Abstract
The methanol–glycerol co-feeding during the induction stage for heterologous protein production in Pichia pastoris has shown significant productive applications. Available model analysis applied to this dual-limited condition is scarce and normally does not consider the interaction effects between the substrates. In this work, a dual-limited growth model of P. pastoris considering an interactive kinetic effect was applied to an optimised fed-batch process production of heterologous Rhizopus oryzae lipase (ROL). In the proposed model, the growth kinetics on glycerol is fully expressed, whereas methanol kinetics is modulated by the co-metabolisation of glycerol, resulting in an enhancing effect of glycerol-specific growth rate. The modelling approach of fed-batch cultures also included the methanol volatilisation caused by the aeration that was found to be a not-negligible phenomenon. The model predicts the ability of P. pastoris to keep control of the methanol concentration in the broth during ROL-optimised production process in fed batch and fits satisfactorily the specific cell growth rate and ROL production. Implications of interaction effect are discussed applying the general procedure of modelling approach.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pichia pastoris is a methylotrophic yeast that has gained importance during last two decades as an expression system for heterologous protein production. It combines the advantage of being able to grow on simple culture media with a moderate eukaryotic glycosylation pattern appropriate for the production of therapeutic proteins [1]. In P. pastoris, methylotrophic metabolism is used by coupling the expression of the gene of interest with the AOX1 inducible promoter (pAOX1) that also controls the expression of native alcohol oxidase, the first enzymatic step in the methanol metabolic pathway.
In bioprocess research, different strategies of methanol feeding have been applied to improve the heterologous protein production in P. pastoris, (See [2, 3] for comprehensive reviews). One of these strategies is the mixed feeding methanol–glycerol operation, in which glycerol assimilation provides carbon and energy for efficient biomass formation, as well as decreasing the methanol demand for cellular growth. Co-metabolism of glycerol also allows methanol to act as the inducer rather than substrate, as a secondary effect. Several works have confirmed the productive potential of the methanol–glycerol mixture feeding strategy for the heterologous production of proteins such as avidin [4] and angiostatin [5, 6]. These favourable productive results were obtained despite the well-known repressor effect of glycerol on the pAOX1 [7]. To avoid this repression effect, the mixture feeding strategy is designed to operate under glycerol-limited growth, thus keeping minimal glycerol concentration in the cultivation broth [8], as methanol is automatically controlled around 2–5 g l−1 which is the range of methanol concentration for full induction of AOX1 promoter [9,10,11].
In general, the research using the methanol–glycerol mixture feeding strategy has mainly focused on improving productive results. Nevertheless, fundamental kinetic analysis of P. pastoris growth under dual-substrate limitation, which is the actual kinetic condition during protein production, has been scarcely reported. Zhang et al. [12] obtained satisfactory predictions of Mut+ P. pastoris growth applying a growth model based on the additive kinetics of each substrate, and further used it in process optimisation of methanol–glycerol mixture feeding for intracellular production of heavy-chain fragment of C. botulinum toxin. Çelik et al. [13] used sorbitol instead of glycerol as co-substrate, modelling cell growth as the unmodified additive expression of each substrate kinetics, considering steady state for the methanol concentration in the broth, given by the assumption of full match between methanol feeding and methanol microbial uptake. However, diverse evidence accumulated since early works with methylotrophic yeasts suggest that dual-limited growth of P. pastoris is the result of a more complex substrate interaction and not the simple addition of the performance on the single substrates. For example, Egli et al. [14] using methanol–glucose mixtures in chemostat reported that individual kinetic full expression is dependent on the dilution rate (D) and observed methanol consumption at D beyond Dcrit on methanol (0.19 h−1). Similar behaviour has been reported in more recent years with P. pastoris Mut+ phenotype. Paulova et al. [15] studied the production of tripsinogen using methanol–glucose mixture feeding in continuous culture observing that methanol fed is completely consumed at D = 0.14 h−1, value that is higher than the specific growth rate on methanol as the only carbon source (0.12 h−1). In our previous work [16] dealing with the optimisation of heterologous production of Rhizopus oryzae lipase using methanol–glycerol mixture feeding, the methanol uptake was observed at a dilution rate equal to 0.10 h−1, i.e. higher than 0.06 h−1, the µmax of this strain growing on methanol as single carbon source. Substrate interactions have been also revealed by the intracellular flux analysis of P. pastoris. These metabolic studies show that simultaneous assimilation of methanol–glycerol modifies the carbon flux causing redistribution through the pathways of central metabolism [17, 18].
In the theory of multiple-limited microbial growth, the interactive relationship between substrates is one of the kinetic modelling options proposed. In this approach, the resulting specific growth rate is the consequence of changes in the concentration of any of the substrates, and thus the model may be structured by multiplying the kinetic expressions of each substrate involved [19]. Another kinetic structure is the additive-enhanced model, proposed by Tsao and Hanson [20], in which the growth rate is the result of the individual contribution of every substrate. This relationship is mathematically represented by the simple addition of each single-substrate kinetics. These two kinds of relationships are the basis for data analysis of microbial growth under dual-limited conditions which, even in its fundamental form, has shown to be reliable for application to several microbial process of interest for modern biotechnology [21]. Derivations from these basic interactive or additive kinetics, such as weighted average model [22] and multiple carbon sources [23], have been proposed to represent more complex nutritional situations. One interesting model has been proposed by Neeleman et al. [24] for the growth kinetics of B. pertrussis. In their work, the dual limitation is caused by an essential substrate (glutamate) assimilated as carbon and energy sources and a non-essential enhancing substrate (lactate). The model structure is a mathematical combination of both the basic relationships, additive and interactive approaches, that showed satisfactory prediction of cell growth and the dynamics of the substrates in batch experiments.
The aim of this work was to develop a dual-limited model of P. pastoris Mut+ producer of Rhizopus oryzae lipase (ROL) based on the theory of additive–interactive kinetics and applying this model to predict cell growth, substrate consumption and production of ROL during optimal fed-batch induction process using methanol–glycerol mixture feeding.
Materials and methods
Microorganism, cultivation media and inoculum preparation
The microorganism used was P. pastoris Mut+X-33 containing the vector pPICZαA-ROL for extracellular expression of R. oryzae lipase (ROL). The strain is a wild type with a single copy of the gene encoding ROL, under the control of the pAOX1 [25]. The strain was kindly provided by Dr. Pau Ferrer (Universitat Autònoma de Barcelona). For activation of the cells, a cryovial taken from the cell bank was thawed at room temperature and inoculated in 250-ml flasks with 50 ml of BMG media containing 100 mM of phosphate buffer, 10 g yeast extract, 20 g peptone, 0.4 mg biotin, and 10 g glycerol per litre. The culture was incubated at 30 °C and 200 rpm for 24 h. The cells were separated by centrifugation at 5000 rpm for 10 min, washed twice with distilled water and inoculated on 50 ml of YNB medium which composition per litre was 1.7 g yeast nitrogen base reagent (Difco BD) without amino acids and nitrogen source, 5 g ammonium sulphate, and 5 g of methanol or glycerol. After 18 h of cultivation, cells were centrifuged, washed twice and used for propagation or kinetic growth experiments.
Bioreactor setup and fed-batch operation
Media for fed-batch operation contained per litre YNB reagent 5.5 g, ammonium sulphate 15 g, and glycerol 2 g. Cells were cultivated in a 3-L BioFlo 115 Celligen bioreactor, equipped with polarographic dissolved oxygen probe Oxyferm FDA (Hamilton, Switzerland) and pH probe EasyFerm Plus (Hamilton, Switzerland). The reactor containing the solution of ammonium sulphate and glycerol was autoclaved at 121 °C for 20 min; YNB reagent solution was sterilised by microfiltration and added aseptically to the reactor. The operation was at 30 °C, pH 5.5, automatically controlled by the addition of 3 N NaOH. Initial volume was 1.2 l, agitation rate 500 rpm and aeration rate was 1.3 vvm which at final stages of operation was increased to 1.5 vvm to maintain the saturation of oxygen in the broth at 20% or higher. The process started with glycerol 2 g l−1 as the only carbon source for growth of P. pastoris in batch phase during 8 h. When glycerol was exhausted, the culture was kept without feeding in transition phase during 6 h. The induction phase was carried out by feeding a mixture of substrates methanol 85 g l−1 and glycerol 35 g l−1, equivalent to 71% methanol and 29% glycerol of the total mass of the carbon source in the inlet stream. It was implemented an exponential mixed substrate feeding rate using a pre-programmed flow profile. This was made by the addition of increasing volumes of feed at each time interval throughout the induction stage. Feeding was operated by a Watson Marlow 120U peristaltic pump, connected to an on–off control system based on the Arduino programmable microcontroller plate. Fed-batch induction was designed for 48 h of induction and 1.65 l of final volume of the broth (∆V 0.45 l). In the present work, µ is a system variable that must be assessed experimentally and thus, the proper design of fed-batch induction with methanol–glycerol mixed feeding was one of the objectives of this investigation. Therefore, a preliminary estimation of µ was required to adjust the substrate consumption to the substrate feeding rate. For simplicity, exponential feeding function was designed assuming only glycerol as carbon and energy sources for an expected µ. Although this assumption is not real since contribution of methanol is not negligible, it was useful for the experimental exploration of the region around the µ of interest (0.05 h−1) for ROL productivity according to our previous factorial study performed in chemostat [16]. Assuming total consumption of glycerol, feeding rate was
Volume variation during fed batch can be described by an algebraic equation assuming similar densities of liquid phases, the stream feeding and the broth inside the bioreactor, thus
Analytical methods
Biomass was measured by optical density at 600 nm and converted to biomass concentration by means of a calibration with dry weight of cell. In this procedure, 1 g l−1 of dry cell weight is equivalent to 2.4 optical density units. Methanol was measured by GC-FID chromatograph Clarus 600, PerkinElmer, using a capillary column Supelco Equity-1, with 5 ml min−1 of N2 as mobile phase, operated at 200, 80 and 200 °C of temperature for the injector, oven and detector, respectively. Glycerol was determined by HPLC PerkinElmer Series 200 equipped with autosampler and refractive index detector at 45 °C using an Aminex HPX-87H column at 35 °C of oven temperature, H2SO4 4 mM as mobile phase and flow of 0.6 ml min−1. Extracellular lipase activity was measured from hydrolysis of p-nitrophenylbutyrate 4 mM at 30 °C, pH 7.0, measuring online the formation of product at 348 nm [26]. One unit of activity (UA) is defined as the amount of enzyme which liberates 1 µmol of p-nitrophenol in 1 min.
Model development
As carbon and energy sources, both substrates (methanol and glycerol) can support growth of P. pastoris independently of each other, and they are consumed by two independent metabolic pathways. Thus, when methanol and glycerol are simultaneously metabolised at carbon-limited cultivation, the observed specific growth rate is the result of the additive contribution of the consumption of each one of the single substrates. Since glycerol is an easily assimilable substrate, it is assumed that basic kinetic behaviour of P. pastoris on glycerol is not significantly modified by the simultaneous methanol assimilation. In contrast, the effects of the co-metabolism of the carbon and energy sources are assumed to have a significant effect on the methanol kinetic behaviour. In this dual-limited growth, methanol consumption contributes to enhance the specific growth rate on glycerol. Assuming known individual kinetic behaviour, the model proposed for this kind of growth is
Since both carbon sources contribute to cell growth, it is necessary to distinguish the fraction of growth attributable to each one to quantify the consumption rates of the individual substrates. At the metabolic level, the model is a representation of the growth based on the functioning of two active pathways, in which subscript 1 describes the growth due to glycerol utilisation pathway, while 2 represents the enhancing contribution that is caused by the interaction effect between the methanol and glycerol pathways. According to Neeleman et al. [24], this analysis leads to
Description of specific substrate consumption rate contains yield parameters associated with each one of the substrate utilisation pathways. These parameters are calculated considering the overall cell yield, which is measured as the total biomass formation per single substrate consumed. For methanol, the procedure leads to
For glycerol consumption, yields are given by
Furthermore, the following assumptions about the bioreactor operation are applied for fed-batch description including well-mixed liquid phase inside the stirred tank reactor; oxygen supply is not a limiting factor; and that methanol volatilisation is not a negligible phenomenon which can be represented by first-order expressions at cultivation conditions. Considering these assumptions, the following set of mass balance equations around the bioreactor describes the basic operation of fed-batch system during the production of ROL:
In our previous work [16] about ROL production in continuous culture using response surface methodology, it was determined the dependency of specific ROL productivity with dilution rate (D) and methanol % feeding as factors, obtaining an optimal response at 71% of methanol feed. For fed-batch process, it is supposed that D = µ; therefore,
Parameter determination
The growth of P. pastoris on both substrates methanol and glycerol was evaluated by means of a set of separated experiments for the independent study of each of the kinetics involved in the model description. The cell growth rate on methanol was determined measuring initial growth of biomass in batch cultures at different initial concentrations of methanol, ranging from 0.5 to 20 g l−1. The method is based on using a low inoculum concentration [27] that allows an increase in the duration of exponential phase with minimal substrate consumption and negligible methanol volatilisation at the conditions of P. pastoris cultivation in shake flask. These features configure an approximation to the condition of cell growth at constant substrate concentration, proper for reliable kinetic parameter determination. Specific cell growth rate was determined as the slope of lnX vs. t. Kinetic experiments were performed in triplicate in 500-ml shake flask with 100 ml of YNB-M medium, at 30 °C and 200 rpm in orbital shaker. The µ data were correlated with initial methanol concentration by means of the substrate inhibition kinetics of Andrews [28].
The kinetic growth model on glycerol was determined in two sets of experiments depending on the substrate concentration range tested. At low glycerol concentration where limited growth is observed (µ < µmax), fed-batch operation with exponential programmed glycerol substrate feeding was implemented. Shake flask experiments were implemented to assess growth at glycerol concentration above 1 g l−1. µ was determined by calculating the slope of lnX vs. t. The µ data were correlated with glycerol concentration according to Monod kinetic behaviour. Methanol volatilisation during bioreactor operation was assessed in not-inoculated assays at cell growth conditions, pH 5.5 and 30 °C. The changes of methanol concentration from liquid phase were determined for a range of process conditions considering agitation between 400 and 700 rpm, and aeration rate 0.5, 1, 1.5 and 2 vvm. Decrease in methanol concentration during operation time was modelled by the first-order kinetics. Fitting equations to experimental data was carried out using lsqcurvefit command of MATLAB (MathWorks, Inc).
The overall yield coefficients \(Y_{{X/M}}^{{Ov}}\) and \(Y_{{X/G}}^{{Ov}}\) were measured as the slope of linear regression of the biomass production vs. substrate consumed, during the exponential growth in fed batch operated with methanol–glycerol mixed feeding. The biomass yield coefficient on glycerol \({Y_{X/G1}}\) was determined by interpolating the specific growth rate 0.05 h−1 on the linear regression of the inverses \({\raise0.7ex\hbox{$1$} \!\mathord{\left/ {\vphantom {1 {{Y_{X/S}}}}}\right.\kern-0pt}\!\lower0.7ex\hbox{${{Y_{X/S}}}$}}\) vs. \({\raise0.7ex\hbox{$1$} \!\mathord{\left/ {\vphantom {1 \mu }}\right.\kern-0pt}\!\lower0.7ex\hbox{$\mu $}}\) curve that was obtained during the study of growth kinetics on glycerol. Parameter µin was estimated by a least squares fitting. This was done minimising simultaneously the data differences of the three variables methanol concentration, glycerol concentration and biomass according to \({\text{Min}}:\sum\nolimits_{{j=1}}^{n} {\left[ {{{\left( {{{\overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\frown}$}}{M} }_j} - {M_j}} \right)}^2}+{{\left( {{{\overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\frown}$}}{G} }_j} - {G_j}} \right)}^2}+{{\left( {\widehat {{X{V_j}}} - X{V_j}} \right)}^2}} \right]} ,\) between the predicted (^) and each one of the j-observed values.
Results and discussion
Feeding mixtures of methanol–glycerol is a strategy that can improve the productivity of heterologous protein production by means of moving upward the optimal specific growth rate for specific protein productivity that is obtained at relatively low µ values (0.02–0.03 h−1) when using methanol-only feedings [29, 30]. This kinetic effect of co-metabolism is of great interest for bioprocess research. Accordingly, in the process design applied in this work, the active production of both biomass and ROL simultaneously takes place during the mixed methanol–glycerol feeding operation, allowing the assessment of modelling with the fed-batch induction stage running with significant biomass production.
Model parameter determination: kinetics of individual substrates
Cell growth on methanol shows a well-defined exponential growth initial phase, while methanol concentration slowly decreased. Figure 1 shows an example of the initial growth kinetics at methanol concentration of 4 g l−1. The cultivation time was 30–40 h, the required time for two–three cell generations, in which methanol consumption varied 5–10%. This time is suitable to satisfy the condition of constant initial methanol concentration during growth. Volatilisation of methanol in flasks is only noticeable at longer incubation times (> 100 h), so over the period of growth calculation the methanol loss is negligible (Fig. 1). The relationship of µ with the methanol initial concentration is shown in Fig. 2, and parameters of Andrews model are presented in Table 1 along with previously reported kinetic data. The value of µmax is similar to the previously reported value 0.087 h−1 [31]. However, K and Ki showed higher substrate affinity and lower inhibitory effect, respectively. Different experimental culture systems used in parameter determination may be the cause of the divergences in kinetic characterisation of the microbial strains [32], and in P. pastoris cultures, methanol volatilisation is an additional variation factor to be considered. Biological factors such as a metabolic burden caused by the genetic transformation made in this strain cannot be discarded as a possible cause, in a similar way to the µ decrease that has been observed in heterologous protein expression in S. cerevisiae [33].
Considering a longer growth time, when methanol consumption reached 30–50%, it was possible to calculate the methanol-into-biomass yield (YX/M) for each of the µ values (data not shown). From these calculations, the maintenance coefficient was estimated from the slope of the inverse plots, obtaining 0.124 g gX−1 h−1. This value is considerably higher than 0.013 g gX−1 h−1, obtained in high cell density cultures [35], but close to the boundary (0.03 g g wet cell weight−1 h−1) of the variation range observed within the methanol inhibitory concentration range (> 10 g l−1) [34]. The maximal experimental µ measured in this work, 0.06 h−1 (Fig. 2), is almost half of the declared value (0.1 h−1) for P. pastoris Mut+ in the manual of commercial transformation kit (Invitrogen, Pichia fermentation process guidelines). The growth kinetics of glycerol fits Monod model within the glycerol concentration range evaluated (Fig. 3). The fed-batch condition used in this determination allowed the evaluation of µ at lower and controlled glycerol concentration. This is not possible to be done in a regular batch system because of the high substrate affinity (low half-saturation constant) of glycerol. The maintenance coefficient on glycerol was 0.008 g gX−1 h−1 and a quantitative relationship between the observed yield coefficient and µ was obtained. The data for designing the exponential feeding function with methanol–glycerol mixtures used in the fed-batch induction operation were obtained from this kinetic characterisation.
Methanol volatilisation in bioreactor
Methanol thermodynamic properties show a tendency to form a gas phase at cultivation temperature. Vaporisation enthalpy (ΔvapH) is 35.24 kJ mol−1 and vapour pressure is 0.22 bar at 30 °C causing possible losses of this substrate that would affect quantification of fermentation parameters such as substrate consumption rate or substrate yield. Despite this, methanol volatilisation is almost ignored in process characterisation of heterologous protein production with P. pastoris [37]. Unlike what was observed in shake flasks, methanol volatilisation during the bioreactor operation is a relevant phenomenon caused mainly by the aeration rather than agitation rate (Table 2). This suggests a dragging effect of methanol from liquid phase directly related to superficial gas velocity. As it is revealed by the first-order constant k, at aeration rates higher than 1 vvm the volatilisation of methanol is the same magnitude order than µ. Thus, the decrease of methanol concentration by this cause can be as high as that attributable to cell consumption, and therefore it has to be considered for the process modelling.
Validation of P. pastoris growth kinetics in bioreactor operation
Prior to fed-batch production of ROL using methanol–glycerol mixture feeding, the methanol kinetic model considering the methanol volatilisation effect was evaluated during P. pastoris growth on methanol as the sole carbon and energy source in batch bioreactor. In this operation, aeration was progressively increased as the methanol demand rises. Figure 4 shows the results of modelling, and it is observed that cell growth and methanol concentration variations were predicted satisfactorily. Evaluation on batch bioreactor is a necessary intermediary validation step of the mathematical description of growth, even more considering that methanol kinetic parameters were determined in a different experimental system. The results obtained in the present work show that shake flask system is a valid option to characterise the basic kinetic behaviour of P. pastoris on methanol in a bioreactor. Its application has particular advantageous features for the kinetic description of methanol of the recombinant P. pastoris, including simplicity, repetitiveness and minimisation of methanol volatilisation, making possible to determine the intrinsic kinetic behaviour of the strain [38].
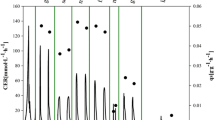
Batch fermentation process of P. pastoris and modelling of cell growth and methanol concentration: (▀) biomass, (●) methanol, (▲) ROL; model prediction represented by lines. Methanol-specific consumption rate (– – –) and methanol concentration model prediction assuming negligible the methanol volatilisation (-----)
In P. pastoris as protein expression systems, basic kinetic data must usually be taken from published reports of protein production process carried out for an aim that is different from modelling itself. Other source of basic kinetic data is the manual of commercial transformation kit provided by the manufacturer, but it must be only considered as a general orientation guide for standard cultivation [39]. The influence of biological factors restricts the kinetic characterisation of P. pastoris to a specific protein-expression system, and this would make it highly strain dependent.
Determination of mixture feeding function for fed-batch cultures
In fed-batch cultures, if only one limiting substrate is exponentially fed then µ is the expected response of the cell growth. Therefore, the given value of µ by the feeding function designed should match the actual µ value measured during the microbial growth. When applied to P. pastoris growth, fed-batch induction operates with methanol as the only substrate, and there is a unique µ value [40, 41], and thus is considered as a system constant. In contrast, in the present operation with dual-carbon source limitation, the kinetic interaction makes that µ cannot be used for the feeding function design, unless the interactive effect is quantitatively determined. To achieve this, an empirical approach was applied with the aim of evaluating the variation of µ, considering the specific glycerol consumption rate as the criterion for designing the feeding function of the methanol–glycerol mixture. The growth of P. pastoris with different feeding functions is presented in Fig. 5. Experimental µ obtained correlates well with the chosen criterion. From this set of assays, the design parameters qGdes and µdes were obtained. They are needed for the modelling evaluation of the fed-batch operation with P. pastoris growing at the optimal µ.
Growth kinetics on fed-batch and continuous cultures
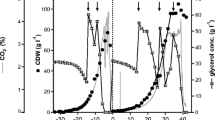
Based on the evaluation of the effect of mixture feedings on P. pastoris growth, a feeding function at glycerol-specific consumption rate of 0.077 g X−1 h−1 (equivalent to 0.045 h−1 for the specific cell growth rate, µdes) was implemented. The initial conditions and the setting of model parameters are shown in Table 3. Because of the implementation of separated experimental studies of the kinetic phenomena involved, µin was the unique parameter determined by data fitting during the mixed feeding induction operation. As shown in Fig. 6, the model predicts the general dynamic tendencies of ROL production and the complementary variables biomass and substrates. Glycerol was not detected in the broth during the whole fed-batch operation, in accordance with the cell growth kinetic controlled by the substrate feeding. Cellular concentration variation is well predicted by the dual-limited growth model (R2 = 0.96). Prediction of biomass formation based on substrate consumption and individual coefficient yields for both methanol and glycerol (0.27 and 0.60 gX g−1, respectively) only differs 5% from the observed biomass (8.5 g predicted vs. 9.0 measured) at 34 h. Considering total process time, the mean difference between predicted and measured biomass was less than 10%, showing a good agreement with stoichiometric biomass prediction. This invariable behaviour either on individual or mixed substrates has been used for the prediction of P. pastoris MutS growth on methanol–glycerol mixed feeding avoiding the use of explicit kinetic expressions of µ [8]. It is observed that methanol concentration remained at low level during most of the time of the production process. In these process conditions, the microbial dynamics results as a control system by the generation of an incremental biomass with each incremental portion of substrate feeding. Therefore, expression of the kinetic properties of P. pastoris keeps the methanol controlled around the optimal induction level. The model fits experimental measurements of ROL activity predicting a stable production during fed-batch induction at specific ROL productivity around 1.2 UA gX−1 h−1 (Fig. 6b) which is slightly higher than that obtained in chemostat system [16]. The specific substrate consumption rates measured are well predicted by the model (Fig. 7A). However, in contrast to glycerol, a decreasing trend is observed in the methanol-specific consumption rate (ranging 0.11–0.07 g gX−1 h−1) which may explain the methanol accumulation at the end of the culture. Zalai et al. [42] also observed methanol accumulation at the final stage during mixed methanol–glycerol fed-batch induction operating at high specific glycerol consumption rates. These authors propose that there would be a repressor effect on AOX1 promoter caused by the glycerol flux, higher than the maximal methanol-specific consumption rate of the strain. Because of the smoothing process conditions implemented in the present operation (low cell density, feeding of dilute methanol), the methanol accumulation was an unexpected result; despite this, the model predicts a slight accumulation during process induction time. Causes of methanol accumulation are not clear, and it is also observed in operations with methanol-only feeding. It seems not to be related to the overcoming of maximal consumption rate, as shown by Spadiut et al. [43] who observed methanol accumulation at induction process designed for methanol-specific consumption of 0.056 g gX−1 h−1, lower than the qM in the present work 0.085 g gX−1 h−1.
Model predicts variation of µ tending gradually towards the value 0.048 h−1 (Fig. 7b). The initial trend to higher µ values is explained by a slight accumulation of glycerol at the early stage of fed-batch operation. After 30 h of growth, it can observe that ROL production is reduced, decreasing specific cell growth rate and accumulation of methanol. The model fails to predict this final stage of P. pastoris growth in fed-batch process, probably because of the appearance of factors not considered in this work, such as intracellular formaldehyde accumulation or cell death.
To verify if our model was able to predict the culture behaviour in a wider range of experimental conditions, we applied the interactive-enhancing kinetic model to our previous study [16] using methanol–glycerol mixed feeding performed on chemostat operation. During continuous culture, steady-state substrate balances for methanol and glycerol are
and from biomass balance, it can be obtained as
Thus, the model was used for the prediction of glycerol and methanol concentrations over the whole set of conditions from the experimental design (D = 0.03–0.1 h−1, and methanol proportion in the feeding stream 45–90%) [16], as shown in Table 4. Model predicts glycerol concentration lower than 0.04 g l−1, while glycerol was not detected experimentally. Methanol prediction is closely comparable to measured values, as shown in Fig. 8. This result is relevant considering that half of the experimental conditions were performed at µ higher than the µmax for methanol as the only carbon source, and confirms the flexible structure of the interactive-enhanced model to fit different metabolic conditions and kinetic behaviour under different culture mode operations.
Methanol concentration at steady state measured in continuous culture using methanol–glycerol feeding previously reported [16] vs. the predicted values obtained from the model in this work
Although the original interactive-enhancing kinetic model was formulated to describe the microbial growth on essential–heterologous substrate relationship such as as glutamate and lactate, similar kinetic structure was applied in the present work to model the dual-limited growth of homologous substrates (methanol and glycerol), also replaceable by each other as carbon and energy sources [44]. In the context of P. pastoris growth for heterologous protein production using methanol–glycerol mixture feeding, the interaction represented by µin must be interpreted as a modulating parameter of the growth response on each substrate caused by the co-metabolism of them. Acting as modulator, the interaction suits the growth rate during dual-substrate limitation depending on growth conditions. In the additive model used by Zhang et al. [12], it was observed an outline of the interactive effect of methanol–glycerol mixture feeding. These authors found out that co-metabolism increases the specific growth rate on glycerol in a proportion quantified as 18% of the µ expected on glycerol only. Therefore, they empirically corrected the mixed kinetic model initially proposed to fit the experimental data. This interaction effect should also explain changes observed on intracellular carbon flux analysis using methanol–glycerol mixture feeding [17, 18].
The complete production process of ROL implemented in this work, which considered simple batch, transition and induction stages, was not only an experimental approach to assess the kinetic growth modelling, but also, compared to standard process, represents an operational design that contributes to the improvement of the productivity of the global process. This is because it transfers the biomass formation to the induction stage and shortens the time of batch and transition unproductive stages. The induction of heterologous protein production using mixtures methanol–glycerol in P. pastoris is a strategy that has shown significant productive potential and deserves bioprocess research for deeper insights. Although a few modelling proposals have been previously reported to either describe or predict both P. pastoris fermentation and protein production (See [45] for a comprehensive review), the model strategy applied in this work represents a new unpublished kinetic modelling approach for this microorganism. Increasing the productivity of heterologous protein with P. pastoris can be considered as a kinetic issue that represents one of the main challenges for bioprocess engineering, and rational application of the methanol–glycerol mixed feeding based on quantitative understanding of fundamental kinetics, as the presented in this work, can contribute towards this goal.
Conclusion
The mixed feeding application for induction of heterologous protein production in P. pastoris requires careful process design to take advantage of the productive potential of the expression system. In this work, fundamental information useful for the design of methanol–glycerol mixture feeding operation is reported. Based on our results, it is concluded that the kinetic description based on the additive-enhancing model represents the growth of P. pastoris using methanol–glycerol under dual-limited condition. Modelling based on interactive-enhancing kinetics properly predicts the general tendencies of biomass and ROL production, the consumption of substrates methanol–glycerol, and variation of µ in a narrow range with a clear trend to the value of interest (0.05 h−1) in the fed-batch operation. The model is also able to successfully predict the culture behaviour in a wide range of experimental conditions performed in continuous cultures. The step-wise research approach including an intermediate validation stage applied in this work to study the main kinetic phenomena provides reliable data to perform the quantitative description of factors affecting the dynamics of methanol during heterologous protein production with P. pastoris in fed-batch using methanol–glycerol mixture feeding. Methanol volatilisation also contributes to the variation of methanol concentration in the broth that becomes a relevant factor caused mainly by aeration rate in bioreactor operation.
Abbreviations
- µ:
-
Specific growth rate (h−1)
- µin :
-
Enhancing growth rate parameter (h−1)
- D:
-
Dilution rate (h−1)
- X:
-
Biomass concentration (g l−1)
- V:
-
Volume of broth (l)
- S:
-
Substrate concentration (g l−1)
- KS :
-
Half-saturation constant (g l−1)
- KI :
-
Methanol inhibition constant (g l−1)
- q:
-
Specific substrate consumption rate (g gX−1 h−1)
- YX/G :
-
Glycerol yield coefficient (gX gG−1)
- YX/M :
-
Methanol yield coefficient (gX gM−1)
- \({Y_{X/G}}^{{Ov}}\) :
-
Overall glycerol yield coefficient (gX gG−1)
- \({Y_{X/M}}^{{Ov}}\) :
-
Overall methanol yield coefficient (gX gM−1)
- kev :
-
Volatilisation first-order constant (h−1)
- qP :
-
Specific ROL productivity (UA gX−1 h−1)
- F:
-
Feed rate (l h−1)
- β(0, 1, 2, 3, 4, 5) :
-
Polynomial coefficients
- Max:
-
Maximal
- 0:
-
Initial
- M:
-
Methanol
- G:
-
Glycerol
- 1:
-
Metabolic glycerol pathway
- 2:
-
Metabolic methanol pathway
- f:
-
Feed
- Des:
-
Designed
References
Macauley-Patrick S, Fazenda ML, McNeil B, Harvey LM (2005) Heterologous protein production using the Pichia pastoris expression system. Yeast 22:249–270. https://doi.org/10.1002/yea.1208
Potvin G, Ahmad A, Zhang Z (2012) Bioprocess engineering aspects of heterologous protein production in Pichia pastoris: a review. Biochem Eng J 64:91–105. https://doi.org/10.1016/j.bej.2010.07.017
Looser V, Bruhlmann B, Bumbak F et al (2015) Cultivation strategies to enhance productivity of Pichia pastoris: a review. Biotechnol Adv 33:1177–1193
Jungo C, Marison I, von Stockar U (2007) Mixed feeds of glycerol and methanol can improve the performance of Pichia pastoris cultures: a quantitative study based on concentration gradients in transient continuous cultures. J Biotechnol 128:824–837. https://doi.org/10.1016/j.jbiotec.2006.12.024
Xie J, Zhang L, Ye Q et al (2003) Angiostatin production in cultivation of recombinant Pichia pastoris fed with mixed carbon sources. Biotechnol Lett 25:173–177
Lin J, Panigraphy D, Trinh LB et al (2000) Production process for recombinant human angiostatin in Pichia pastoris. J Ind Microbiol Biotechnol 24:31–35
Hellwig S, Emde F, Raven N et al (2001) Analysis of single-chain antibody production in Pichia pastoris using on-line methanol control in fed-batch and mixed-feed fermentations. Biotechnol Bioeng 74:344–352
D’Anjou MC, Daugulis AJ (2001) A rational approach to improving productivity in recombinant Pichia pastoris fermentation. Biotechnol Bioeng 72:1–11
Katakura Y, Zhang W, Zhuang G et al (1998) Effect of methanol concentration on the production of human B2-glycoprotein I domain V by a recombinant Pichia pastoris: a simple system for the control of methanol concentration using a semiconductor gas sensor. J Ferment Bioeng 86:482–487
Ferrer P, Alarcón M, Ramón R et al (2009) Recombinant Candida rugosa LIP2 expression in Pichia pastoris under the control of the AOX1 promoter. Biochem Eng J 46:271–277. https://doi.org/10.1016/j.bej.2009.05.018
Zhou X-S, Lu J, Fan W-M, Zhang Y-X (2002) Development of a responsive methanol sensor and its application in Pichia pastoris fermentation. Biotech Lett 24:643–646
Zhang W, Hywood KJ, Plantz BA et al (2003) Pichia pastoris fermentation with mixed-feeds of glycerol and methanol: growth kinetics and production improvement. J Ind Microbiol Biotechnol 30:210–215. https://doi.org/10.1007/s10295-003-0035-3
Çelik E, Çalık P, Oliver SG (2009) A structured kinetic model for recombinant protein production by Mut+ strain of Pichia pastoris. Chem Eng Sci 64:5028–5035. https://doi.org/10.1016/j.ces.2009.08.009
Egli T, Käppeli O, Fiechter A (1982) Mixed substrate growth of methylotrophic yeasts in chemostat culture: Influence of dilution rate on the utilisation of a mixture of glucose and methanol. Arch Microbiol 131:8–13
Paulová L, Hyka P, Branská B et al (2012) Use of a mixture of glucose and methanol as substrates for the production of recombinant trypsinogen in continuous cultures with Pichia pastoris Mut+. J Biotechnol 157:180–188. https://doi.org/10.1016/j.jbiotec.2011.10.010
Canales C, Altamirano C, Berrios J (2015) Effect of dilution rate and methanol–glycerol mixed feeding on heterologous Rhizopus oryzae lipase production with Pichia pastoris Mut+ phenotype in continuous culture. Biotechnol Prog 31:707–714. https://doi.org/10.1002/btpr.2069
Solá A, Jouhten P, Maaheimo H et al (2007) Metabolic flux profiling of Pichia pastoris grown on glycerol/methanol mixtures in chemostat cultures at low and high dilution rates. Microbiology 153:281–290. https://doi.org/10.1099/mic.0.29263-0
Unrean P (2013) Pathway analysis of Pichia pastoris to elucidate methanol metabolism and its regulation for production of recombinant proteins. Biotechnol Prog 30:28–37. https://doi.org/10.1002/btpr.1855
Bader FG (1978) Analysis of double-substrate limited growth. Biotechnol Bioeng 20:183–202. https://doi.org/10.1002/bit.260200203
Tsao GT, Hansson TP (1975) Extended Monod equation for batch cultures with multiple exponential phases. Biotechnol Bioeng 17:1591–1598. https://doi.org/10.1002/bit.260171104
Schmitt E, Bura R, Gustafson R, Ehsanipour M (2016) Kinetic modeling of Moorella thermoacetica growth on single and dual-substrate systems. Bioprocess Biosyst Eng 39:1567–1575. https://doi.org/10.1007/s00449-016-1631-8
Mankad T, Bungay HR (1988) Model for microbial growth with more than one limiting nutrient. J Biotechnol 7:161–166
Egli T, Lendenmann U, Snozzi M (1993) Kinetics of microbial growth with mixtures of carbon sources. Antonie Van Leeuwenhoek 63:289–298
Neeleman R, Joerink M, Beuvery C, Van Boxtel T (2001) Dual-substrate utilization by Bordetella pertussis. Appl Microbiol Biotechnol 57:489–493. https://doi.org/10.1007/s002530100811
Minning S, Serrano A, Ferrer P et al (2001) Optimization of the high-level production of Rhizopus oryzae lipase in Pichia pastoris. J Biotechnol 86:59–70
Cabrera Z, Gutarra MLE, Guisan JM, Palomo JM (2010) Highly enantioselective biocatalysts by coating immobilized lipases with polyethyleneimine. Catal Commun 11:964–967. https://doi.org/10.1016/j.catcom.2010.04.010
Lee J, Chang HL, Parulekar SJ, Hong J (1991) An alternate method for estimation of cell growth kinetics from batch cultures. Biotechnol Bioeng 37:26–34. https://doi.org/10.1002/bit.260370106
Andrews JF (1968) A mathematical model for the continuous culture of microorganisms utilizing inhibitory substrates. Biotechnol Bioeng 10:707–723
Zhang T, Gong F, Peng Y, Chi Z (2009) Optimization for high-level expression of the Pichia guilliermondii recombinant inulinase in Pichia pastoris and characterization of the recombinant inulinase. Process Biochem 44:1335–1339. https://doi.org/10.1016/j.procbio.2009.07.008
Trinh LB, Phue JN, Shiloach J (2003) Effect of methanol feeding strategies on production and yield of recombinant mouse endostatin from Pichia pastoris. Biotechnol Bioeng 82:438–444. https://doi.org/10.1002/bit.10587
Zhou X, Zhang Y (2002) Decrease of proteolytic degradation of recombinant hirudin produced by Pichia pastoris by controlling the specific growth rate. Biotech Lett 24:1449–1453
Kovárová-Kovar K, Egli T (1998) Growth kinetics of suspended microbial cells: from single-substrate-controlled growth to mixed-substrate kinetics. Microbiol Mol Biol Rev 62:646–666
Görgens JF, van Zyl WH, Knoetze JH, Hahn-Hägerdal B (2001) The metabolic burden of the PGK1 and ADH2 promoter systems for heterologous xylanase production by Saccharomyces cerevisiae in defined medium. Biotechnol Bioeng 73:238–245
Zhang W, Bevins MA, Plantz BA et al (2000) Modeling Pichia pastoris growth on methanol and optimizing the production of a recombinant protein, the heavy-chain fragment C of botulinum neurotoxin, serotype A. Biotechnol Bioeng 70:1–8
Jahic M, Rotticci-Mulder J, Martinelle M et al (2002) Modeling of growth and energy metabolism of Pichia pastoris producing a fusion protein. Bioprocess Biosyst Eng 24:385–393. https://doi.org/10.1007/s00449-001-0274-5
Curvers S, Linnemann J, Klauser T et al (2002) Recombinant protein production with Pichia pastoris in continuous fermentation—kinetic analysis of growth and product formation. Eng Life Sci 2:229–235
Mendoza Muñoz DF, Algecira Enciso NA, Córdoba Ruiz H, Barrera Avellaneda LA (2008) A simple structured model for recombinant IDShr protein production in Pichia pastoris. Biotech Lett 30:1727–1734. https://doi.org/10.1007/s10529-008-9750-1
Grady CJ, Smets B, Barbeau D (1996) Variability in kinetic parameter estimates: a review of possible causes and a proposed terminology. Water Res 30:742–748
Cos O, Ramón R, Montesinos JL, Valero F (2006) Operational strategies, monitoring and control of heterologous protein production in the methylotrophic yeast Pichia pastoris under different promoters: a review. Microb Cell Fact 5:17. https://doi.org/10.1186/1475-2859-5-17
Schenk J, Balazs K, Jungo C et al (2008) Influence of specific growth rate on specific productivity and glycosylation of a recombinant avidin produced by a Pichia pastoris Mut+ strain. Biotechnol Bioeng 99:368–377. https://doi.org/10.1002/bit
Jacobs PP, Inan M, Festjens N et al (2010) Fed-batch fermentation of GM-CSF-producing glycoengineered Pichia pastoris under controlled specific growth rate. Microb Cell Fact 9:93. https://doi.org/10.1186/1475-2859-9-93
Zalai D, Dietzsch C, Herwig C, Spadiut O (2012) A dynamic fed batch strategy for a Pichia pastoris mixed feed system to increase process understanding. Biotechnol Prog 28:878–886. https://doi.org/10.1002/btpr.1551
Spadiut O, Zalai D, Dietzsch C (2014) Quantitative comparison of dynamic physiological feeding profiles for recombinant protein production with Pichia pastoris. Bioprocess Biosyst Eng 37:1163–1172. https://doi.org/10.1007/s00449-013-1087-z
Zinn M, Witholt B, Egli T (2004) Dual nutrient limited growth: models, experimental observations, and applications. J Biotechnol 113:263–279. https://doi.org/10.1016/j.jbiotec.2004.03.030
Theron S, Berrios J, Delvigne F, Fickers P (2018) Integrating metabolic modeling and population heterogeneity analysis into optimizing recombinant protein production by Komagataella (Pichia) pastoris. Appl Microbiol Biotechnol 102:63–80
Acknowledgements
This work was supported by Project N°11110486 from Fondo Nacional de Ciencia y Tecnología (FONDECYT, Chile). C. Canales wishes to thank Comisión Nacional de Investigación Científica y Tecnológica (CONICYT, Chile) for scholarship support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Canales, C., Altamirano, C. & Berrios, J. The growth of Pichia pastoris Mut+ on methanol–glycerol mixtures fits to interactive dual-limited kinetics: model development and application to optimised fed-batch operation for heterologous protein production. Bioprocess Biosyst Eng 41, 1827–1838 (2018). https://doi.org/10.1007/s00449-018-2005-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-018-2005-1