Abstract
We investigated how foraging habits vary among three ecologically distinct wide-ranging seabirds. Using amino acid δ15N proxies for nutrient regime (δ15NPhe) and trophic position (Δδ15NGlu-Phe), we compared Newell’s shearwater (Puffinus newelli) and Laysan albatross (Phoebastria immutabilis) foraging habits over the past 50–100 years, respectively, to published records for the Hawaiian petrel (Pterodroma sandwichensis). Standard ellipses constructed from the isotope proxies show that inter-population and interspecific foraging segregation have persisted for several decades. We found no evidence of a shift in nutrient regime at the base of the food web for the three species. However, our data identify a trophic decline during the past century for Newell’s shearwater and Laysan albatross (probability ≥ 0.97), echoing a similar decline observed in the Hawaiian petrel. During this time, Newell’s shearwaters and Hawaiian petrels have experienced population declines and Laysan albatross has experienced range extension and apparent population stability. Counting other recent studies, a pattern of trophic decline over the past century has now been identified in eight species of pelagic seabirds that breed in the Hawaiian Islands. Because our study species forage broadly across the North Pacific Ocean and differ in morphological and behavioral traits and feeding methods, the identified trophic declines suggest a pervasive shift in food web architecture within the past century.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A variety of pelagic predators ranging from sharks to tuna to seabirds have experienced population declines in recent decades (Myers and Worm 2003; Myers et al. 2007; Paleczny et al. 2015). Among these wide-ranging marine predators, seabirds are unusual in their reliance on terrestrial habitats for breeding, where they are more accessible for ecological study. While this creates valuable opportunities for researchers, an inherent consequence is that most studies of pelagic seabirds take place at breeding colonies, and not at sea where the birds typically spend 90% of their lives (Ballance et al. 2001). Even with the advantage of animal tracking technology (Kays et al. 2015), characterizing at-sea behavior is a challenge for seabirds that regularly fly thousands of kilometers across the open ocean in search of food. Moreover, current tracking efforts cannot reconstruct foraging from decades ago when oceanic ecosystems differed from today (Jackson 2008). Understanding seabird foraging ecology is a first step toward determining whether changes in oceanic food webs have affected various ecological parameters like seabird population size. Furthermore, the degree to which food web alterations induced by overfishing of large marine vertebrates, or other potential anthropogenic effects such as climate change (Jackson 2008), have differentially influenced endangered vs. abundant species or ecologically distinct species is poorly known.
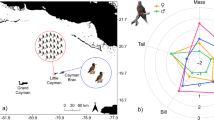
Hawaiian seabirds are an ecologically diverse assemblage of species, several of which are now endangered. Newell’s shearwaters (Puffinus newelli), once thought to be extinct on the Hawaiian Islands (Mitchell et al. 2005), are capable of diving to catch prey at least 10 m beneath the surface and often feed in association with tuna schools or in mixed-species flocks (Ainley et al. 1997, 2014). In contrast, Laysan albatross (Phoebastria immutabilis) feed by sitting on the water surface and frequently scavenge from fishing vessels where they are often killed as bycatch (Cousins et al. 2000; Awkerman et al. 2009). Together with the endangered Hawaiian petrel (Pterodroma sandwichensis), a surface seizer that uses several feeding strategies including foraging in association with tuna, these three species of the North Pacific Ocean exhibit ecological contrasts (Simons and Hodges 1998). Of the three species, Newell’s shearwaters have the most constrained marine distribution and shortest foraging trips during the breeding season—25% and 33% of the trip distance of the Hawaiian petrel and Laysan albatross, respectively (Fig. 1; Spear et al. 1995; Fernández et al. 2001; Adams and Flora 2010). Laysan albatross have an extensive marine range that encompasses nearly the entirety of the North Pacific Ocean (Fig. 1; Awkerman et al. 2009). Their population has rebounded from the early 1900s to a stable estimated population of at least 800,000 individuals and they are expanding their breeding range (Naughton et al. 2007; Arata et al. 2009). Unlike Laysan albatross, both Newell’s shearwaters and Hawaiian petrels have experienced steep population declines in recent decades, with the population of Newell’s shearwaters on Kaua`i estimated to have plummeted 94% between 1993 and 2013 (Raine et al. 2017). Threats on land (e.g., collisions with power lines, light attraction, predation, habitat change) are implicated in the fatalities of these birds (Cooper and Day 1998; Raine et al. 2017). Yet, the effects of concurrent changes to oceanic food webs on seabird foraging are largely unknown.
N* distribution, at-sea distributions of Newell’s shearwater (NESH), Laysan albatross (LAAL) and Hawaiian petrel (HAPE), and δ15N (‰) values of sinking particles and core top sediments. The color gradient is the distribution of N* defined and gridded by Sherwood et al. (2014), where N* = N−16P + 2.9 μmol kg−1, P = phosphorus. Positive N* values are interpreted as an increase in nitrogen fixation and negative values as net denitrification (Sherwood et al. 2014). Curved lines delineate seabird distributions as labeled in the figure (Ainley et al. 1997; Simons and Hodges 1998; Awkerman et al. 2009). The data for sinking particles and sediments mark an area of elevated δ15N in the eastern tropical north Pacific Ocean and illustrate an isotopic gradient that extends in all directions from the area of elevated δ15N. Filled black points represent δ15N of sinking particles and open black points represent δ15N of core top sediments (Altabet and Francois 1994)
As pelagic seabirds forage over large oceanic expanses, they integrate molecular information on trophic dynamics and nutrient regimes in their tissues (Bearhop et al. 2006; Hinke et al. 2015). This information can be accessed through compound-specific nitrogen isotope analysis of tissues to provide a spatiotemporal perspective on seabird foraging (Gagne et al. 2018). This technique is based on the observation that the δ15N of “trophic” amino acids, like glutamic acid (δ15NGlu), becomes 15N-enriched with increasing trophic level due to deamination and other metabolic reactions involving nitrogen transformation while the δ15N of “source” amino acids, like phenylalanine (δ15NPhe), fractionates to a smaller degree with each trophic transfer (Gaebler et al. 1963; McClelland and Montoya 2002; Chikaraishi et al. 2007; Ohkouchi et al. 2017). Consequently, δ15NPhe primarily reflects the isotope value of source nitrogen at the base of the food web and, as such, can be used as a nutrient proxy. The difference between the δ15NGlu and δ15NPhe of a consumer is a trophic proxy (Δδ15NGlu-Phe) and is used to calculate trophic position (TP): TP = (δ15NGlu−δ15NPhe−β)/TDF + 1, where β is the δ15NGlu−δ15NPhe difference in primary producers and TDF is the trophic discrimination factor, or the net elevation of δ15NGlu relative to δ15NPhe per trophic step (McMahon and McCarthy 2016). Estimates of TDF among species are highly variable and not available for many pelagic seabirds, including our study species. Thus, we refrained from calculating trophic position and, as has been done in other studies, rely on Δδ15NGlu-Phe as a trophic proxy (Lorrain et al. 2009; Ellis 2012; Choy 2013; Ostrom et al. 2017).
δ15NPhe from seabirds can establish spatial constraints on foraging because the δ15N of source nitrogen varies across the Pacific Ocean. For example, there is an isotopic gradient southeast of the Hawaiian Islands with a conspicuous localized region of 15N-enriched waters (4–10°N and 135–140°W) (Fig. 1; Altabet and Francois 1994; Graham et al. 2010). Such δ15N gradients result from the prevalence of different biogeochemical processes. The δ15N of the primary nitrogen source of oceans, nitrate, is 5–6 ‰ and becomes elevated due to denitrification and phytoplankton uptake (Karl et al. 1997; Sigman et al. 2000). Nitrogen fixation produces ammonium with δ15N values of 0 ‰ (Karl et al. 1997; Casciotti et al. 2008). Because the δ15N at the base of the food web is transferred to consumers, δ15NPhe values identify the nutrient regime in which seabirds forage.
Spatial variation in nitrogen added to or removed from the ocean by biogeochemical processes is identified as N*. N* refers to the excess or deficit in nitrogen (N) relative to phosphorus (P) from the expected Redfield N:P stoichiometry of 16:1, where N* = N − 16P + 2.9 μmol kg−1 (Deutsch et al. 2001). Average marine nitrate corresponds to an N* of 0, and nitrogen fixation and denitrification increase or decrease N* from 0, respectively (Gruber and Sarmiento 1997; Deutsch et al. 2001). Anthropogenic atmospheric nitrogen deposition also produces positive N* values (Kim et al. 2014). However, atmospheric nitrogen deposition is greatest in coastal regions of the western North Pacific Ocean and is not likely a major influence on the food webs of pelagic seabirds that do not frequent coastal waters (Kim et al. 2014). While we cannot dismiss the possibility that atmospheric nitrogen deposition affects δ15NPhe gradients, we consider δ15NPhe in terms of the relative importance of denitrification and phytoplankton uptake vs. nitrogen fixation on nutrient δ15N.
Amino acid-specific data from ecologically diverse trans-Pacific seabirds can help us to expand our understanding of seabird foraging ecology and develop a more comprehensive representation of oceanic food webs, but it is important to carefully select tissue types. While the timing of body contour molt is sometimes protracted and often poorly constrained, remiges record information related to a specific time period in the annual cycle (Warham 1996; Edwards and Rohwer 2005). In contrast to feathers, the slow turnover time of bone collagen means that collagen records isotopic information over a period of a year or longer (Rucklidge et al. 1992). Thus, collagen isotope data spatially integrate dietary information from across the birds’ annual marine distribution, rather than reflecting a single season or foraging location.
In this study, we developed δ15NPhe and Δδ15NGlu-Phe chronologies based on the analysis of Newell’s shearwater and Laysan albatross bone collagen. We used δ15NPhe data to assess variation in nutrient regime use and Δδ15NGlu-Phe data as an indicator of trophic position differences to ask if foraging ecology differed within and between three ecologically distinct species. Using decadal and centennial records, we also asked if, like the Hawaiian petrel, the foraging ecology of Newell’s shearwater and Laysan albatross changed during the recent past.
Materials and methods
Sample acquisition
Samples dating from 2001 to present were collected from salvaged carcasses (Supplementary Table 1). Acquisition of Hawaiian petrel samples from Haleakalā National Park (Maui) and Hawai`i Volcanoes National Park (Hawai`i) between 2001 and 2010 is described in Wiley et al. (2013). Most of the salvaged Newell’s shearwaters from Kaua`i were found dead after grounding by light attraction or being killed by introduced predators between 2013 and 2016. Laysan albatross were salvaged from the Hawaii longline fisheries between 2003 and 2014. Samples from prior to 2000 are from museum study skins housed in collections at the National Museum of Natural History, the Bernice Bishop Museum, and the California Academy of Sciences (Supplementary Table 1). Samples from after 2000 are designated as the modern time period in each species.
Sample sizes
Samples from modern and historical specimens were derived from after hatch-year birds. Hawaiian petrels were aged previously by Wiley et al. (2013). We determined Newell’s shearwater ages using the color and shape of primaries (Pyle 2008). For historical Laysan albatross, we report the age recorded with the specimen in museum collections. Modern Laysan albatross were aged based on bursa size; the absence of a bursa indicates the bird was likely greater than 4 years of age at death (Broughton 1994). We obtained bone samples from 22 Laysan albatross divided evenly between two time periods corresponding to before and after the onset of industrialized fishing in the North Pacific Ocean: pre-1950 (1902–1937; the historical sample) and post-2000 (2003–2014; the modern sample). Of the 11 historical Laysan albatross, 8 were from Laysan Island, the remaining 3 were from Lisianski Island, the Aleutians West Census, and Midway Island. For Newell’s shearwaters, we divided 24 samples into three time periods (Supplementary Table 1). The oldest available museum samples were from 1964 to 1966 (n = 5); we compared those to two more recent time periods: 9 birds from 1983 to 1998 and 10 birds from 2013 to 2016. Newell’s shearwaters prior to 2000 originated from Kaua`i (n = 4), Hawai`i (n = 4), Oahu (n = 3), and the North Pacific Ocean within 500 miles of the Hawaiian Islands (n = 3) (Supplementary Table 1). Ancient and modern Hawaiian petrel data are from Ostrom et al. (2017) and include Maui individuals from the Foundation (1000–1400 CE 550–950 y B.P., n = 5) and Modern (1950–2010, n = 7) time periods and birds from the island of Hawai`i from the Late Expansion (1400–1800 CE 150–550 y B.P., n = 8) and Modern (1950–2010, n = 8) time periods. Hawaiian petrel time periods are defined by Kirch (1990) and represent archaeological periods in the development of human societies in the Hawaiian Islands.
Sample preparation
Collagen was isolated and purified according to Stafford et al. (1988) as modified by Wiley et al. (2013). Modern and ancient bone fragments (50–200 mg) were scraped with a razor blade, rinsed with ultrapure distilled water (E-pure, Barnstead), demineralized with quartz-distilled 1 N hydrochloric acid (HCl) and soaked in 0.05 N potassium hydroxide overnight to remove humate contaminants. The resulting collagen was lyophilized, gelatinized (0.05 N HCl at 105 °C for 1–8 h), filtered (0.22 or 0.45 μm Millipore Millex GV filter), lyophilized again and frozen prior to lipid removal.
Gelatin (0.5–1.2 mg) was hydrolyzed (0.5 mL, 12 N quartz-distilled HCl at 105 °C for approximately 20 h). Lipids were removed from the resultant filtrate with n-hexane/dichloromethane (3:2, v/v), and evaporated to dryness in methanol under a gentle N2 stream at 50 °C. Amino acids in the lipid-extracted hydrolysate were esterified and acylated with N-pivaloyl/isopropyl derivatization (Chikaraishi et al. 2009). Samples were esterified with thionyl chloride/2-propanol (1:4, v/v) at 105 °C for 2 h then acylated with pivaloyl chloride/dichloromethane (1:4, v/v) at 105 °C for 2 h. The amino acid derivatives were extracted with n-hexane/dichloromethane (3:2, v/v) and stored at − 25 °C.
δ15N amino acid analysis
The δ15N of individual amino acids was determined by gas chromatography/combustion/isotope ratio mass spectrometry using an Isoprime isotope ratio mass spectrometer (IRMS; Elementar, UK) coupled to a 7890 gas chromatograph (GC; Agilent Technologies, USA) via a combustion and reduction furnace. Combustion and reduction were performed in a glass capillary tube with CuO, NiO, and Pt wires at 950 °C. The amino acids were injected splitless on column at 250 °C and separated on a BPX-5 capillary column (60 m × 0.32 mm inner diameter, 1.0 μm film thickness; SGE Analytical Science, USA). The GC oven temperature program was: initial temperature 40 °C for 2 min, ramp of 10 °C min−1 to 280 °C and hold for 10 min, ramp of 10 °C min−1 to 325 °C and hold for 25 min. Carrier gas (He) flow was 1.6 ml min−1. CO2 and H2O were removed from the sample stream using a liquid nitrogen trap.
Stable isotope values are expressed in per mil (‰) as δ15Na = [(15N/14Nsample/15N/14Nstandard)-1] × 103 relative to the standard, atmospheric N2. Accuracy was evaluated by daily analysis of external reference mixtures consisting of NP/iPr derivatives of several isotopically characterized amino acids (Gly, Val, Leu, Pro, Asp, Met, Glu, Phe). Reproducibility of the standards was 0.7 ‰ or better. Samples were analyzed in duplicate or triplicate. If duplicates of a sample differed by no more than 0.5 ‰ the average of the two was reported. If not, the analysis was repeated a third time. The standard deviation of triplicates was less than 0.7 ‰.
Nitrogen isotope analyses for Laysan albatross and Newell’s shearwater were conducted at Michigan State University. Laboratory analyses for the previously published Hawaiian petrel study (Ostrom et al. 2017) were performed by Yoshito Chikaraishi at the Japan Agency for Marine-Earth Science and Technology using a Delta-plus XP IRMS (Thermo Fisher Scientific) coupled to a 6890 GC (Agilent Technologies) via combustion and reduction furnaces. For this reason, we performed a blind inter-laboratory comparison of amino acid-specific results on Laysan albatross bone collagen between the two laboratories. This enabled us to determine whether it is reasonable to compare data from the two studies.
Model selection
We evaluated the effect of large-scale climatic phenomena (i.e., El Niño Southern Oscillation, ENSO) on Newell’s shearwater and Laysan albatross temporal δ15NPhe and Δδ15NGlu-Phe data by including the Multivariate ENSO Index (MEI) and time period as independent variables in regression models. The model was evaluated in a Bayesian framework with hierarchical levels to account for analytical error. δ15NPhe and Δδ15NGlu-Phe were evaluated separately. Model selection was conducted based on a generalization of Akaike’s Information Criterion (AIC), deviance information criterion (DIC; Gelman et al. 2004). DIC does not penalize models with extra parameters as intensely as AIC does. Based on an estimate that avian bone collagen has a half-life of approximately 6 months (Hobson and Clark 1992), we assigned MEI values based on the average NOAA MEI ranking for the 18 months prior to the bird’s date of death.
Statistical analysis
We developed a hierarchical method that accounts for analytical variation and reduces type 1 errors in hypothesis testing. The method consists of two sub-models. The first, the observational model, estimates variation associated with the measurement process (analytical error) using replicate samples. The second, the ecological model, estimates population means and covariance between δ15NPhe and Δδ15NGlu-Phe given uncertainty due to analytical error. The observational model is \(y[i,j,t,s, 1:r] \sim {\text{mvnorm}}\left( {\mu_{{\left( {i,t,s} \right)}} ,\sigma } \right)\) and the ecological model is \(\mu_{{\left( {i,t,s} \right)}} \sim {\text{mvnorm}}\left( {M_{{\left( {t,s} \right)}} ,\varSigma_{(t,s)} } \right)\) where \(y[i,j,t,s,1:r]\) is the data vector indexed by individual (i), replicate (j), time bin (t), and species (s), and consisting of isotope values 1:r, where r is the number of amino acid variables used, \(\mu_{{\left( {i,t,s} \right)}}\) is the mean vector of individual i, in time bin t for species s. \(\sigma\) is the covariance matrix (dimensions r by r) associated with the total analytical error shared across all individuals, time bins, and species. \(M_{\left( t \right)}\) is the population mean vector for time t and species s and \({{\varSigma }}_{(t,s)}\), the covariance matrix associated with naturally occurring isotopic variation, was assumed constant within (but not between) time bin t and species s.
The model parameters were estimated in a Bayesian framework using the program JAGS (Plummer 2003) interfaced to R (R Development Core Team 2013). Minimally informative priors were used for estimated parameters (Rossman et al. 2016). The model was fit in JAGS using a Markov Chain Monte Carlo for 100,000 iterations with a 10,000 iteration burn in and three chains. The posterior distributions were thinned at a rate of saving one iteration in every three. Convergence was ensured through monitoring traceplots and Rhat values (Gelman and Hill 2007). The probability that two parameters were different was calculated by summing the number of posterior estimates in which one parameter was larger than the other dividing by the total number of posterior estimates. Specifically, we report probabilities that δ15NPhe and Δδ15NGlu-Phe differ between species and colonies and that they declined over time. In this method, using true probabilities, both high and low values indicate what is traditionally thought of as “significance”. Given our sample sizes, we consider probabilities higher than 90% and lower than 10% to be ecologically significant.
We also used posterior estimates to visually represent the data and associated uncertainty. Standard ellipses characterize the foraging ecology of each population. Estimated population δ15NPhe and Δδ15NGlu-Phe means define the center of each standard ellipse and a covariance matrix determines the shape and orientation (Jackson et al. 2011; Rossman et al. 2015). We generated probability density distributions for Newell’s shearwater and Laysan albatross δ15NPhe and Δδ15NGlu-Phe values, similar to those for the Hawaiian petrel in Ostrom et al. (2017).
To further characterize the significance of temporal shifts in Δδ15NGlu-Phe for Newell’s shearwater and Laysan albatross, we calculated effect sizes from raw data. For comparison, we did the same for Hawaiian petrel (Ostrom et al. 2017) and the five species identified by Gagne et al. (2018) as having experienced significant trophic declines. The Gagne et al. (2018) Δδ15NGlu-Phe data were grouped into historical and modern time bins based on sample availability. For Bulwer’s petrel, white-tailed shearwater, and sooty tern, we constructed two groups of individuals from prior to and after 1950. Brown booby and white tern samples were divided such that groups were separated by more than 25 years (prior to 1965 and after 1989 for brown booby and prior to 1924 and after 1968 for white tern). Effect sizes were calculated according to the equation
as described by Coe (2002).
Results
Model selection
The models with and without the effect for MEI were similar; the DIC values for NESH were 201.1 with MEI vs. 201.3 without MEI, and for LAAL were 314.3 for both models. In addition, both slopes for the effect of MEI overlapped zero. Based on these two outcomes, we selected the most parsimonious models without the effect MEI.
Inter-laboratory comparison
Between laboratories, the average difference in the absolute values of δ15NGlu and δ15NPhe from Laysan albatross bone collagen was 0.5 ‰ and 0.3 ‰, respectively. This similarity ensures the comparability of previously published Hawaiian petrel data and our Newell’s shearwater and Laysan albatross datasets.
Interspecific and inter-colony variation in amino acid δ15N
Our results indicate that all three species, including Hawaiian petrels breeding on two separate islands, likely have significantly different δ15NGlu-Phe means in modern as well as historical time periods (P ≥ 0.99; Table 1a; Fig. 2), although the historical time period of each species does not reflect the same chronological time. Relative to Newell’s shearwaters, Maui Hawaiian petrels likely have a lower δ15NPhe mean in the historical time period (P = 0.95) but not in the modern time period (P = 0.70). In modern and historical time periods, both Hawaiian petrels from Maui and Newell’s shearwaters likely have a lower δ15NPhe mean than Hawaiian petrels from the island of Hawai`i and Laysan albatross (P ≥ 0.95). There is also high probability that Hawaiian petrels from the island of Hawai`i have a lower δ15NPhe mean than Laysan albatross in the historical time period (P = 0.995).
Isotope standard ellipses representing variation in δ15NPhe and Δδ15NGlu-Phe (‰) values for (a) historical and (b) post-2000 Newell’s shearwater (NESH, purple), Laysan albatross (LAAL, blue) and Hawaiian petrels (HAPE) breeding on Maui (red) and Hawaii (green). Model estimated mean δ15NPhe and Δδ15NGlu-Phe values for each population define the center of the ellipse and a covariance matrix determines the shape and orientation
Temporal variation in δ15NPhe and δ15NGlu-Phe
The probability that δ15NPhe declined in Newell’s shearwaters over the past 50 years was low (P = 0.27; Table 1b; Fig. 3). There was a 0.79 probability that δ15NPhe declined in Laysan albatross over the past 100 years. We did not consider this difference to be ecologically or statistically significant. Our results indicated a high probability that δ15NGlu-Phe declined in Newell’s shearwater (P = 0.97) and Laysan albatross (P = 0.98) over the past 50 and 100 years, respectively. In Newell’s shearwaters, the majority of this decline occurred between the 1960’s and 1980’s–1990’s (P = 0.91) rather than between the 1980’s–1990’s and post-2000 time periods (0.85).
Effect sizes calculated from raw Δδ15NGlu-Phe data for the comparison of historical and modern time periods are 1.1 for Laysan albatross, 1.9 for Newell’s shearwater, 2.4 for Hawaii colony Hawaiian petrel, and 4.3 for Maui colony Hawaiian petrel. These effect sizes reflect a 71 to greater than 99.9% probability that an individual could be correctly assigned to its time period by its Δδ15NGlu-Phe value (Coe 2002). Effect sizes calculated for the study species of Gagne et al. (2018) are as follows: 0.17 for sooty tern (Onychoprion fuscatus), 0.34 for Bulwer’s petrel (Bulweria bulwerii), 0.40 for white tern (Gygis alba), 0.57 for brown booby (Sula leucogaster), and 0.95 for wedge-tailed shearwater (Ardenna pacifica). These effect sizes reflect a 53–68% probability that an individual could be successfully grouped by its Δδ15NGlu-Phe value (Coe 2002).
Discussion
Our isotope results identify trophic segregation among three ecologically distinct pelagic seabirds that breed in the Hawaiian Islands: Laysan albatross, Newell’s shearwater and Hawaiian petrel. It is striking that the relative position of the three seabirds in isotope space is similar between the modern time period and the historical time periods (Fig. 2). We also infer a decline in Δδ15NGlu-Phe for each of the species, a shift that indicates a trophic decline (Fig. 3). In our subsequent discussion, we examine the ecological implications of our modern data for each species prior to exploring factors that could account for the observed trophic decline.
Modern populations
The inter-laboratory comparison justified including the Hawaiian petrel data (Ostrom et al. 2017) in our analyses. The two Hawaiian petrel populations occupy unique isotopic niches despite the close proximity of their breeding colonies and their high degree of morphological similarity (Fig. 2a; Wiley et al. 2012, 2013; Ostrom et al. 2017). The low δ15NPhe of the Maui Hawaiian petrel colony indicates these birds have a greater tendency to utilize waters characterized by nitrogen fixation and positive N* than their Hawai`i Island counterparts (Fig. 2a; Ostrom et al. 2017). The possibility that Hawaiian petrel δ15NPhe is strongly influenced by differences in nutrient regime is supported by observational and tagging data, which show that Hawaiian petrels forage over areas where the influence of nitrogen fixation varies (as given by N* in Fig. 1; Spear et al. 1995; Adams and Flora 2010; VanZandt 2012). Moreover, Hawaiian petrels distributed southeast of the islands overlap an area of 15N-enriched water (Altabet and Francois 1994). Differential foraging, likely a change in foraging location, within a δ15N gradient could drive inter-colony δ15NPhe segregation in Hawaiian petrels.
Hawaiian petrels from Maui appear to occupy a higher trophic position than their Hawai`i Island conspecifics (Fig. 2; Ostrom et al. 2017). While this may indicate that food chains supplied by nitrogen fixation are longer than those supplied by nitrate, other factors may influence this intra-species trophic disparity. The slight morphological differences between individuals from the islands of Maui and Hawai`i (e.g., culmen and tarsus length, 8% larger; Judge et al. 2014) are unlikely to explain the observed trophic separation. Ballance et al. (2001) showed that even a 1000-fold range in seabird body mass does not explain segregation in the size of dietary items. Thus, it is likely that Hawaiian petrels from Maui and Hawai`i Islands specialize on distinct suites of prey items, which contribute notably to differences in their isotope data.
Our interspecies comparison shows that the Newell’s shearwater standard ellipse does not overlap with those of the Hawaiian petrel colonies (Fig. 2a). The mean δ15NPhe of Newell’s shearwaters is similar to that of Maui Hawaiian petrels, likely resulting from a similar reliance on oceanographic regions characterized by nitrogen fixation. Observational data show that Newell’s shearwaters are dispersed across the isotopic gradient extending east from the area of 15N-enriched waters southeast of the Hawaiian Islands (Spear et al. 1995). This could contribute to the population’s wide δ15NPhe range of 3.7‰. The Δδ15NGlu-Phe data suggest that Newell’s shearwaters occupy a lower trophic position than either of the two Hawaiian petrel populations. The diet of Newell’s shearwater is poorly known. Ainley et al. (2014) reported on Newell’s shearwater diet using stomach contents from carcasses of recently fledged individuals. The stomach contents were dominated by squid (e.g., ommastrephids) with few fish, mainly flying fish. This appears to be similar to Hawaiian petrel diet, but with limited data for both species it is unclear how much the relative proportion of diet items differs between the two (Simons 1985). The Δδ15NGlu-Phe differences might also be driven by interactions in mixed-species feeding flocks. Although foraging in large diverse aggregations can improve foraging success, evidence also suggests that intense interference competition can occur, especially among seabirds employing distinct foraging methods (Schreiber and Burger 2001). For example, surface seizers, like Hawaiian petrels, might reduce access to prey by diving birds, like Newell’s shearwaters, resulting in Newell’s shearwaters eating smaller or lower trophic level prey (Schreiber and Burger 2001).
Laysan albatross occupy a distinct niche position. Their δ15NPhe mean indicates little reliance on food webs supplied by nitrogen fixation (Fig. 2). Additionally, Laysan albatross appears to occupy the lowest trophic position of the three species. Consistent with this, recent amino acid-specific isotope data from contour feathers and remiges place Laysan albatross at the same or a lower trophic position than smaller seabirds (e.g., Bulwer’s petrel, Gagne et al. 2018). Yet, albatrosses can be considered top predators. The low trophic position of Laysan albatross forces us to think more broadly about the trophic dynamics of our species. It prompted us to ask if dietary studies, metabolic effects or diet quality might provide insight in our understanding of Laysan albatross Δδ15NGlu-Phe, and how it relates to that of our other two study species.
Information available on the diet of Laysan albatross does not offer a clear explanation of their low trophic position. As is true of our other study species, squid and fish are important components of the stomach remains of Laysan albatross, including glass and neon flying squids, sauries, and sardines (Harrison et al. 1983; Gould et al. 1997; Pitman et al. 2004; Bisson 2008). Offal can also be common in Laysan albatross diets along with smaller portions of non-cephalopod invertebrates (Gould et al. 1997; Bisson 2008). However, stomach content data are particularly difficult to obtain, especially when seabirds are not at a colony. At sea data from birds obtained from fisheries can be heavily influenced by offal discarded by the fishery (e.g., Gould et al. 1997). Stomach content data can also be influenced by more rapid digestion of soft bodied prey, they are often conducted on a specific age class of birds and have a tendency to contain recently ingested prey (often in boluses), with the hard remains from prior meals perhaps present in pellets (e.g., Harrison et al. 1983, Pitman et al. 2004). Such biases complicate the use of stomach content analyses to interpret isotope data (e.g., Gagne et al. 2018). The possibility that Laysan albatross feed at a lower trophic level than expected from stomach content data has been proposed previously (Gould et al. 1997). Supporting this, a recent Laysan albatross lipid profile shows an important contribution of low trophic level prey items (crustaceans and fish eggs) to the diet that might have otherwise been missed (Conners et al. 2018).
Metabolic effects (type of nitrogen excretion, amino acid transformations) and diet quality (e.g., protein content) have the potential to influence isotope values of amino acids (McMahon and McCarthy 2016). Because our study species are wide-ranging predators, have some overlap in prey taxa consumed, and belong to the same taxonomic order, we do not anticipate large differences in amino acid transformation pathways and associated isotopic discrimination. Further, because their diet appears to be highly proteinaceous, diet quality likely does not account for the Δδ15NGlu-Phe differences we observe. Thus, our current data appear to bolster other findings that point to the possibility that Laysan albatross consumes a significant proportion of lower trophic level prey and this lowers their Δδ15NGlu-Phe values.
Temporal data
Our data show that the foraging segregation observed among our study species in modern time (post-2000) is not a recent phenomenon but has endured over the course of several decades. This is illustrated by the observation that the relative position of each population’s isotopic niche is similar between historical and modern time periods (Fig. 2). We observe this despite differences among our species in the dates of the historical time periods. This observation implies long-term stability in the pattern of differential foraging ecology exhibited by the wide-ranging predators (Fig. 2).
While δ15NPhe provides no evidence of a shift in source nitrogen at the base of our study species’ food webs, δ15NGlu-Phe indicates that, like the Hawaiian petrel (Wiley et al. 2013; Ostrom et al. 2017), Newell’s shearwater and Laysan albatross experienced a significant decline in trophic position within the past century. The trophic decline we observed in Laysan albatross is in contrast to the findings of Gagne et al. (2018), who did not observe a significant temporal trend in this species. This disparity could be related to the fact that Gagne et al. (2018) analyzed feathers that, unlike bone collagen, integrate over only a short portion of the individual’s annual cycle (Edwards and Rohwer 2005).
Our isotope chronologies augment the current picture of a trophic decline among predatory seabirds in the North Pacific Ocean. There are now eight Hawaiian seabirds that show a trophic decline including our three study species plus Bulwer’s petrel, wedge-tailed shearwater, brown booby, white tern, and sooty tern (Ostrom et al. 2017; Gagne et al. 2018). Our study species represent two avian families and diverse foraging strategies and grant extensive spatial coverage of the world’s largest ocean. They also tend to differ in the oceanographic parameters that they favor (e.g., sea surface temperature and salinity; Schreiber 1984; Spear et al. 1995; Block et al. 2011) and their population dynamics. The observed trophic declines coincide with both severe population declines (Hawaiian petrel and Newell’s shearwater) and population stability (Laysan albatross; Arata et al. 2009; Raine et al. 2017). The study species of Gagne et al. (2018) similarly exhibit numerous ecological differences. For example, brown boobies often make shallow dives (up to 2 m) and pursue prey underwater whereas sooty and white terns feed at the surface without submerging (Niethammer and Patrick 1998; Schreiber and Norton 2002; Schreiber et al. 2002). The white tern has a small population of about 15,000 breeding pairs in the Hawaiian Archipelago whereas the wedge- tailed shearwater’s population in Hawaii is at least 270,000 breeding pairs (Whittow 1997; Niethammer and Patrick 1998). Thus, our study expands the perception that a trophic decline was ecologically widespread among North Pacific seabird species, a primary question of our study, and builds evidence for a pervasive shift in food web architecture across the North Pacific Ocean.
Understanding how the trophic decline exhibited by pelagic seabirds influences the flow of materials and energy in food webs could be greatly advanced by amino acid-specific isotope studies. This will require accurate estimates of trophic position for a wide variety of taxonomic groups and trophic levels, and better understanding of the origin of nutrients at the base of the food web. The determination of trophic position via the amino acid-specific nitrogen isotope approach requires an estimate of trophic discrimination factor, which does not exist for most organisms, including many pelagic seabirds. Thus, defining trophic discrimination factors and understanding the variability among them is one challenge for future studies. Data sets on essential amino acids that must be obtained from diet, rather than synthesized, will assist in identifying sources of nutrients at the food web bases. These data can be complimented by the carbon isotope composition of amino acids which reflects that of the primary production at the food web base (Larsen et al. 2013).
Currently, climate change and fisheries are considered important drivers of oceanic ecosystem change and trophic alteration (Ward and Myers 2005; Jackson 2008; Cheung et al. 2012; Woodworth-Jefcoats et al. 2013; Ostrom et al. 2017; Gagne et al. 2018). While we did not observe an influence of MEI on our data, this is in contrast to Gagne et al. (2018) who found that climate indicators ranked 6th or lower after foraging distance and dietary indicators in terms of their influence on trophic position. Thus, it would not be appropriate to exclude climate from consideration as a factor that could influence trophic decline. As previously discussed, fishing is another major anthropogenic force capable of imposing a trophic decline on pelagic seabirds (Ostrom et al. 2017; Gagne et al. 2018). With industrial fisheries, humans have the power to modify vast oceanic ecosystems to a degree once thought impossible (Jackson 2008). While characterizing ecosystem interactions for the vast pelagic realm is arduous, it is even more difficult to predict the long-term consequences of large-scale fisheries. Yet, there is mounting evidence that fisheries compete with marine predators for food and alter the distributions, abundances, and average sizes of marine organisms (Myers and Worm 2003; Ward and Myers 2005; Polovina and Woodworth-Jefcoats 2013). Both target and non-target species are subject to take by fisheries, and some fisheries (e.g., mid-water trawls, purse seines) are non-discriminant when it comes to forage fish (Worm and Lenihan 2013). Pacific Ocean commercial fisheries have been estimated to discard millions of tonnes annually (Kelleher 2005). The continued use of non-discriminant fishing methods and the vast volume of non-target species taken emphasizes the far-reaching nature of fisheries beyond target catch. Interactions and consequences of salient drivers of ecosystem change must be understood so we can prevent alterations to the pelagic realm that will further compromise trophic structure and the welfare of marine life.
References
Adams J, Flora S (2010) Correlating seabird movements with ocean winds: linking satellite telemetry with ocean scatterometry. Mar Biol 157:915–929
Ainley DG, Telfer TC, Reynolds MH (1997) Newell’s Shearwater (Puffinus newelli), version 2.0. In: Rodewald PG (ed) The Birds of North America. Cornell Lab of Ornithology, Ithaca, New York
Ainley DG, Walker WA, Spencer GC, Holmes ND (2014) The prey of Newell’s shearwater Puffinus newelli in Hawaiian waters. Mar Ornithol 44:69–72
Altabet M, Francois R (1994) Sedimentary nitrogen isotopic ratio as a recorder for surface ocean nitrate utilization. Glob Biogeochem Cycles 8:103–116
Arata JA, Sievert PR, Naughton MB (2009) Status assessment of Laysan and black-footed albatrosses, North Pacific Ocean, 1923–2005. US Geological Survey, Reston, Virginia, pp 2009–5131
Awkerman JA, Anderson DJ, Whittow GC (2009) Laysan Albatross (Phoebastria immutabilis), version 2.0. In: Rodewald PG (ed) The birds of North America. Cornell Lab of Ornithology, Ithaca, New York
Ballance LT, Ainley DG, Hunt GL Jr (2001) Seabird foraging ecology. In: Steele JH, Thorpe SA, Turekian KK (eds) Encyclopedia of ocean sciences, vol 5. Academic Press, London, pp 2636–2644
Bearhop S, Phillips RA, McGill R, Cherel Y, Dawson DA, Croxall JP (2006) Stable isotopes indicate sex-specific and long-term individual foraging specialization in diving seabirds. Mar Ecol Prog Ser 311:157–164
Bisson JR (2008) Diet dynamics and trophic relations of Laysan and black-footed albatrosses associated with pelagic longline fishing. MS thesis, University of Hawaii at Manoa, Hawaii, USA
Block BA, Jonsen ID, Jorgensen SJ, Winship AJ, Shaffer SA, Bograd SJ, Hazen EL, Foley DG, Breed GA, Harrison AL, Ganong JE, Swithenbank A, Castleton M, Dewar H, Mate BR, Shillinger GL, Schaefer KM, Benson SR, Weise MJ, Henry RW, Costa DP (2011) Tracking apex marine predator movements in a dynamic ocean. Nature 475:86–90
Broughton JM (1994) Size of the bursa of Fabricius in relation to gonad size and age in Laysan and Black-footed Albatrosses. Condor 96:203–207
Casciotti KL, Trull TW, Glover DM, Davies D (2008) Constraints on nitrogen cycling at the subtropical North Pacific Station ALOHA from isotopic measurements of nitrate and particulate nitrogen. Deep Res Part II 55:1661–1672
Cheung WW, Meeuwig JJ, Feng M, Harvey E, Lam VW, Langlois T, Slawinski D, Sun C, Pauly D (2012) Climate-change induced tropicalisation of marine communities in Western Australia. Mar Freshw Res 63:415–427
Chikaraishi Y, Kashiyama Y, Ogawa NO, Kitazato H, Ohkouchi N (2007) Metabolic control of nitrogen isotope composition of amino acids in macroalgae and gastropods: implications for aquatic food web studies. Mar Ecol Prog Ser 342:85–90
Chikaraishi Y, Ogawa NO, Kashiyama Y, Takano Y, Suga H, Tomitani A, Miyashita H, Kitazato H, Ohkouchi N (2009) Determination of aquatic food- web structure based on compound-specific nitrogen isotopic composition of amino acids. Limnol Oceanogr 7:740–750
Choy CA (2013) Pelagic food web connectivity in the north Pacific subtropical gyre: a combined perspective from multiple biochemical tracers and diet. PhD dissertation, University of Hawaii at Manoa, Hawaii, USA
Coe R (2002) It’s the effect size, stupid: What effect size is and why it is important. A paper presented at the Annual Conference of the British Educational Research Association, University of Exeter, Exeter, Devon, England
Conners MG, Goetsch C, Budge SM, Walker WA, Mitani Y, Costa DP, Shaffer SA (2018) Fisheries exploitation by albatross quantified with lipid analysis. Front Mar Sci. 5:113
Cooper BA, Day RH (1998) Summer behavior and mortality of dark-rumped Petrels and Newell’s Shearwaters at power lines on Kaua`i. Colon Waterbirds 21:11–19
Cousins KL, Dalzell P, Gilman E (2000) Managing pelagic longline-albatross interactions in the North Pacific Ocean. Marine Ornithol 28:159–174
Deutsch C, Gruber N, Key RM, Sarmiento JL, Ganachaud A (2001) Denitrification and N2 fixation in the pacific ocean. Glob Biogeochem Cycles 15:483–506
Development Core Team R (2013) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Edwards AE, Rohwer S (2005) Large-scale patterns of molt activation in the flight feathers of two albatross species. Condor 107:835–848
Ellis G (2012) Compound-specific stable isotopic analysis of protein amino acids: ecological implications in modern and ancient systems. PhD dissertation, University of South Florida, Florida, USA
Fernández P, Anderson DJ, Sievert PR, Huyvaert KP (2001) Foraging destinations of three low-latitude albatross (Phoebastria) species. J Zool 254:391–404
Gaebler OH, Choitz HC, Vitti TG, Vukmirovich R (1963) Significance of N15 excess in nitrogenous compounds of biological origin. Can J Biochem Physiol 41:1089–1097
Gagne TO, Hyrenbach KD, Hagemann ME, Van Houtan KS (2018) Trophic signatures of seabirds suggest shifts in oceanic ecosystems. Sci Adv 4:eaao3946
Gelman A, Hill J (2007) Data analysis using regression and multilevel/hierarchical models. Cambridge University Press, New York
Gelman A, Carlin JB, Stern HS, Rubin DB (2004) Bayesian data analysis: second edition. Texts in statistical science. CRC Press, Boca Raton. ISBN 1-58488-388-X
Gould P, Ostrom P, Walker W (1997) Trophic relationships of albatrosses associated with squid and large-mesh drift-net fisheries in the North Pacific ocean. Can J Zool 75:549–562
Graham BS, Koch PL, Newsome SD, McMahon KW, Aurioles D (2010) Using isoscapes to trace the movements and foraging behavior of top predators in oceanic ecosystems. In: West JB, Bowen GJ, Dawson TE, Tu KP (eds) Isoscapes: understanding movement, pattern, and process on earth through isotope mapping. Springer Science, Dordrecht, pp 299–318
Gruber N, Sarmiento JL (1997) Global patterns of marine nitrogen fixation and denitrification. Glob Biogeochem Cycles 11:235–266
Harrison CS, Hida TS, Seki MP (1983) Hawaiian seabird feeding ecology. Wildl Monogr 85:1–71
Hinke JT, Polito MJ, Goebel ME, Jarvis S, Reiss CS, Thorrold SR, Trivelpiece WZ, Watters GM (2015) Spatial and isotopic niche partitioning during winter in chinstrap and Adélie penguins from the South Shetland Islands. Ecosphere 6:125
Hobson KA, Clark RG (1992) Assessing avian diets using stable isotopes I: turnover of 13C in tissues. Condor 94:181–188
Jackson JB (2008) Ecological extinction and evolution in the brave new ocean. Proc Natl Acad Sci 105:11458–11465
Jackson AL, Inger R, Parnell AC, Bearhop S (2011) Comparing isotopic niche widths among and within communities: SIBER—stable isotope bayesian ellipses in R. J Anim Ecol 80:595–602
Judge SW, Hu D, Bailey CN (2014) Comparative analyses of Hawaiian Petrel Pterodroma sandwichensis morphometrics. Marine Ornithol 42:81–84
Karl D, Letelier R, Tupas L, Dore J, Christian J, Hebel D (1997) The role of nitrogen fixation in biogeochemical cycling in the subtropical North Pacific Ocean. Nature 388:533–538
Kays R, Crofoot MC, Jetz W, Wikelski M (2015) Terrestrial animal tracking as an eye on life and planet. Science 348:aaa2478
Kelleher K (2005) Discards in the world’s marine fisheries: an update. FAO Fisheries Technical Paper No. 470
Kim IN, Lee K, Gruber N, Karl DM, Bullister JL, Yang S, Kim TW (2014) Increasing anthropogenic nitrogen in the North Pacific Ocean. Science 346:1102–1106
Kirch PV (1990) The evolution of sociopolitical complexity in prehistoric Hawaii: an assessment of the archaeological evidence. J World Prehist 4:311–345
Larsen T, Ventura M, Anderson N, O’Brien DM, Piatkowski U, McCarthy M (2013) Tracing carbon sources through aquatic and terrestrial food webs using amino acid stable isotope fingerprinting. PLoS One 8:e73441
Lorrain A, Graham B, Ménard F, Popp B, Bouillon S, Van Breugel P, Cherel Y (2009) Nitrogen and carbon isotope values of individual amino acids: a tool to study foraging ecology of penguins in the Southern Ocean. Mar Ecol Prog Ser 391:293–306
McClelland JW, Montoya JP (2002) Trophic relationships and the nitrogen isotopic composition of amino acids in plankton. Ecology 83:2173–2180
McMahon KW, McCarthy MD (2016) Embracing variability in amino acid δ15N fractionation: mechanisms, implications, and applications for trophic ecology. Ecosphere 7:e01511
Mitchell C, Ogura C, Meadows DW, Kane A, Strommer L, Fretz S, Leonard D, McClung A (2005) Hawaii’s comprehensive wildlife conservation strategy. Department of Land and Natural Resources, Honolulu
Myers RA, Worm B (2003) Rapid worldwide depletion of predatory fish communities. Nature 423:280–283
Myers RA, Baum JK, Shepherd TD, Powers SP, Peterson CH (2007) Cascading effects of the loss of apex predatory sharks from a coastal ocean. Science 315:1846–1850
Naughton MB, Romano MD, Zimmerman TS (2007) A conservation action plan for Black-footed Albatross (Phoebastria nigripes) and Laysan Albatross (P. immutabilis). Ver. 1.0
Niethammer KR, Patrick LB (1998) White tern (Gygis alba), version 2.0. In: Poole AF, Gill FB (eds) The birds of North America. Cornell Lab of Ornithology, Ithaca, New York
Ohkouchi N, Chikaraishi Y, Close HG, Fry B, Larsen T, Madigan DJ, McCarthy MD, McMahon KW, Nagata T, Naito YI, Ogawa NO, Popp BN, Steffan S, Takano Y, Tayasu I, Wyatt ASJ, Yamaguchi YT, Yokoyama Y (2017) Advances in the application of amino acid nitrogen isotopic analysis in ecological and biogeochemical studies. Org Geochem 113:150–174
Ostrom PH, Wiley AE, James HF, Rossman S, Walker WA, Zipkin EF, Chikaraishi Y (2017) Broad-scale trophic shift in the pelagic North Pacific revealed by an oceanic seabird. Proc R Soc B 284:20162436–20162442
Paleczny M, Hammill E, Karpouzi V, Pauly D (2015) Population trend of the world’s monitored seabirds, 1950–2010. PLoS One 10:e0129342
Pitman RL, Walker WA, Everett WT, Gallo-Reynoso JP (2004) Population status, foods and foraging of Laysan albatrosses Phoebastria immutabilis nesting on Guadalupe Island, Mexico. Marine Ornithol 32:159–165
Plummer M (2003) JAGS: a program for analysis of Bayesian graphical models using Gibbs Sampling. In: Hornik K, Leisch F, Zeileis A (eds) Proceedings of the third international workshop on distributed statistical computing. R Project for Statistical Computing, Technische Universität Wien, Vienna
Polovina JJ, Woodworth-Jefcoats PA (2013) Fishery-induced changes in the subtropical Pacific pelagic ecosystem size structure: observations and theory. PLoS One 8:e62341
Pyle P (2008) Molt and age determination in Procellariiformes. Identification guide to North American birds, part 2. Slate Creek Press, Point Reyes Station, pp 248–260
Raine AF, Holmes ND, Travers M, Cooper BA, Day RH (2017) Declining population trends of Hawaiian petrel and Newell’s Shearwater on the island of Kaua`i, Hawaii, USA. Condor 119:405–415
Rossman S, Ostrom PH, Stolen M, Barros NB, Gandhi H, Stricker CA, Wells RS (2015) Individual specialization in the foraging habits of female bottlenose dolphins living in a trophically diverse and habitat rich estuary. Oecologia 178:415–425
Rossman S, Ostrom PH, Gordon F, Zipkin EF (2016) Beyond carbon and nitrogen: guidelines for estimating three-dimensional isotopic niche space. Ecol and Evol 6:2405–2413
Rucklidge GJ, Milne G, McGaw BA, Milne E, Robins SP (1992) Turnover rates of different collagen types measured by isotope ratio mass spectrometry. Biochim Biophys Acta 1156:57–61
Schreiber RW (1984) Tropical seabird biology. Cooper Ornithological Society, Kansas
Schreiber EA, Burger J (2001) Biology of marine birds. CRC Press, Florida
Schreiber EA, Norton RL (2002) Brown Booby (Sula leucogaster), version 2.0. In: Poole AF, Gill FB (eds) The birds of North America. Cornell Lab of Ornithology, Ithaca, New York
Schreiber EA, Feare CJ, Harrington BA, Murray BA Jr., Robertson WB Jr., Robertson MJ, Woolfenden GE (2002) Sooty tern (Onychoprion fuscatus), version 2.0. In: Poole AF, Gill FB (eds) The birds of North America. Cornell Lab of Ornithology, Ithaca, New York
Sherwood OA, Guilderson TP, Batista FC, Schiff JT, McCarthy MD (2014) Increasing subtropical North Pacific Ocean nitrogen fixation since the Little Ice Age. Nature 505:78–81
Sigman DM, Mccorkle DC, Francois R, Fischer G (2000) The δ15N of nitrate in the Southern Ocean: nitrogen cycling and circulation in the ocean interior. J Geophys Res 105:19599–19614
Simons TR (1985) Biology and behavior of the endangered Hawaiian dark-rumped petrel. Condor 87:229–245
Simons TR, Hodges CN (1998) Hawaiian Petrel (Pterodroma sandwichensis), version 2.0. In: Rodewald PG (ed) The birds of North America. Cornell Lab of Ornithology, Ithaca, New York
Spear LB, Ainley DG, Nur N, Howell SNG (1995) Population size and factors affecting at-sea distributions of four endangered Procellariids in the tropical pacific. Condor 97:613–638
Stafford TW, Brendel K, Duhamel RC (1988) Radiocarbon, 13C and 15N analysis of fossil bone: removal of humates with XAD-2 resin. Geochim Chosmochim Acta 52:2257–2267
VanZandt ML (2012) Distribution and habitat selection of the endangered Hawaiian Petrel (Pterodroma sandwichensis), from the island of Lāna’i. PhD dissertation, University of Hawaii at Hilo, Hawaii, USA
Ward P, Myers RA (2005) Shifts in open-ocean fish communities coinciding with the commencement of commercial fishing. Ecology 86:835–847
Warham J (1996) The behavior, population biology and physiology of the petrels. Academic Press, University of Canterbury, Christchurch
Whittow GC (1997) Wedge-tailed Shearwater (Ardenna pacifica), version 2.0. In: Poole AF, Gill FB (eds) The birds of North America Editors. Cornell Lab of Ornithology, Ithaca, New York
Wiley AE, Welch AJ, Ostrom PH, James HF, Stricker CA, Fleischer RC, Gandhi H, Adams J, Ainley DG, Duvall F, Holmes N (2012) Foraging segregation and genetic divergence between geographically proximate colonies of a highly mobile seabird. Oecologia 168:119–130
Wiley AE, Ostrom PH, Welch AJ, Fleischer RC, Gandhi H, Southon JR, Stafford TR Jr, Penniman JR, Hu D, Duvall FP, James HF (2013) Millennial-scale isotope records from a wide-ranging predator show evidence of recent human impact to oceanic food webs. Proc Natl Acad Sci 110:8972–8977
Woodworth-Jefcoats PA, Polovina JJ, Dunne JP, Blanchard JL (2013) Ecosystem size structure response to 21st century climate projection: large fish abundance decreases in the central North Pacific and increases in the California Current. Glob Chang Biol 19:724–733
Worm B, Lenihan HS (2013) Threats to marine ecosystems: overfishing and habitat degradation. In: Bertness MR, Silliman BJ, Stachowicz J (eds) Marine Community Ecology and Conservation, Chapter 20. Sinauer Press, Sunderlands, Massachusetts, pp 449–476
Acknowledgements
We thank the Bird Division, National Museum of Natural History, the Bernice Bishop Museum, and the California Academy of Sciences for the loan of specimens and permission to sample them. We are grateful to the National Marine Fisheries Service, Alaska Fisheries Science Center, Pacific Islands Regional Office—Fisheries Observer Program staff and observers, the vessels and crew of the Hawaii longline fisheries that supported the observers, Oikonos and Shannon Fitzgerald, Bill Walker, and Hannah Nevins, in particular, for facilitating sampling of salvaged Laysan albatross. We acknowledge the Hawaii Department of Land and Natural Resources (Division of Forestry and Wildlife) and the Kaua`i Endangered Seabird Recovery Project for facilitating sampling of salvaged Newell’s shearwaters. Funding was generously provided by the National Science Foundation DEB 0,745,604, Michigan State University, and the Wetmore Fund of the Bird Division, Smithsonian Institution.
Author information
Authors and Affiliations
Contributions
KEM, PHO, HFJ, and AEW formulated the idea. KEM, PHO, and SR wrote the manuscript. All authors engaged in intellectual exchange and manuscript editing. KEM, PHO, YC, and HG developed methodology and performed laboratory analyses. SR and KEM performed statistical analyses. AR and JB facilitated access to salvaged birds. KEM, AEW, HFJ, AR, and PHO collected samples.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Seth Newsome.
This study is the first to develop decadal-scale amino acid nitrogen isotope chronologies on bone collagen of threatened and endangered seabirds. We identify a pattern of long-term stability in trophic segregation coupled with recent trophic decline in three species of Hawaiian seabirds.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Morra, K.E., Chikaraishi, Y., Gandhi, H. et al. Trophic declines and decadal-scale foraging segregation in three pelagic seabirds. Oecologia 189, 395–406 (2019). https://doi.org/10.1007/s00442-018-04330-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00442-018-04330-8