with monosodium glutamate (1 g/L) and malic acid (1 g/L). However, we found a down regulation of AdoMet production, possibly due to the regulatory effects of these metabolic products, when present in excess, on AdoMet yielding pathways. This also matches with the earlier findings that glutamine synthase downregulates AdoMet production even though it was identified as overexpression targets [6]. This adds to the sense of caution that all the overexpression targets predicted from FSEOF need not produce the anticipated outcomes.
From the predicted set of overexpression targets (Table 4), we were interested in looking into a specific gene that has not been much studied in relation to AdoMet production. In this regard, the roles of methionine adenosyltransferase are very obvious. Moreover, adenylate kinase has already been confirmed to promote AdoMet production experimentally in our earlier work [11]. In addition, the potential metabolic outcomes of glutamine synthetase and glutamate synthase were also tested by the supplementation with l-glutamine and monosodium glutamate, respectively, in our earlier work [11]. Interestingly, IPP has been shown to have central role in the energy metabolism of yeast [36]. Moreover, we anticipated manipulation of IPP could also impact the cycling of malate, a product of fumarase (also predicted by our FSEOF analysis), between Malate/phosphate exchanger and malate/α-ketoglutarate antiporter. In this perspective, the increased availability of inorganic phosphate by the action of IPP may promote the export of α-ketoglutarate into the cytosol for its conversion into glutamate by glutamate synthase (Fig. 1), an additional overexpression target predicted by our FSEOF analysis. Adding to the scarcity of literature connecting IPP expression with AdoMet, we looked into the experimental outcomes of IPP overexpression on AdoMet yield.
Cloning of SAM2, ADK, MUP and IPP Gene in pGAP(M)
Multiple transformants appeared on YPD-Zeocin selection plate which were screened by PCR and positive clones were preserved for further analysis (expression of protein, activity and product yield). The pGAP(M)-SAM2, pGAP(M)-ADK, pGAP(M)-MUP, and pGAP(M)-IPP recombinant constructs were made. The P. pastoris AQ15X33 has been modified to overexpress single genes, resulting in the following variants: X33-pGAP(M)-SAM2, X33-pGAP(M)-ADK, X33-pGAP(M)-MUP, and X33-pGAP(M)-IPP, respectively.
Cloning of double-gene construct mutant pGAP(M)-ADK-SAM2
As the gene ADK contains BglII site, the pGAP(M)-ADK was linearized with BamH1 and the construct of pGAP(M)-SAM2-TT was popped out by digesting the plasmid pGAP(M)-SAM2 with BglII and BamH1. As the BamH1 and BglII have compatible cohesive ends, the linearized pGAP(M)-ADK and pGAP(M)-SAM2-TT gene construct was ligated and transformed into E. coli. The transformants were screened and this named as pGAP(M)-ADK-SAM2. The pGAP(M)-ADK-SAM2 was linearized with EcoN1 and integrated into P. pastoris X33 and the positive clones were named as X33-pGAP(M)-ADK-SAM2 [37].
Construction of triple-gene plasmid pGAP(M)-ADK-SAM2-MUP
MUP gene contains BamH1 site so it was not possible to get gene construct of pGAP(M)-MUP-TT by restriction digestion. Therefore, here, we amplified the whole cassette of pGAP(M)-MUP-TT with GAP forward and TT Reverse. Both forward and reverse primer have BglII site which replace the BamH1 site at the end of TT. This cassette was digested with BglII and the plasmid pGAP(M)-ADK-SAM2 was linearized with BamH1 and ligated and transformed in E. coli. The transformed were screened and the plasmid extracted from the positive clones was named as pGAP(M)-ADK-SAM2-MUP. PCR conformation has been done with specific gene forward and gene reverse for ADK, SAM2, MUP genes (Fig. 2). The pGAP(M)-ADK-SAM2-MUP was linearized with EcoN1 and integrated into P. pastoris X33 and the positive clones were named as X33-pGAP(M)-ADK-SAM2-MUP.
Shake flask expression studies on single-, double- and triple-gene constructs
The expression studies on various clones and Host X33, a renowned expression host, were carried out to assess their efficiency in producing S-adenosylmethionine (AdoMet). Five different clones, X33-pGAP(M)-SAM2, X33-pGAP(M)-ADK, X33-pGAP(M)-MUP, X33-pGAP(M)-ADK-SAM2 and X33-pGAP(M)-ADK-SAM2-MUP designated as C1, C2, C3, C4, and C5, respectively, were examined for their AdoMet production capabilities. The expression studies were carried out in buffered complex glycerol medium (BMGY) with the addition of 2% glucose and 2% glycerol as sole carbon sources every 24 h for a duration of 96 h. Furthermore, l-methionine (L-Met) was supplemented at a concentration of 1% every 24 h.
The results demonstrate that the single-gene insertion clone C1 exhibited a higher production of 201 mg/L AdoMet compared to clones C2 and C3. The sole expression of the precursor-enhancing genes, ADK (adenylate kinase) and MUP (methionine uptake protein), did not yield a significant increase in AdoMet levels when compared to the insertion of the SAM2 gene alone. To further enhance AdoMet production, a new clone designated as C4 was constructed by cloning the ADK gene along with the SAM2 gene. Clone C4 showed higher productivity of 246 mg/L of AdoMet than C1, indicating the synergistic effect of these two genes on AdoMet biosynthesis. Finally, clone C5, which harbored all three genes (SAM2, MUP, and ADK), exhibited the highest AdoMet production of about 275 mg/L compared to the other clones studied.
These findings highlight the importance of gene combinations and their impact on AdoMet production in Host X33. The results suggest that the simultaneous expression of SAM2, MUP, and ADK genes results in a significant improvement in AdoMet biosynthesis which is also already proved by our team with AOX promoter (Fig. 3).
However, reactor scale experiments in fed batch mode yielded 4.4 g/l (Fig. 4) suggesting scope for yield optimization. It is observed that after 72 h the AdoMet accumulation in the cell was reduced though the cell mass was increasing. This may be due to the depletion of ATP inside the cell which is used for its growth and metabolism.
Strain construction and confirmation of IPP
As the model predicts IPP as one of the overexpression targets. We amplified the IPP gene from the X33 genome and subcloned it into the pGAP(M) expression vectors from P. pastoris (modified vector). pGAP(M)-IPP recombinant plasmid constructs were transformed into P. pastoris host X33 to form X33-pGAP(M)-IPP. Antibiotic marker plates were used to screen colonies. We used PCR with GAP forward primer and gene-specific reverse primer to confirm the presence of genes. All the positive clones were expressed, and AdoMet production was tested in shake flasks using BM(D/G)Y medium with L-met supplementation. Expression studies revealed that AdoMet accumulation was increased by 16% and 14% when glucose and glycerol is used as sole carbon source, respectively (Table 5).
Considering the yield from IPP overexpression of 24.27 mg/l and 25.0 mg/l in glucose and glycerol, respectively, it is still low to those observed in studies focused on the production of AdoMet towards industrial scale. For example, yeast strains have been engineered to produce around 10 g/l of AdoMet [14, 22]. The yields from our study are not comparable to these yields since our work is only focused on predicting and validating the potential gene targets for increasing AdoMet under shake flask conditions. For example, our work on pGAP(M)-ADK-SAM2-MUP overexpression in P. pastoris X33 host under yielded 275.20 mg/l of AdoMet under shake flask condition in the presence of glucose. However, reactor scale experiments in fed batch mode yielded 4.4 g/l suggesting scope for yield optimization. Further, combination of IPP overexpression along ADK-SAM2-MUP overexpression can also be considered for future studies.
Conclusion
Pichia pastoris is a promising methylotrophic yeast that produces recombinant protein and other valuable products. Using the reconstructed model iAUKM, we investigated P. pastoris’ ability to produce S-Adenosyl-l-methionine (AdoMet) and predicted various gene targets that could increase AdoMet levels. One of the candidate enzymes that was predicted to be overexpressed was inorganic phosphatase. When compared to the X33 host, the IPP integrated P. pastoris accumulated 16% and 14% more AdoMet in glucose and glycerol containing medium, respectively. IPP plays a role in ATP synthesis by hydrolyzing inorganic pyrophosphate (PPi) into two molecules of inorganic phosphate (Pi). During certain biochemical reactions, such as DNA synthesis, RNA synthesis, or other reactions that involve the formation of high-energy phosphate bonds, PPi is released as a byproduct. However, PPi is not a stable molecule and has a tendency to hydrolyze into two molecules of Pi. This hydrolysis reaction is catalyzed by inorganic pyrophosphatase. The hydrolysis of PPi into two Pi molecules is an energetically favorable process and can be coupled to the synthesis of ATP. In ATP synthesis, the energy released by the hydrolysis of PPi is used to drive the formation of ATP from adenosine diphosphate (ADP) and Pi. This process is commonly referred to as substrate-level phosphorylation. Inorganic pyrophosphatase acts as a catalyst in this hydrolysis reaction, ensuring the efficient conversion of PPi into Pi. By removing PPi from the reaction, inorganic pyrophosphatase helps maintain the thermodynamic driving force necessary for ATP synthesis. Therefore, inorganic pyrophosphatase plays a crucial role in ATP synthesis by facilitating the hydrolysis of inorganic pyrophosphate into inorganic phosphate, which is essential for the formation of ATP from ADP and Pi. In summary, in silico findings reveal key factors limiting AdoMet production as well as rationalized approaches to improve AdoMet production.
References
Cereghino GPL, Cereghino JL, Ilgen C, Cregg JM (2002) Production of recombinant proteins in fermenter cultures of the yeast Pichia pastoris. Curr Opin Biotechnol 13(4):329–332
Zhan C, Wang S, Sun Y, Dai X, Liu X, Harvey L et al (2016) The Pichia pastoris transmembrane protein GT1 is a glycerol transporter and relieves the repression of glycerol on AOX1 expression. FEMS Yeast Res 16(4):1–10
Zepeda AB, Pessoa A, Farías JG (2018) Carbon metabolism influenced for promoters and temperature used in the heterologous protein production using Pichia pastoris yeast. Brazilian J Microbiol 49:119–127. https://doi.org/10.1016/j.bjm.2018.03.010
Bottiglieri T (2002) S-Adenosyl-l-methionine (SAMe): from the bench to the bedside - molecular basis of a pleiotrophic molecule. Am J Clin Nutr 76(5):1151S
Zweier (2014) S-adenosylmethionine metabolism and liver disease. Bone 23(1):1–7
Qin X, Lu J, Zhang Y, Wu X, Qiao X, Wang Z et al (2020) Engineering Pichia pastoris to improve S-adenosyl-l-methionine production using systems metabolic strategies. Biotechnol Bioeng. https://doi.org/10.1002/bit.27300
Gawel LJ, Turner JR, Parks LW (1961) Accumulation of s-adenosylmethionine. 1–3
Shiozaki S, Shimizu S, Yamada H (1984) Unusual intracellular accumulation of s-adenosyl-l-methionine by microorganisms. Agric Biol Chem 48(9):2293–2300
Thomas D, Rothstein R, Rosenberg N, Surdin-Kerjan Y (1988) SAM2 encodes the second methionine S-adenosyl transferase in Saccharomyces cerevisiae: physiology and regulation of both enzymes. Mol Cell Biol 8(12):5132–5139
Thomas D, Surdin-Kerjan Y (1991) The synthesis of the two S-adenosyl-methionine synthetases is differently regulated in Saccharomyces cerevisiae. MGG Mol Gen Genet 226(1–2):224–232
Chan SY, Appling DR (2003) Regulation of S-Adenosylmethionine levels in Saccharomyces cerevisiae. J Biol Chem 278(44):43051–43059. https://doi.org/10.1074/jbc.M308696200
Hu H, Qian J, Chu J, Wang Y, Zhuang Y, Zhang S (2009) DNA shuffling of methionine adenosyltransferase gene leads to improved S-adenosyl-l-methionine production in Pichia pastoris. J Biotechnol 141(3–4):97–103
Hu H, Qian J, Chu J, Wang Y, Zhuang Y, Zhang S (2009) Optimization of l-methionine feeding strategy for improving S-adenosyl-l-methionine production by methionine adenosyltransferase overexpressed Pichia pastoris. Appl Microbiol Biotechnol 83(6):1105–1114
Liu W, Tang D, Shi R, Lian J, Huang L, Cai J et al (2019) Efficient production of S-adenosyl-l-methionine from dl-methionine in metabolic engineered Saccharomyces cerevisiae. Biotechnol Bioeng 116(12):3312–3323
Chen Y, Xu D, Fan L, Zhang X, Tan T (2015) Manipulating multi-system of NADPH regulation in Escherichia coli for enhanced S-adenosylmethionine production. RSC Adv 5(51):41103–41111. https://doi.org/10.1039/C5RA02937F
Chen H, Wang Z, Cai H, Zhou C (2016) Progress in the microbial production of S-adenosyl-l-methionine. World J Microbiol Biotechnol 32(9):1–8
Zhao W, Hang B, Zhu X, Wang R, Shen M, Huang L et al (2016) Improving the productivity of S-adenosyl-l-methionine by metabolic engineering in an industrial Saccharomyces cerevisiae strain. J Biotechnol 236:64–70. https://doi.org/10.1016/j.jbiotec.2016.08.003
He J, Deng J, Zheng Y, Gu J (2006) A synergistic effect on the production of S-adenosyl-l-methionine in Pichia pastoris by knocking in of S-adenosyl-l-methionine synthase and knocking out of cystathionine-β synthase. J Biotechnol 126(4):519–527
Hayakawa K, Matsuda F, Shimizu H (2016) Metabolome analysis of Saccharomyces cerevisiae and optimization of culture medium for S-adenosyl-l-methionine production. AMB Express. https://doi.org/10.1186/s13568-016-0210-3
Hu XQ, Chu J, Zhang SL, Zhuang YP, Wang YH, Zhu S et al (2007) A novel feeding strategy during the production phase for enhancing the enzymatic synthesis of S-adenosyl-l-methionine by methylotrophic Pichia pastoris. Enzyme Microb Technol 40(4):669–674
Wang Y, Wang D, Wei G, Wang C (2013) Improved co-production of S-adenosylmethionine and glutathione using citrate as an auxiliary energy substrate. Bioresour Technol 131:28–32. https://doi.org/10.1016/j.biortech.2012.10.168
Ren W, Cai D, Hu S, Xia S, Wang Z, Tan T et al (2017) S-Adenosyl-l-methionine production by Saccharomyces cerevisiae SAM 0801 using DL-methionine mixture: from laboratory to pilot scale. Process Biochem 62(18):48–52. https://doi.org/10.1016/j.procbio.2017.07.014
Ravi Kant H, Balamurali M, Meenakshisundaram S (2014) Enhancing precursors availability in Pichia pastoris for the overproduction of S-adenosyl-l-methionine employing molecular strategies with process tuning. J Biotechnol 188:112–121. https://doi.org/10.1016/j.jbiotec.2014.08.017
Orth JD (2010) Ines Thiele BØP. What is flux balance? Nat Biotechnol 28(3):245–248
Edwards JS, Covert M, Palsson B (2002) Metabolic modelling of microbes: the flux-balance approach. Environ Microbiol 4(3):133–140
Wang H, Marcišauskas S, Sánchez BJ, Domenzain I, Hermansson D, Agren R et al (2018) RAVEN 2.0: A versatile toolbox for metabolic network reconstruction and a case study on Streptomyces coelicolor. PLoS Comput Biol 14(10):1–17
Agren R, Liu L, Shoaie S, Vongsangnak W, Nookaew I, Nielsen J (2013) The RAVEN toolbox and its use for generating a genome-scale metabolic model for penicillium chrysogenum. PLoS Comput Biol 9(3):e100298
Wang W, Kramer PM, Yang S, Pereira MA, Tao L (2001) Reversed-phase high-performance liquid chromatography procedure for the simultaneous determination of S-adenosyl-L-methionine and S-adenosyl-L-homocysteine in mouse liver and the effect of methionine on their concentrations. J Chromatogr B Biomed Sci Appl 762(1):59–65. https://doi.org/10.1016/s0378-4347(01)00341-3
Tomàs-Gamisans M, Ferrer P, Albiol J (2016) Integration and validation of the genome-scale metabolic models of Pichia pastoris: a comprehensive update of protein glycosylation pathways, lipid and energy metabolism. PLoS ONE 11(1):1–24
Tomàs-Gamisans M, Ferrer P, Albiol J (2018) Fine-tuning the P. pastoris iMT1026 genome-scale metabolic model for improved prediction of growth on methanol or glycerol as sole carbon sources. Microb Biotechnol 11(1):224–237
Ye R, Huang M, Lu H, Qian J, Lin W, Chu J et al (2017) Comprehensive reconstruction and evaluation of Pichia pastoris genome-scale metabolic model that accounts for 1243 ORFs. Bioresour Bioprocess. https://doi.org/10.1186/s40643-017-0152-x
Hartner FS, Glieder A (2006) Regulation of methanol utilisation pathway genes in yeasts. Microb Cell Fact 5:1–21
Sahu U, Rajendra VKH, Kapnoor SS, Bhagavat R, Chandra N, Rangarajan PN (2017) Methionine synthase is localized to the nucleus in Pichia pastoris and Candida albicans and to the cytoplasm in Saccharomyces cerevisiae. J Biol Chem 292(36):14730–14746
Guillamón JM, Van Riel NAW, Giuseppin MLF, Verrips CT (2001) The glutamate synthase (GOGAT) of Saccharomyces cerevisiae plays an important role in central nitrogen metabolism. FEMS Yeast Res 1(3):169–175
Bakker BM, Overkamp KM, Van Maris AJA, Kötter P, Luttik MAH, Van Dijken JP et al (2001) Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol Rev 25(1):15–37
Serrano-Bueno G, Hernández A, López-Lluch G, Pérez-Castiñeira JR, Navas P, Serrano A (2013) Inorganic pyrophosphatase defects lead to cell cycle arrest and autophagic cell death through NAD+ depletion in fermenting yeast. J Biol Chem 288(18):13082–13092
Cregg JM (2007) Pichia protocols. Methods Mol Biol 389:1–10
Acknowledgements
The authors are thankful to Department of Biotechnology (DBT) for providing fellowship to Kabilan S and DBT-Builder Program for providing facility and Dr. Anand Ramaian Santhaseela, Anna University for supporting us.
Funding
This study was supported by Department of Biotechnology, Ministry of Science and Technology, India, BT/PR12153/INF/22/200/2014, BT/PR7605/FNS/20/732/2013.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
We affirm that there are no financial, personal, or professional affiliations that might be perceived as influencing the content of this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
449_2023_2913_MOESM2_ESM.xml
Supplementary file2 iAUKM model in SBML format. Pichia pastoris GEM model in SBML format generated with the RAVEN toolbox (XML 3397 KB)
449_2023_2913_MOESM3_ESM.xlsx
Supplementary file3 GEMs performance comparison. Analysis of the prediction capabilities of the model iAUKM for identifying the over expression targets for S-adenosyl L-methionine (XLSX 21 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Subash Chandra Bose, K., Shah, M.I., Krishna, J. et al. Genome-scale metabolic model analysis of Pichia pastoris for enhancing the production of S-adenosyl-l-methionine. Bioprocess Biosyst Eng 46, 1471–1482 (2023). https://doi.org/10.1007/s00449-023-02913-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-023-02913-1