Abstract
Industrial production of lignocellulosic ethanol requires a microorganism utilizing both hexose and pentose, and tolerating inhibitors. In this study, a hydrolysate-cofermenting Saccharomyces cerevisiae strain was obtained through one step in vivo DNA assembly of pentose-metabolizing pathway genes, followed by consecutive adaptive evolution in pentose media containing acetic acid, and direct screening in biomass hydrolysate media. The strain was able to coferment glucose and xylose in synthetic media with the respective maximal specific rates of glucose and xylose consumption, and ethanol production of 3.47, 0.38 and 1.62 g/g DW/h, with an ethanol titre of 41.07 g/L and yield of 0.42 g/g. Industrial wheat straw hydrolysate fermentation resulted in maximal specific rates of glucose and xylose consumption, and ethanol production of 2.61, 0.54 and 1.38 g/g DW/h, respectively, with an ethanol titre of 54.11 g/L and yield of 0.44 g/g. These are among the best for wheat straw hydrolysate fermentation through separate hydrolysis and cofermentation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lignocellulosic biomass, such as oil palm empty fruit bunch (OPEFB) and wheat straw (WS), does not compete with humans for food and is a sustainable source of sugars for expanding biofuel and chemical production [1, 2]. Lignocellulosic biomass hydrolysates, especially those produced from agricultural wastes, herbaceous and hardwood biomass, are rich in pentoses, such as d-xylose and l-arabinose [2, 3]. To maximize the yield of lignocellulosic ethanol, all carbohydrate fractions including hexoses (i.e. glucose and galactose, etc.) and pentoses (d-xylose and l-arabinose) should be fully utilized by the microorganisms [3, 4]. The budding yeast Saccharomyces cerevisiae commonly used for industrial ethanol production cannot metabolize xylose and arabinose naturally. Pentose-fermenting yeasts have been developed by intensive genetic engineering and prolonged evolutionary engineering [4, 5]. However, for industrial application in lignocellulosic hydrolysate fermentation, tolerance to inhibitory chemicals in the hydrolysates is important [6].
For xylose utilization, either xylose isomerase (XI) pathway or xylose reductase/xylitol dehydrogenase (XR/XDH) pathway was introduced into yeast strains [7, 8]. Through the XI pathway, xylose is directly converted to xylulose and it does not require any cofactor and this leads to high theoretical ethanol yield [9, 10]. On the other hand, the XR/XDH pathway offers higher metabolic fluxes than the XI pathway; however, it accumulates xylitol due to the uncoupled cofactor consumption of NADPH-preferred XR and NAD+-preferred XDH [11]. Numerous genetic manipulations have been applied to S. cerevisiae expressing XR and XDH to solve the co-factor imbalance problems and improve ethanol yield [10]. For l-arabinose utilization, three enzymes from bacterial arabinose operon (araBAD) were overexpressed, which include genes encoding l-arabinose isomerase (araA), L-ribulokinase (araB), and L-ribulose-5-phosphate 4-epimerase (araD) [12, 13]. Efficient alcoholic fermentation of these two sugars requires combinatorial engineering of yeast strains to enhance the non-oxidative pentose phosphate pathway and prolonged evolutionary cultivation to accumulate spontaneous beneficial mutation [4, 5].
Commercial production of lignocellulosic ethanol is still challenging due to the inhibitory compounds in lignocellulose hydrolysates. Acetic acid is a primary inhibitor in the hydrolysates of various lignocellulosic materials [14,15,16], which negatively influences cell growth and sugar fermentation, whereas 5-hydroxymethyl-2-furaldehyde and furfural are below the inhibitory concentration [17]. It was shown that ~ 7 g/L acetic acid delayed xylose utilization in the evolved xylose-fermenting yeast strains [17, 18]. Although introducing heterologous acetaldehyde dehydrogenase can generate a yeast strain that converts acetate to ethanol, it is still necessary to make the yeast strain tolerant to high concentration of acetic acid for initial cell propagation [19].
Traditional method in pentose-fermenting yeast construction involves multiple steps of pentose-metabolizing pathway genetic engineering in the host strain followed by prolonged evolutionary engineering [4, 5, 17, 20]. It takes years to develop a successful pentose-fermenting yeast strain for industrial lignocellulose hydrolysate fermentation. Recently, xylose-metabolizing pathway genes were refactored in S. cerevisiae through combinatorial CRISPR-Cas9-mediated rational metabolic engineering and evolutionary engineering [3]. This allows rapid xylose-fermenting yeast construction with glucose and xylose cofermentation capabilities. Besides CRISPR-Cas9, DNA assembler is an effective method to assemble an entire biochemical pathway in a single step in S. cerevisiae via in vivo homologous recombination [21].
In this study, a hydrolysate-cofermenting yeast strain was rapidly constructed through one-step in vivo DNA assembly, followed by consecutive evolutionary engineering in pentose media containing acetic acid, and lignocellulose hydrolysate medium screening. A series of gene expression cassettes containing varied xylose isomerase (XI) genes and arabinose-metabolizing pathway genes were in vivo assembled and integrated into the genome of a uracil auxotrophic S. cerevisiae strain JUK36α [8] using an integrative and replicative plasmid [22]. This allowed the rapid recombination of xylose- and arabinose-metabolizing pathway genes in the host strain with optimal pentose-metabolizing pathway combination. The obtained engineered yeast strain, 36aS1, was subsequently enhanced for pentose utilization and hydrolysate-cofermentation through consecutive adaptive cultivation alternatively in complex media containing xylose and arabinose in the presence of 6 g/L acetic acid until no further improvement in cell growth was observed. This endowed the strain library, 36aS1.10, with optimal and stable genome integration of pentose-metabolizing pathway genes.
Two lignocellulose hydrolysate samples were collected from industries. A potential hydrolysate-cofermenting yeast strain was isolated on lignocellulose hydrolysate agar plates and liquid media. This generated an engineered yeast strain, 36aS1.10.4, which displayed superior glucose and xylose cofermentation capabilities in both synthetic and industrial lignocellulose hydrolysate media. High ethanol titre, productivity and yield were obtained from both lignocellulose hydrolysate samples. This strain is promising in efficient fermentation of industrial lignocellulose hydrolysates and can be applied as a workhorse in lignocellulosic ethanol industries.
Materials and methods
Materials
The S. cerevisiae strains used in this work are listed in Table 1. Diploid yeast strain S. cerevisiae ATCC 24860 was obtained from American Type Culture Collection (ATCC, Rockville, MD, USA). S. cerevisiae haploid strain JUK36α was isolated from strain S. cerevisiae ATCC 24860 [8]. It was overexpressed with the non-oxidative pentose phosphate pathway (PPP) genes and xylulokinase gene, XKS1. The auxotrophic marker gene ura3 and the aldose reductase gene gre3 were disrupted in JUK36α. Strain Escherichia coli DH5α was obtained from Life Technologies (Rockville, MD, USA) and grown in Luria Bertani (LB) broth containing ampicillin (50 mg/L) at 37 °C for plasmid construction and maintenance.
All chemicals were of analytical grade and were obtained from Sigma-Aldrich (Singapore) unless otherwise stated. The restriction enzymes and Phusion DNA polymerase were purchased from the New England Biolabs (Ipswich, MA, USA).
Oil palm empty fruit bunch (OPEFB) hydrolysate (hydrolysate I) was prepared by Teck Guan Holdings Sdn Bhd (Tawau, Malaysia) through hydrothermal pretreatment followed by enzymatic hydrolysis using the enzyme mixture of cellulast 1.5 L and Novozym 188 (Novozymes A/S, Bagsværd, Denmark) according to the protocols described in earlier reports [23, 24]. Wheat straw hydrolysate (hydrolysate II) was prepared by Inbicon A/S (Fredericia, Denmark) in a similar manner also according to the previous report [23]. The composition of lignocellulose hydrolysate samples and their fermentation media is listed in Table 2.
Microorganism cultivation media and lignocellulose hydrolysate fermentation media
Synthetic medium contained 1.7 g/L yeast nitrogen base w/o amino acids (YNB), 5 g/L ammonium sulphate, and specified concentration of glucose (YNBG), xylose (YNBX) or arabinose (YNBA), or sugar mixture. YP medium contained 10 g/L yeast extract, 20 g/L peptone, supplemented with glucose (YPD), xylose (YPX) or arabinose (YPA) at specified concentration with an initial pH 5.5. All engineered yeast strains were stored at − 80 °C in stock medium (YPX or YPD) containing 0.9% sodium chloride and 20% (w/v) glycerol.
Lignocellulose hydrolysate samples were supplemented with 10 g/L yeast extract, 20 g/L peptone, and sugar if necessary to generate hydrolysate fermentation media with an initial pH of 5.5. The composition of lignocellulose hydrolysate fermentation media is listed in Table 2.
Twenty grams per liter Bacto agar was added to each medium and was autoclaved at 121 °C for 15 min. Subsequently, the autoclaved mixture was spread into petri dishes to generate its corresponding agar plates.
In vivo DNA assembly of pentose-metabolizing pathway genes
The plasmids used in this work are listed in Table 1. The primers used in this study are listed in Table S1. The codon-optimized XI genes, BvuXylA (Bacteroides vulgatus XylA, GenBank accession number KY362292), XIqXylA (Orpinomyces sp. ukk1 XylA, GenBank accession number KY348519), TAAXylA (Tannerella sp. 6_1_58FAA_CT1 XylA, GenBank accession number KY362293) were synthesized by Bio Basic Asia Pacific Pte Ltd (Singapore) [8]. The arabinose pathway genes (araA, araB and araD) were amplified from the genome of Lactobacillus plantarum through polymerase chain reaction (PCR) using Phusion DNA polymerase (New England Biolabs, Ipswich, MA, USA). The yeast expression plasmids pJFE11 and pPY1 were constructed from pYES2 previously in our laboratory [8]. All the XI genes were individually cloned into the XhoI/XbaI sites of plasmid pPY1 between PGK1 promoter and CYC1 terminator to generate the XI gene expression cassettes (Fig. 1b; Table 1). Promoters and terminators were PCR amplified from the genomic DNA of S. cerevisiae ATCC 24,860. The araB-araD-araA fragment was amplified from L. plantarum genome directly and was cloned into the BamH/XhoI sites of plasmid pJFE11 between TEF1 promoter and CYC1 terminator to generate plasmid pJFE11-BDA. The arabinose-metabolizing gene cassettes araD-PGK1t-PGK1p-araB-ADH1t-TPH3p-araA were constructed via overlap extension PCR (OE-PCR) with BamH/XhoI linearized plasmid pJFE11 to generate plasmid pJFE11-ABD (Table 1). A high effective autonomous replicative sequence in S. cerevisiae genome, ARS315 [22], and the homologous arms of rDNA gene in S. cerevisiae, rDNA1 and rDNA2, were amplified from the genomic DNA of S. cerevisiae ATCC 24,860. The yeast selection marker KanMX was amplified from pUG6 [25]. ARS315, rDNA1, rDNA2 and KanMX were cloned into the plasmid pUC19 [26] to generate the integrative and replicative plasmid pNTS2AK (Fig. 1a; Table 1). Pentose-metabolizing gene cassettes with their respective flanked arms were individually amplified from their respective plasmids. Each xylose isomerase gene expression cassette was mixed with the BamHI/XbaI linearized plasmid pNTS2AK and was then cotransformed with each arabinose-metabolizing gene cassette into JUK36α using LiAc/SS carrier DNA/PEG method [27]. This generated six sets of xylose and arabinose metabolic pathway gene combination. The seventh was created by transforming the mixture of all the five pentose-metabolizing pathway gene cassettes in equal amount and then cotransforming them with one part of the linearized plasmid pNTS2AK. The overlapping regions were 45–50 bp between the gene cassettes and the linearized backbone of plasmid pNTS2AK (Table S1) [21].
Flowchart for rapid yeast strain development. a Linearized integrative and replicative plasmid pNTS2AK; b xylose and arabinose pathway gene cassettes; c best pentose-utilizing strain 36aS1; d adaptive evolution of strain 36aS1 alternatively in xylose and arabinose liquid media containing acetic acid; e adapted strain library 36aS1.10 with optimal pentose-metabolizing gene integration; f strain 36aS1.10.4 screening on OPEFB hydrolysates agar plates and in OPEFB hydrolysate liquid media
Strain screening for optimal cell growth in pentose media
After gene cassette transformation, recombinant yeast strain libraries with six possible combinations were generated (Fig. 1b). The transformed yeast cells were plated on the YPD agar plates containing 300 µg/L G418. After 3 days, seven colonies from each set of transformants with the largest colony size were selected and transferred to YP agar plates containing xylose (YPX) or arabinose (YPA). Afterwards, functional strains were screened on synthetic medium agar plates supplemented with 67 mg/L uracil and 20 g/L xylose (YNBX) or 20 g/L arabinose (YNBA). They were further evaluated in 12-mL tubes containing 5 mL synthetic medium supplemented with 50 g/L xylose (YNBX). Cell growth was measured at 72 h and the strain with the highest cell density at 600 nm (OD600) was selected for further strain adaptive evolution (Fig. 2).
Adaptive evolution and screening of hydrolysate-cofermenting yeast strain
Strain 36aS1 was selected due to its best growth in xylose media (Figs. 1c, 2). Series transfer with 48 h-intervals was performed alternatively in YPX (50 g/L xylose) and YPA (20 g/L arabinose) media containing 6 g/L acetic acid with an initial OD600 of 0.2 for each transfer. Every transferred culture was considered as one generation and all generation strains were harvested at 48 h and then individually stored at − 80 °C until use. They were later on grown in 24-well plate containing YNBX (50 g/L xylose) and YNBA (20 g/L arabinose) media with an initial OD600 of 0.2. Cell growth, OD600, was continuously monitored in a microplate reader (Infinite 200 PRO, Tecan Group, AG, Männedorf, Switzerland) and the specific cell growth rate at 24 h was calculated (Fig. 3). Strain library obtained after 10 rounds of transfer, 36aS1.10, was further transferred in YPD medium (20 g/L dextrose) to test its genetic stability (Figs. 1d, e, 3). In the meantime, it was spread on YP agar plates containing industrial hydrolysate I (YPH1, Table 2). Fermentation performance of colonies with larger size was evaluated in YP liquid medium containing hydrolysate I (YPH1) (Fig. 1f). The best performing strain, 36aS1.10.4, was isolated according to its final OD600 value and ethanol titre in YPH1 (Fig. 4). The assembled pentose-metabolizing pathways in strain 36aS1.10.4 were confirmed by PCR analysis.
Fermentation in both synthetic and lignocellulose hydrolysate fermentation media
Seed culture of 36aS1.10.4 was prepared by reviving the stock culture from − 80 °C in 12 mL tube containing 3 mL YPX (50 g/L xylose) at 30 °C and 200 rpm for 24 h. About 10 mL of the revived culture was transferred into 500 mL flasks containing 100 mL YPX medium (50 g/L xylose) and then incubated at 30 °C and 200 rpm for 24 h. This seed culture was subsequently harvested, washed with sterile double distilled water, and later on used to inoculate fermentation medium at an initial OD600 of 4.0 (approximately 1 g DW/L of yeast cells) unless otherwise stated. Fermentation was performed in 125 mL flasks containing 25 mL synthetic medium (YNB media) and hydrolysate fermentation media (Table 2) at 30 °C and 100 rpm under oxygen-limited conditions with the initial pH at 5.5. The oxygen-limited conditions in the flasks were maintained by capping the flasks with rubber stoppers pierced with a needle to allow the release of CO2. All fermentation experiments were conducted in duplicate and average results were reported.
Cell dry weight measurement and metabolite analysis
OD600 was monitored using a UV–visible spectrophotometer (Shimadzu, Tokyo, Japan). For determination of cell dry weight, cell cultures (50 mL) with varied OD600 (1–5) were filtered through 0.22 µm filter paper using Aspirator A-3S (Fisher Scientific, Tokyo, Japan). Cells were washed twice with distilled water, dried at 105 °C in an oven for 24 h, and weighed. One OD600 unit corresponded to 0.241 g/L cell dry weight. Concentrations of glucose, xylose, xylitol, acetate, glycerol, ethanol, furfural, and hydroxymethylfurfural (HMF) in lignocellulose hydrolysates or produced during fermentation were determined by Agilent 1200 series HPLC system (Agilent Technologies, Santa Clara, CA, USA) equipped with a refractive index detector RID-10A using an Aminex HPX-87H ion exchange column (Bio-Rad Laboratories, Woodinville, WA, USA). The column was eluted with 5 mM of sulphuric acid at a flow rate of 0.6 mL/min and 60 °C.
Results and discussion
Pentose-utilizing yeast strain construction
In our previous study, a background strain JUK36α with enhanced xylulokinase and non-oxidative pentose phosphate pathway was constructed by overexpressing xylulokinase (XKS1), and the nonoxidative pentose phosphate pathway genes, transaldolase (TAL1), transketolase (TKL1), ribose-5-phosphate isomerase (RKI1), and ribulose 5-phosphate 3-epimerase (RPE1), and disrupting the aldose reductase gene gre3 [8]. In this study, gene cassettes containing codon-optimized XI genes (BvuXylA, XIqXylA and TAAXylA) and l-arabinose-metabolizing pathway genes (araA, araB and araD) were individually or co-transformed into strain JUK36α. This was conducted by in vivo assembly of gene cassettes containing above pentose-metabolizing genes in S. cerevisiae with a replicative and integrative plasmid (Fig. 1a) [22]. In the end, fermentation performance of 49 transformants was evaluated in synthetic medium with 50 g/L xylose (YNBX). A colony (36aS1) with superior cell growth was selected (Figs. 1b, c, 2). It harboured B. vulgatus XylA (BvuXylA) and TEF1p-araB-araD-araA-CYC1t operon (Table 1). In addition, strain 36aS1 also exhibited growth on YNBA agar plates with 20 g/L arabinose (data not shown).
Metabolic pathway engineering requires expression of gene cassettes containing all pathway genes with their own promoters and terminators. In vivo DNA assembly is efficient in random assembling multiple genes to generate a targeted metabolic pathway in yeast rapidly [21, 28]. It has an advantage in that the pathway can be directly created in vivo through a single transformation, whereas other methods require multiple steps of plasmid construction and transformation [29, 30]. In this study, a series of xylose isomerase and arabinose-metabolizing gene cassettes were in vivo assembled in S. cerevisiae JUK36α and the best functional strain 36aS1 was isolated with optimal combination of xylose and arabinose-metabolizing pathway genes (Fig. 2). It harboured codon-optimized B. vulgatus XylA (BvuXylA) and TEF1p-araB-araD-araA-CYC1t operon (Table 1). This has not been reported previously [4, 12, 13].
Adaptive evolution and screening of hydrolysate-cofermenting yeast strain
To improve pentose utilization and hydrolysate fermentation capability, strain 36aS1 was alternatively cultivated in YPX (50 g/L xylose) and YPA (20 g/L arabinose) media in the presence of 6 g/L acetic acid (Fig. 1d). Cell growth was monitored in both YNBX and YNBA medium after each round of adaptive cultivation. As can be seen, cell growth on xylose was significantly enhanced until seventh round of cultivation (Fig. 3). Although cell growth on arabinose was also improved, it was not as significant as that on xylose. After round seven, no further improvement in cell growth was observed on both sugars, suggesting that the incorporation of the pentose-metabolizing genes in the yeast genome has reached its optimum. The 10th -generation culture showed a maximum specific growth rate of ~ 0.14 1/h in YNBX medium and ~ 0.07 1/h in YNBA medium. The stability of the 10th-generation culture was further assessed by consecutive transfer and cultivation in YPD medium (20 g/L glucose). Cell growth ability on xylose and arabinose did not deteriorate (Fig. 3). As a result, a stable pentose-metabolizing yeast strain library 36aS1.10 was obtained. The above strain evolution process was rapid in assembling the pentose-metabolizing-pathway gene cassettes with stable and optimal genome integration.
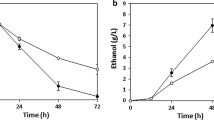
To isolate strains for efficient lignocellulose hydrolysate fermentation, strain library 36aS1.10 was spread on OPEFB hydrolysate YP agar plates (YPH1, Table 2). In total, 23 clones with larger colony size were selected for cell growth and fermentation performance evaluation in liquid OPEFB hydrolysate fermentation medium YPH1 (Fig. 4). While majority of these clones displayed weak growth after 3-day cultivation, six clones showed moderately good growth (Fig. 4a). However, only one clone (clone 4) accumulated ethanol in higher titre than the rest, indicating glucose and xylose cofermentation (Fig. 4b). This clone was denoted as 36aSa.10.4 and was selected for further characterization (Fig. 1e, f).
Fermentation performance of strain 36aS1.10.4 in glucose and xylose media
Fermentation performance of strain 36aSa.10.4 was assessed in the YNB media with xylose as the sole carbon source, and with the mixture of glucose and xylose (Fig. 5; Table 3). Although strain 36aS1.10.4 harboured the arabinose-metabolizing pathway genes (Fig. 1; Table 1) and was able to utilize arabinose (Fig. 3), here only glucose and xylose fermentation results were presented as both industrial hydrolysate samples contained no arabinose (Table 2). In xylose medium (Fig. 5a), xylose was rapidly converted to ethanol in about 40 h with an ethanol yield of 0.40 g/g xylose (Table 3). The maximal specific xylose consumption rate and maximal ethanol production rate were 1.47 g/g DW/h and 0.60 g/g DW/h, respectively. In glucose and xylose mixture medium (Fig. 5b), glucose was almost consumed and xylose was slightly utilized at 24 h; a considerable xylose consumption was observed afterwards. Xylose concentration was reduced to 2.25 g/L at 48 h with about 98% total sugar conversion. Under this condition, strain 36aS1.10.4 had the maximal specific rates of glucose and xylose consumption of 3.47 g/g DW/h and 0.38 g/g DW/h, respectively. In this case, the maximal ethanol production rate was 1.62 g/DW/h and the ethanol yield was 0.42 g/g sugars (Table 3).
It can be observed that when xylose was used as the sole carbon source its consumption rate (1.47 g/g DW/h) was higher than majority of the reported results obtained for xylose fermentation (Table 3) except that reported by Zhou and his colleagues [31]. However, in the presence of glucose, xylose consumption rate was greatly reduced to 0.38 g/g DW/h (Table 3). Such result was lower than that reported by Demeke and his colleagues, 1.1 g/g DW/h [17] and that by Liu and her coworkers, 0.53 g/g DW/h [32] for glucose and xylose cofermentation. However, it was higher than or equal to those reported in majority of other research where xylose-specific consumption rates were in the range of 0.05–0.38 g/g DW/h (Table 3). Such difference was largely attributed to the different adaptive evolution procedures and duration for each strain development. In addition, it was also associated with the fermentation media composition. Generally, fermentation conducted in complex media, such as YP media, presented higher xylose consumption rates [17, 32]. It is worth to note that when glucose and xylose mixture was used as the carbon source, the maximal ethanol-specific production rate (1.62 g/g DW/h) was much higher than that obtained using xylose as the sole carbon source (0.60 g/g DW/h) (Table 3). While the latter is higher than majority of the reported data for xylose fermentation, the former was the highest among those in most of the earlier reports for glucose and xylose cofermentation even though synthetic media was used (Table 3). Such promising results were obtained with only 18-day strain adaptive evolution, demonstrating the high efficiency of strain development approach adopted in this study.
Furthermore, it is worthwhile to compare the ethanol yield obtained from recombinant yeast strains harbouring XI or XR/XDH pathways. Strain 36aS1.10.4 obtained an ethanol yield of 0.40 g/g for xylose fermentation and 0.42 g/g for glucose and xylose cofermentation (Table 3). Such results were comparable with those obtained for strains with XI pathway (Table 3). In general, strains harbouring XI pathway exhibited higher ethanol yield that those harbouring XR/XDH pathway (Table 3). This is due to the cofactor imbalance problems in XR/XDH strains [10, 11]. However, with the engineering of XR, ethanol yield could be considerably improved [10]. Moreover, the presence of glucose [16, 34, 35] or the high initial cell density [32] enhanced the ethanol yield (Table 3).
Fermentation performance of strain 36aS1.10.4 in industrial lignocellulose hydrolysate fermentation media
As the original OPEFB hydrolysate (YPH1) had low sugar content and was readily fermentable (Fig. 4), additional sugar was supplemented to Hydrolysate I to observe the difference in fermentation performance by strains 36aS1 and 36aS1.10.4. Fermentation was conducted in YP medium containing sugar-enriched hydrolysate I (YPH1.1, Table 2). Under this condition, strain 36aS1 only consumed half amount of xylose in 72 h with the maximal specific xylose consumption rate of 0.30 g/g DW/h (Fig. 6a; Table 4). The final ethanol titre of 43 g/L was obtained with the maximal specific ethanol productivity of 0.91 g/g DW/h. On the other hand, the evolved strain 36aS1.10.4 almost consumed all xylose in 72 h with the maximal xylose consumption rate of 0.54 g/g DW/h (Fig. 6b; Table 4). This was 1.8-fold that of strain 36aS1. It is notable that strain 36aS1.10.4 exhibited not only higher xylose utilization rate but also higher glucose consumption rate than the parent strain, 36aS1 (2.82 versus 1.96 g/g DW/h). This demonstrated that the sugar utilization ability of strain 36aS1.10.4 in lignocellulose hydrolysate was improved after consecutive evolution in pentose media containing 6 g/L acetic acid.
In the end, fermentation performance of strain 36aS1.10.4 was evaluated in an original industrial wheat straw hydrolysate (YPH2, Table 2). In this case, strain 36aS1.10.4 was inoculated with an initial cell density of 2.5 g DW/L. About 98% of total sugars was converted and an ethanol titre of 54.11 g/L at 46 h was obtained (Fig. 6c). The maximal specific rates for glucose and xylose consumption were 2.61 and 0.54 g/g DW/h, respectively. Interestingly, the latter was higher than that obtained for glucose and xylose cofermentation in synthetic media (Table 3), further confirming that YP media are more favourable for sugar cofermentation. The maximal specific ethanol productivity was 1.38 g/g DW/h, which was higher than those in other reports (Table 4), however, consistent with that in our earlier report [32].
Among all the hydrolysate samples reported, wheat straw hydrolysate obtained from Inbicon (YPH2) exhibited the highest acetic acid content, 7.42 g/L (Table 4). Strain 36aS1.10.4 was able to ferment this hydrolysate with the maximal specific xylose consumption rate of 0.54 g/g DW/h, which was among the highest compared to those in earlier reports (Table 4). Strain 36aS1.10.4 yielded an ethanol volumetric productivity of 1.18 g/L/h in YPH2, which was also among the highest (Table 4) except that reported by Lee and his colleagues (1.63 g/L/h) when an engineered industrial S. cerevisiae yeast strain was used [37]. The above analysis suggests that strain 36aS1.10.4 is promising in lignocellulose hydrolysate fermentation.
Formic acid, acetic acid, furfural, hydroxymethylfurfural (HMF), and biomass-derived phenolic compounds such as vanillin, syringaldehyde, 4-hydroxybenzaldehyde, and phenol are the main lignocellulosic biomass-derived inhibitory compounds. Both evolutionary engineering [17, 39, 40] and rational metabolic engineering [41] were successfully applied to enhance inhibitor tolerance and, therefore, hydrolysate capability of engineered yeast strains. In this study, the combination of adaptive evolution in complex media containing pentoses and acetic acid, and further screening in lignocellulose hydrolysate media allowed the rapid isolation of a stable hydrolysate-cofermenting recombinant yeast strain 36aS1.10.4.
Further fermentation in sugar-enriched industrial hydrolysate media (Table 2) demonstrated that strain 36aS1.10.4 exhibited much better performance than the unevolved strain 36aS1. (Fig. 6a, b; Table 4). This could be attributed to its evolution in pentose media containing acetic acid (Fig. 3) and its isolation from hydrolysate media (Fig. 4). In addition, glucose and xylose cofermentation was observed for strain 36aS1.10.4 and this could be due to its isolation in hydrolysate media based on its superior ethanol production capabilities. The exceptionally higher ethanol titre obtained by strain 36aS1.10.4 than those by the rest isolates indicates glucose and xylose cofermentation (Fig. 4b).
Notably, acetic acid content did not decrease in all hydrolysate fermentation experiments (Fig. 6). Adaptive evolution of strain 36aS1 in pentose media containing acetic acid generated pentose-utilizing strain library 36aS1.10 (Fig. 3) with enhanced hydrolysate fermentation capacity (Fig. 6a, b). However, such evolution process did not endow the strain with acetic acid utilization capability. Among the inhibitory compounds in lignocellulose hydrolysate, acetic acid is not the strongest inhibitor to glucose and xylose cofermentation [42]. However, it was the primary inhibitory compound present in lignocellulose hydrolysate (Table 2), in agreement with many earlier reports [14,15,16,17]. Adaptive evolution of strain 36aS1 in pentose media containing acetic acid successfully generated a hydrolysate-cofermenting strain 36aS1.10.4 (Fig. 4b). The above analysis suggests that the strain development methods adopted in this study are potential in generating yeast strains for cellulosic ethanol industries.
OPEFB is a potential lignocellulosic biomass available in Southeast Asia for biofuel production [1]. It has been extensively investigated for cellulosic ethanol production [43,44,45,46] with the highest ethanol titre of 83.6 g/L obtained through formiline pretreatment followed by simultaneous saccharification and fermentation [45]. For these studies [43,44,45,46], pentoses were almost removed in biomass pretreatment processes and primarily glucose was converted to ethanol using the wild-type S. cerevisiae strains. More recently, a diploid xylose-fermenting S. cerevisiae strain STXQ was successfully used to convert both glucose and xylose in a laboratory-generated OPEFB hydrolysate sample to ethanol and an ethanol titre of 28.4 g/L was obtained [32]. In this study, OPEFB hydrolysate (hydrolysate I) contained 6.07 g/L acetic acid comparable to those reported for other lignocellulose hydrolysate samples (Table 4) although its sugar content was low (Table 2). The parent strain 36aS1 was able to ferment OPEFB hydrolysate most likely through glucose and xylose cofermentation (Fig. 4). Both the parent strain 36aS1 and the evolved strain 36aS1.10.4 were able to ferment sugar-enriched OPEFB hydrolysate (YPH1.1). Ethanol titre of 50.36 g/L was obtained by strain 36aS1.10.4 with an ethanol yield of 0.41 g/g, both higher than those obtained by the parent strain 36aS1 (Table 4). This demonstrated the enhanced glucose and xylose cofermentation capabilities and potential of strain 36aS1.10.4 in OPEFB hydrolysate fermentation compared to strain 36aS1.
Conversion of wheat straw to ethanol has achieved tremendous advancement in recent years with the focus on high-gravity ethanol production. Through phosphoric acid plus hydrogen peroxide (PHP) pretreatment and high-solid loading separate hydrolysis and fermentation (SSF), up to 71.2 g/L ethanol titre was obtained with ethanol yield of 0.33–0.46 g/g consumed sugar [47, 48]. However, a commercial S. cerevisiae yeast strain from Angel Yeast Co., Ltd. (Yichang, China) was used and only glucose was fermented. Recently, recombinant xylose-fermenting yeast strains have been frequently used in conversion wheat straw hydrolysate to ethanol. Using a continuous membrane bioreactor, ethanol titre of 44 g/L was obtained with an ethanol yield of about 0.42 g/g [49]. In batch separate hydrolysis and cofermentation (SHCF) of alkaline preextracted and steam pretreated wheat straw, ethanol titre and yield of 54.5 g/L and 0.46 g/g were obtained, respectively [38]. Compared to SHCF, simultaneous saccharification and cofermentation (SSCF) is more favourable to high-titre ethanol production [35, 50, 51]. Through hybrid SSCF/SHCF processing of SO2 pretreated wheat straw, ethanol titre and yield reached 40 g/L and 0.39 g/g, respectively [50]. By a temperature-profiled multifeed SSCF of wheat straw, ethanol titre of 65 g/L and yield of 0.36 g/g were obtained [51]. The highest ethanol titre of 71 g/L was obtained through SSCF using a flocculating xylose-fermenting yeast strain with ethanol yield of 0.45 g/g [35]. In this study, an ethanol titre of 54.11 g/L and yield of 0.44 g/g were obtained for industrial wheat straw hydrolysate sample (YPH2) using strain 36aS1.10.4 (Table 4). These results are quite comparable to the best results obtained through SHCF reported so far [38]. They further confirm that the rapid strain development methods reported in this work is effective and strain 36aS1.10.4 is potential in industrial lignocellulosic ethanol production, in particular, when both cellulose and hemicellulose hydrolysates should be utilized. Strain 36aS1.10.4 may be potentially used for fermentation of various industrial lignocellulose hydrolysates and, therefore, can be the workhorse in lignocellulosic ethanol industries.
Conclusions
The combination of DNA assembly with repetitive genome integration using an integrative and replicative plasmid harbouring an autonomous replicative sequence allowed the pentose-utilizing gene cassettes assembled in vivo and integrated into the yeast genome. It resulted in a pentose-utilizing recombinant yeast strain with optimal combination and integration of pentose-metabolizing pathway genes. By consecutive transfer alternatively in YPX and YPA media containing 6 g/L acetic acid, a genetically stable yeast strain library, 36aS1.10, was obtained with enhanced growth in pentose media. Direct isolation on lignocellulose hydrolysate plates followed by screening in hydrolysate liquid medium generated the evolved strain 36aS1.10.4, which exhibited strong glucose and xylose cofermentation capacity. In the fermentation of an original wheat straw hydrolysate, 54.11 g/L ethanol was obtained with ethanol volumetric productivity of 1.18 g/L/h and ethanol yield of 0.44 g/g. These are quite comparable to the best results obtained so far for wheat straw hydrolysate through SHCF. We, therefore, conclude that strain development methods outlined in this study are efficient in rapid generation of a hydrolysate-cofermenting yeast strain. Strain S. cerevisiae 36aS1.10.4 can be a flexible workhorse for ethanol production from a variety of lignocellulose hydrolysates.
References
Geng AL (2013) Conversion of oil palm empty fruit bunch to biofuels. In: Fang Z (ed) Biofuels, book 3. INTECH Open Access Publisher, Shanghai (ISBN:9535110500)
Kwak S, Jin YS (2017) Production of fuels and chemicals from xylose by engineered Saccharomyces cerevisiae: a review and perspective. Microb Cell Fact 16:82
Tran Nguyen Hoang P, Ko JK, Gong G, Um Y, Lee SM (2018) Genomic and phenotypic characterization of a refactored xylose-utilizing Saccharomyces cerevisiae strain for lignocellulosic biofuel production. Biotechnol Biofuels 11:268
Wang C, Zhao J, Qiu C, Wang S, Shen Y, Du B, Ding Y, Bao X (2017) Coutilization of d-glucose, d-xylose, and l-arabinose in Saccharomyces cerevisiae by coexpressing the metabolic pathways and evolutionary engineering. Biomed Res Int 2017:5318232
Garcia SR, Karhumaa K, Fonseca C, Sanchez NV, Almeida JR, Larsson CU, Bengtsson O, Bettiga M, Hahn-Hagerdal B, Gorwa-Grauslund MF (2010) Improved xylose and arabinose utilization by an industrial recombinant Saccharomyces cerevisiae strain using evolutionary engineering. Biotechnol Biofuels 3:13
Hahn-Hägerdal B, Karhumaa K, Fonseca C, Spencer-Martins I, Gorwa-Grauslund MF (2007) Towards industrial pentose-fermenting yeast strains. Appl Microbiol Biotechnol 74:937–953
Peng B, Shen Y, Li X, Chen X, Hou J, Bao X (2012) Improvement of xylose fermentation in respiratory-deficient xylose-fermenting Saccharomyces cerevisiae. Metab Eng 14:9–18
Peng B, Huang S, Liu T, Geng A (2015) Bacterial xylose isomerases from the mammal gut Bacteroidetes cluster function in Saccharomyces cerevisiae for effective xylose fermentation. Microb Cell Fact 14:70
Bettiga M, Hahn-Hägerdal B, Gorwa-Grauslund MF (2008) Comparing the xylose reductase/xylitol dehydrogenase and xylose isomerase pathways in arabinose and xylose fermenting Saccharomyces cerevisiae strains. Biotechnol Biofuels 1:16
Jo JH, Park YC, Jin YS, Seo JH (2017) Construction of efficient xylose-fermenting Saccharomyces cerevisiae through a synthetic isozyme system of xylose reductase from Scheffersomyces stipitis. Bioresour Technol 241:88–94
Eliasson A, Christensson C, Wahlbom CF, Hahn-Hägerdal B (2000) Anaerobic xylose fermentation by recombinant Saccharomyces cerevisiae carrying XYL1, XYL2, and XKS1 in mineral medium chemostat cultures. Appl Environ Microbiol 66:3381–3386
Wisselink HW, Toirkens MJ, Del Rosario Franco Berriel M, Winkler AA, van Dijken JP, Pronk JT, van Maris AJA (2007) Engineering of Saccharomyces cerevisiae for efficient anaerobic alcoholic fermentation of l-arabinose. Appl Environ Microbiol 73:4881–4891
Wisselink HW, Toirkens MJ, Wu Q, Pronk JT, van Maris AJA (2009) Novel evolutionary engineering approach for accelerated utilization of glucose, xylose, and arabinose mixtures by engineered Saccharomyces cerevisiae strains. Appl Environ Microbiol 75:907–914
Chandel AK, da Silva SS, Singh OV (2011) Detoxification of lignocellulosic hydrolysates for improved bioethanol production. In: dos Santos Bernardes MA (ed) Biofuel production—recent developments and prospects. INTECH Open Access Publisher, Shanghai
Koppram R, Albers E, Olsson L (2012) Evolutionary engineering strategies to enhance tolerance of xylose utilizing recombinant yeast to inhibitors derived from spruce biomass. Biotechnol Biofuels 5:32
Li H, Shen Y, Wu M, Hou J, Jiao C, Li Z, Liu X, Bao X (2016) Engineering a wild-type diploid Saccharomyces cerevisiae strain for second-generation bioethanol production. Bioresour Bioprocess 3:51
Demeke MM, Dietz H, Li Y, Foulquie-Moreno MR, Mutturi S, Deprez S, Den Abt T, Bonini BM, Liden G, Dumortier F et al (2013) Development of a d-xylose fermenting and inhibitor tolerant industrial Saccharomyces cerevisiae strain with high performance in lignocellulose hydrolysates using metabolic and evolutionary engineering. Biotechnol Biofuels 6:89
Chen Y, Stabryla L, Wei N (2016) Improved acetic acid resistance in Saccharomyces cerevisiae by overexpression of the WHI2 gene identified through inverse metabolic engineering. Appl Environ Microbiol 82:2156–2166
Guadalupe-Medina V, Metz B, Oud B, van Der Graaf CM, Mans R, Pronk JT, van Maris AJA (2014) Evolutionary engineering of a glycerol-3-phosphate dehydrogenase-negative, acetate-reducing Saccharomyces cerevisiae strain enables anaerobic growth at high glucose concentrations. Microb Biotechnol 7:44–53
Hahn-Hägerdal B, Karhumaa K, Jeppsson M, Gorwa-Grauslund MF (2007) Metabolic engineering for pentose utilization in Saccharomyces cerevisiae. Adv Biochem Eng Biotechnol 108:147–177
Shao Z, Zhao H, Zhao H (2009) DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res DNA 37:e16
Shen FL, Huang SC, Hou PC, Geng AL, Ruan WQ (2017) A high effective autonomous replicative sequence in Saccharomyces cerevisiae. Food Ferment Ind 3:20–25
Jorgensen H (2009) Effect of nutrients on fermentation of pretreated wheat straw at very high dry matter content by Saccharomyces cerevisiae. Appl Biochem Biotechnol 153:44–57
Wang Z, Ong HX, Geng A (2012) Cellulase production and oil palm empty fruit bunch saccharification by a new isolate of Trichoderma koningii D-64. Proc Biochem 47:1564–1571
Güldener U, Heck S, Fiedler T, Beinhauer J, Hegemann JH (1966) A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res 24:2519–2524
Yanisch-Perron C, Vieira J, Messing J (1985) Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119
Gietz RD, Schiestl RH (2007) Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc 2:38–41
Kim B, Du J, Eriksen DT, Zhao H (2013) Combinatorial design of a highly efficient xylose-utilizing pathway in Saccharomyces cerevisiae for the production of cellulosic biofuels. Appl Environ Microb 79:931–941
Sedlak M, Ho NW (2004) Production of ethanol from cellulosic biomass hydrolysates using genetically engineered Saccharomyces yeast capable of cofermenting glucose and xylose. Appl Biochem Biotechnol 116:403–416
Latimer LN, Lee ME, Medina-Cleghorn D, Kohnz RA, Nomura DK, Dueber JE (2014) Employing a combinatorial expression approach to characterize xylose utilization in Saccharomyces cerevisiae. Metab Eng 25:20–29
Zhou H, Cheng JS, Wang BL, Fink GR, Stephanopoulos G (2012) Xylose isomerase overexpression along with engineering of the pentose phosphate pathway and evolutionary engineering enable rapid xylose utilization and ethanol production by Saccharomyces cerevisiae. Metab Eng 14:611–622
Liu TT, Huang SC, Geng AL (2018) Recombinant diploid Saccharomyces cerevisiae strain development for rapid glucose and xylose co-fermentation. Fermentation 4:59
Klimacek M, Kirl E, Krahulec S, Longus K, Novy V, Nidetzky B (2014) Stepwise metabolic adaption from pure metabolization to balanced anaerobic growth on xylose explored for recombinant Saccharomyces cerevisiae. Microb Cell Fact 13:37
Li YC, Mitsumasu K, Gou ZX, Gou M, Tang YQ, Li GY, Wu XL, Akamatsu T, Taguchi H, Kida K (2016) Xylose fermentation efficiency and inhibitor tolerance of the recombinant industrial Saccharomyces cerevisiae strain NAPX37. Appl Microbiol Biotechnol 100:1531–1542
Novy V, Wang R, Westman JO, Franzén CJ, Nidetzky B (2017) Saccharomyces cerevisiae strain comparison in glucose-xylose fermentations on defined substrates and in high-gravity SSCF: convergence in strain performance despite differences in genetic and evolutionary engineering history. Biotechnol Biofuels 10:205
Papapetridis I, Verhoeven MD, Wiersma SJ, Goudriaan M, van Maris AJA, Pronk JT (2018) Laboratory evolution for forced glucose-xylose co-consumption enables identification of mutations that improve mixed-sugar fermentation by xylose-fermenting Saccharomyces cerevisiae. FEMS Yeast Res 18(6):foy056
Lee YG, Jin YS, Cha YL, Seo JH (2017) Bioethanol production from cellulosic hydrolysates by engineered industrial Saccharomyces cerevisiae. Bioresour Technol 228:355–361
Yuan Z, Li G, Hegg EL (2018) Enhancement of sugar recovery and ethanol production from wheat straw through alkaline pre-extraction followed by steam pretreatment. Bioresour Technol 266:194–202
Wallace-Salinas V, Gorwa-Grauslund MF (2013) Adaptive evolution of an industrial strain of Saccharomyces cerevisiae for combined tolerance to inhibitors and temperature. Biotechnol Biofuels 6:151
Smith J, van Rensburg E, Görgens JF (2014) Simultaneously improving xylose fermentation and tolerance to lignocellulosic inhibitors through evolutionary engineering of recombinant Saccharomyces cerevisiae harbouring xylose isomerase. BMC Biotechnol 14:41
Kim SK, Jin YS, Choi IG, Park YC, Seo JH (2015) Enhanced tolerance of Saccharomyces cerevisiae to multiple lignocellulose-derived inhibitors through modulation of spermidine contents. Metab Eng 29:46–55
Li YC, Gou ZX, Zhang Y, Xia ZY, Tang YQ, Kida K (2017) Inhibitor tolerance of a recombinant flocculating industrial Saccharomyces cerevisiae strain during glucose and xylose co-fermentation. Braz J Microbiol 48:791–800
Jung YH, Kim IJ, Han JI, Choi IG, Kim KH (2011) Aqueous ammonia pretreatment of oil palm empty fruit bunches for ethanol production. Bioresour Technol 102:9806–9809
Jung YH, Kim IJ, Kim HK, Kim KH (2013) Dilute acid pretreatment of lignocellulose for whole slurry ethanol fermentation. Bioresour Technol 132:109–114
Cui X, Zhao X, Zeng J, Loh SK, Choo YM, Liu D (2014) Robust enzymatic hydrolysis of Formiline-pretreated oil palm empty fruit bunches (EFB) for efficient conversion of polysaccharide to sugars and ethanol. Bioresour Technol 166:584–591
Duangwang S, Ruengpeerakul T, Cheirsilp B, Yamsaengsung R, Sangwichien C (2016) Pilot-scale steam explosion for xylose production from oil palm empty fruit bunches and the use of xylose for ethanol production. Bioresour Technol 203:252–258
Qiu J, Ma L, Shen F, Yang G, Zhang Y, Deng S, Zhang J, Zeng Y, Hu Y (2017) Pretreating wheat straw by phosphoric acid plus hydrogen peroxide for enzymatic saccharification and ethanol production at high solid loading. Bioresour Technol 238:174–181
Qiu J, Tian D, Shen F, Hu J, Zeng Y, Yang G, Zhang Y, Deng S, Zhang J (2018) Bioethanol production from wheat straw by phosphoric acid plus hydrogen peroxide (PHP) pretreatment via simultaneous saccharification and fermentation (SSF) at high solid loadings. Bioresour Technol 268:355–362
Mahboubi A, Ylitervo P, Doyen W, De Wever H, Molenberghs B, Taherzadeh MJ (2017) Continuous bioethanol fermentation from wheat straw hydrolysate with high suspended solid content using an immersed flat sheet membrane bioreactor. Bioresour Technol 241:296–308
Cassells B, Karhumaa K, Sànchez I, Nogué V, Lidén G (2017) Hybrid SSF/SHF processing of SO2 pretreated wheat straw-tuning co-fermentation by yeast inoculum size and hydrolysis time. Appl Biochem Biotechnol 181:536–547
Westman JO, Wang R, Novy V, Franzén CJ (2017) Sustaining fermentation in high-gravity ethanol production by feeding yeast to a temperature-profiled multifeed simultaneous saccharification and co-fermentation of wheat straw. Biotechnol Biofuels 10:213
Acknowledgements
This study was funded by the Science and Engineering Research Council of the Agency for Science Technology and Research (A*STAR) Singapore (Grant no. 092 139 0035). The authors are grateful for the industrial lignocellulose hydrolysate samples provided by Teck Guan Holdings Sdn Bhd, Tawau, Malaysia and Inbicon A/S, Fredericia, Denmark. We are also thankful for the internship opportunities provided by Ngee Ann Polytechnic Singapore to Shuangcheng Huang and Tingting Liu.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Huang, S., Liu, T., Peng, B. et al. Enhanced ethanol production from industrial lignocellulose hydrolysates by a hydrolysate-cofermenting Saccharomyces cerevisiae strain. Bioprocess Biosyst Eng 42, 883–896 (2019). https://doi.org/10.1007/s00449-019-02090-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-019-02090-0