Abstract
Sperm carries a reservoir of proteins regulating the molecular functions to attain functional competence. Semen samples collected from buffalo bulls were assessed for sperm functional attributes (n = 11) and proteome profiling (n = 6). Sperm proteins were extracted and profiled by employing LC–MS/MS. Overall, the buffalo sperm contained 1365 proteins, of which 458 were common between the groups. The unique proteins were 477 and 430 in good and poor quality semen, respectively. In the whole proteome of buffalo sperm, sexual reproduction with phosphatidylethanolamine-binding protein1 (PEBP1), fetuin-B (FETUB) and acrosin (ACR) was the most enriched (p = 8.44E−19) biological process, also with thermogenesis (p = 0.003), oocyte meiosis (p = 0.007) and vascular smooth muscle contraction (p = 0.009) apart from metabolic pathways. In good quality semen, mesenchyme migration (p = 1.24E−07) and morphogenesis (p = 0.001) were abundant biological processes. In good quality semen, the fluid shear stress (p = 0.01) and, in poor quality semen, valine, leucine and isoleucine degradation (p = 3.8E−05) pathways were enriched. In good quality semen, 7 proteins were significantly (p < 0.05) upregulated and 33 proteins were significantly (p < 0.05) downregulated. On validating the abundantly expressed sperm proteins, serine protease inhibitor Kazal-type 2-like (SPINK2; 2.17-fold) and neddylin (NEDD8; 1.13-fold) were upregulated and YBX2 was downregulated (0.41-fold) in good quality semen as compared with poor quality semen (1-fold). The present findings revealed the importance of sperm proteins in oocyte maturation, fertilization process and early embryonic development. The variations in the proteomic composition can be used as potential markers for the selection of breeding bulls.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Globally, the reproductive efficiency in humans and dairy animals has been declining for the past 50 years. The reasons for the low reproductive efficiency are multifactorial but male factors and idiopathic causes influence the fertility in approximately 50–60% of the breeding population (Agarwal et al. 2015). Breeding bulls are considered to be superior if the seminal biomolecules aid the sperm to attain functional competence for successful fertilization and embryonic development. Semen samples harbor signature proteome, transcriptome and metabolome for the reproductive success. The bio-molecular compositions of the sperm vary in quantity and quality among the bulls of different fertility status (Somashekar et al. 2017). In this regard, it is imperative to understand the composition and biological functions of all the biomolecules especially proteins in semen samples. Recent developments in new generation omics technologies coupled with bioinformatics have helped researchers in understanding the role of sperm biomolecules in the establishment of fertility.
Sperm are terminally differentiated, transcriptionally and translationally silent but are rich with proteins that determine the success of the sequential events of fertilization in the female reproductive tract. In humans (Jodar et al. 2016) and animals such as bulls (Somashekar et al. 2017), goat (Pinto et al. 2019), sheep (Zhu et al. 2020) and pig (Roca et al. 2020), proteomic profiling has been used for assessing male sperm functional competence and fertility status.
Successful breeding of buffalo is a major concern as there are wide variations in the semen quality among bulls (Verma et al. 2015). The selection of bulls that produce good quality semen is most important as one bull produces thousands of semen doses per year (Bhakat et al. 2011). Although a few buffalo sperm proteomic studies have been carried out in the recent past (Selvaraju et al. 2016a, b; Codognoto et al. 2018; Hou et al. 2019; Aslam et al. 2019; Fu et al. 2019), an in-depth quantitative proteomic profile has not been conducted concerning the semen quality. Only the differentially expressed protein spots between high and low fertile buffalo sperm (MALDI–TOF/MS) analysis revealed that the proteins associated with energy metabolism and capacitation factors are overexpressed in low fertile sperm (Aslam et al. 2019). The whole protein profiling of sperm is needed for elucidating the molecular processes that are important for the establishment of fertilization and pregnancy. Although in the normozoospermic buffalo, sperm protein profiles have been conducted and identified ODF2, AKAP4 and TUBB were the abundant proteins involved in transport, phosphorylation and macromolecule localization (Fu et al. 2019), the proteomic differences between the good and poor quality semen have not been recognize. Identification and quantification of such signature proteins in sperm elucidate not only the proteomic differences between the semen samples varying in quality and also the processes and pathways determining the success of the fertilization. Hence, the present study aims (i) to understand the functional proteomics of sperm proteins in general and (ii) to identify the signature proteins that are determining the sperm functional competence between the good and the poor quality buffalo semen samples.
Materials and methods
Sample collection and grouping
Neat and frozen semen samples were obtained from buffalo bulls (Bubalus bubalis, n = 11), aged between 4 and 6 years (four ejaculates from each bull), maintained at the Central Frozen Semen Production and Training Institute, Hessarghatta, Bengaluru, India. The animals were initially selected based on semen production capacity as determined by semen volume, sperm concentration per milliliter and total sperm production per ejaculate. From the neat semen samples, 0.5 mL was aliquoted for protein profiling. Another aliquot (0.25 mL) was subjected to sperm functional parameters analyses. The remaining semen sample was diluted with Tris-egg yolk extender, frozen using a programmable biological freezer.
In the neat semen samples, plasmalemma integrity (live), acrosomal integrity, mitochondrial membrane potential and chromatin distribution were assessed. In the post-thaw semen samples, sperm kinematics, plasmalemma integrity, acrosome integrity, mitochondrial membrane potential and chromatin distribution were assessed. Based on the sperm functional tests, the semen samples were scored. The samples were grouped according to the percentage of the positive cells obtained in each test. The semen samples with higher than average values for each functional parameter were scored as good quality semen (n = 6) and less than average were scored as poor quality semen (n = 5). The semen samples from three animals having the highest scores from the good quality and three animals having the lowest scores from the poor quality semen producers were selected for the LC–MS/MS study (Fig. 1).
Selection of good and poor semen producer bulls. a The bulls were selected based on sperm production capacity. The sperm production capacity of the selected bulls differs significantly (p < 0.05). b Sperm functional parameter analyses in neat semen and classification of good and poor semen samples. The sperm functional parameters such as live, functional membrane integrity (FMI), acrosomal integrity (AP), mitochondrial membrane potential (MMP) and chromatin distribution (CD) were assessed. The percentages of sperm positive for acrosomal integrity, mitochondrial membrane potential and chromatin distribution were significantly (p < 0.05) higher in good as compared with poor semen samples
Assessment of sperm functional parameters
Sperm kinematic analysis
Sperm kinetic parameters were analyzed using CASA (Sperm Class Analyzer; Microptic, Barcelona, Spain) as described (Selvaraju et al. 2016b). In brief, the cell concentration was maintained at 7 × 106/mL in Tris buffer (Tris, 274.1 mM; glucose, 55.5 mM; citric acid, 87.08 mM; pH, 6.8) for analysis. The diluted sample (8 µL) was placed on a prewarmed glass slide (37 °C) and covered with an 18 × 18 mm coverslip. At least 10 different fields with a minimum of 500 sperm in each field were analyzed with the following set parameters: Speed type - VCL, cell size between 5 and 70 µm2 and sperm with < 10, 10–35, 35–60 and > 60 µm/s were classified as immotile, slow, medium and fast motile, respectively. Total motile - > 10 µm/s, progressive forward motility - > 60% straightness index and circular - < 50% linearity index. All the parameters were analyzed after deleting the non-sperm objects in each field.
Plasmalemma integrity
The percentage of plasma membrane intact cells was examined by using eosin (5%)–nigrosin (10%) stain (Selvaraju et al. 2016a). In brief, the sperm sample and stain were mixed in a 1:1 ratio, allowed to react for 30 s and then smeared onto a glass slide and 200 sperm were assessed from different fields under 40× phase contrast objective (Eclipse 80i, Nikon, Japan). The unstained sperm were considered as a structural membrane intact and stained sperm were considered as membrane-damaged cells.
Functional membrane integrity
The percentage of sperm positive for functional membrane integrity was assessed using the HOS-G test as described earlier (Selvaraju et al. 2008). In brief, 50 µL of the semen samples was added to 450 µL of hypo-osmotic solution (150 mOsm) and 300 mOsm (control). The samples were incubated at 37 °C for 30 min; then, a drop of semen sample was kept on a glass slide and 200 sperm were assessed from different fields under 40× phase contrast objective (Eclipse 80i, Nikon, Japan). A sperm was considered as functional membrane intact if the hairpin bend in the principal piece of the tail was present otherwise considered as functional membrane lost cells.
Acrosome integrity
The semen sample was washed in 1 mL of Tyrode’s albumin lactate pyruvate (SP-TALP) by centrifugation at 1000 rpm for 5 min and the sperm were smeared on the glass slide immediately. The slides were air-dried and fixed with methanol for 10 min, followed by washing thrice (2 min each) in freshly prepared phosphate-buffered saline with 0.6% bovine serum albumin. The staining stock was prepared by adding 8 µL of fluorescein isothiocyanate conjugated with pisum sativum agglutinin (FITC-PSA; 5 µg/mL) and 2 µL of propidium iodide (PI; 75 µg/mL) and volume was made up to 2 mL. The smear was overlaid with 50 µL of freshly prepared staining solution and incubated in the dark for 20 min. Two hundred sperm were counted in a fluorescent microscope (Eclipse 80i, Nikon, Japan) with an excitation filter of 510–560 nm and a barrier filter of 505 nm. The sperm with evenly distributed acrosome were considered as acrosome intact and cells with unevenly distributed acrosome were considered as acrosome lost. The percentage of acrosome intact sperm was calculated from the total cells counted.
Mitochondrial membrane potential
The mitochondrial membrane potential was assessed using JC-1 (5, 5′, 6, 6′-tetrachloro-1, 1′, 3, 3′-tetraethyl benzimidazolyl carbocyanine iodide) stain. One microliter of JC-1 (1.53 mM in dimethyl sulfoxide) was added to 50 µL of Tris buffer (pH 6.8) and mixed well until the stain dissolved completely. Subsequently, 10 µL of the semen sample was added to the stain mixture and incubated for 30 min in the dark at 37 °C. The samples were smeared onto the slide and the midpiece was observed in a fluorescent microscope (Eclipse 80i, Nikon, Japan) fitted with a barrier filter of 505 nm and excitation filter 510–560 nm. The sperm with yellow to orange color at the midpiece was considered as positive for mitochondrial membrane potential and with green color was considered as negative (Selvaraju et al. 2008).
Chromatin distribution
Chromatin distribution was assessed by Feulgen’s staining procedure (Selvaraju et al. 2008). In brief, the semen samples were smeared onto the glass slide and air-dried for 10 min, fixed in 10% neutral buffered formal saline (sodium phosphate monobasic, 33.3 mM; disodium hydrogen phosphate anhydrous, 45 mM; formaldehyde, 37%-100 mL; distilled water, 900 mL) for 30 min and followed by washing in running water. The slides were air-dried and incubated in 5 N HCl for 30 min, washed in running water twice for 5 min each and followed by incubation in Schiff’s reagent for 30 min in the dark. The slides were then washed thrice with freshly prepared sulfite water (5 mL of 10% potassium meta bisulfite and 5 mL of 1 N HCl in 100 mL distilled water) for 2 min each. Two hundred sperm were counted under 1000× in a phase-contrast microscope (Eclipse 80i, Nikon, Japan). Sperm with evenly stained chromatin were considered normal and the unevenly stained ones were considered abnormal.
Protein isolation
The aliquot of neat semen samples designated for protein isolation was immediately added with phenyl methyl sulfonyl fluoride (100 mM) and centrifuged at 7000×g for 10 min at 4 °C to separate the sperm from seminal plasma. The seminal plasma was removed and the sperm pellet was washed twice with TC buffer (Tris; 3.99 mM, CaCl2; 2.61 mM, sodium azide; 1.53 mM, pH 7.2) and resuspended in protein extraction buffer (TC buffer with 0.1 M phenyl methyl sulfonyl fluoride and 0.1% Triton X-100). The sperm-TC buffer with Triton X-100 was incubated at 4 °C for 1 h, with intermittent vortexing for a brief period at 5 min intervals. In the end, the samples were centrifuged at 1764×g for 30 min at 4 °C (D’amours et al. 2012; Somashekar et al. 2017); the supernatant containing protein was recovered and stored at − 80 °C for further LC/MS–MS work.
Sample preparation by tryptic in-solution digestion and mass spectrometry analysis
The procedure employed for LC–MS/MS analysis of trypsin digested peptide fragments of ethanol precipitated protein sample was carried out as per the established procedure (Shevchenko et al. 2007). In brief, 20 µg of the lyophilized sample was reconstituted in 100 mM ammonium bicarbonate buffer with 10 mM dithiothreitol followed by alkylation with 50 mM iodoacetamide (in the dark). The protein was then subjected to in-solution digestion with sequencing grade trypsin (50 ng/µL in 25 mM ammonium bicarbonate containing 10% acetonitrile). These samples were incubated at 37 °C for 16 h. The digested peptides were recovered by vacuum drying and reconstituted in 20 µL of 0.1% (v/v) formic acid in 2% acetonitrile. One microliter of this sample was subjected to nano-UHPLC–MS/MS analysis. The ion source was electron spray ionization (ESI, nano-spray), fragmentation mode was collision-induced dissociation (CID, y and b ions), MS scan mode was FT-ICR/Orbitrap and the MS/MS scan in the range from 500 to 200 m/z using linear ion trap. For collision-induced dissociation (CID) MS/MS analysis, the doubly or triply charged ions were selected. Twenty femtomoles of standard BSA digest were also analyzed in parallel to the sequence to check the performance of the instrument.
Protein identification
Peptide mass fingerprinting was performed on a proteome discoverer (Thermo Fusion V 1.4). For fixed and variable modifications, carbamidomethylation of cysteine residues and oxidation of methionine residues were selected, respectively. The percent mass error tolerance and fragment mass error tolerance were set to 12 ppm and 0.8 Da, respectively. The data were searched against the UniProt Bubalus bubalis database (a non-redundant database with reviewed proteins) and Bos taurus from NCBI. Proteins showing 1 or more than 1 peptide were considered for analysis. The false discovery rate (FDR) was kept very stringent (0.8%), which was verified manually. The corresponding search against the database was obtained for all the three animals from each group with a list of proteins along with its peak area-based quantification values, score, coverage, number of unique peptides, number of peptides, number of peptide sequence matches, molecular weight and the calculated pI.
Label-free protein quantification
Raw LC–MS/MS data for all the replicates were imported into Progenesis QI for Quantitative Proteomics (Nonlinear Dynamics Limited, UK). The imported runs were quality-controlled by visual inspection of the 2D representations and automatically aligned to the most suitable reference run identified by the software. Considering the good initial alignment quality, the data set was not subjected to any further manual correction such as vector editing. Relative quantitation of all peptides was done with default settings of the software. After successful alignment, no further filtering was applied for subsequent quantification steps. After peak picking, no further filtering was applied so that all detected features were normalized to a reference run identified by the software. For peptide identification, data were searched against the combined target–decoy database specified above with the following parameters: digest reagent, trypsin; maximum missed cleavages, three; fixed modifications, carbamidomethyl; and variable modifications, deamidation N, deamidation Q and oxidation M. Search tolerance parameters were as follows: peptide tolerance, ± 1.2 Da; fragment tolerance, ± 0.6 Da; and FDR, < 1%. Default settings (i.e., no protein grouping and quantitation from non-conflicting features only) were used for protein building.
Bioinformatic analysis
To understand the role of proteins on sperm functions, total proteins observed in both groups were subjected to GO analysis and enrichment studies. To identify the functions of unique proteins in good and poor quality semen, the proteins present in at least two animals (to avoid the individual animal error) were selected. The functional profiling of total and unique proteins in sperm was carried out using PANTHER (https://www.pantherdb.org), whereas the enriched functions and pathways were analyzed using ShinyGo (v0.61) and STRING (version 11) with Homo sapiens as background. Enrichment analysis is calculated based on hypergeometric distribution followed by FDR correction. FDR less than 0.05 was considered as statistically enriched.
Validation using western blot
Semen samples from different sets of buffalo bulls (n = 6) were used for the validation study. Semen samples having > 90% acrosome intact sperm in neat semen were considered as good (n = 3), otherwise as poor (n = 3) quality semen samples. Acrosome integrity was considered for validation study as this parameter was significantly different in neat and post-thaw samples of good and poor semen quality. Sperm proteins from good and poor quality semen were isolated for western blotting. The abundantly as well as consistently expressed proteins in good quality sperm, namely SPINK2 and NEDD8 and significantly downregulated protein, YBX2, were validated using western blotting experiments. Since the sequences of the selected proteins were found to have similarities between the Homo sapiens and Bubalus bubalis, the antibodies raised against the proteins of Homo sapiens were used for the present study. The percentages of similarity between human and water buffalo proteins were 100%, 90.3% and 73.1% for NEDD8, YBX2 and SPINK2, respectively. For the SPINK2 protein, the epitope specificity was more than 80%. The protocol followed for western blotting of the sperm proteins is as described earlier (Somashekar et al. 2017). In brief, 20 µg of protein from each animal was electrophoresed in 12% SDS gel. The gel was equilibrated for 20 min in a blotting buffer before electro-transfer and the proteins were transferred to PVDF membrane (0.2 microns) at 20 V, 70 mA at 4 °C for 3 h. After transfer, the membrane was washed with Tris buffer saline with Tween-20 (TBS-T) for 2 min and then incubated in a blocking buffer containing 5% bovine serum albumin at 4 °C for 1 h. The membrane was washed with TBS-T and incubated with primary antibodies such as mouse anti-βactin (DSHB. Cat. no.: 8-7A5, 1:1000 dilution, raised in mouse), rabbit anti-SPINK2 (Sigma, Cat. no.: HPA026813, 1:1000 dilution), rabbit anti-NEED8 (Enzo Cat. no. ENZABS376, 1:2000 dilution) and mouse anti-YBX2 (DSHB, 1:1000 dilution) at 4 °C with constant rocking overnight. After incubation, the membrane was washed with TBS-T thrice within 10 min. Then, the membrane was incubated in HRP conjugated secondary antibodies such as goat anti-mouse for β-actin (1:2000 dilution), goat anti-rabbit for SPINK2 (1:2000 dilution), mouse anti-rabbit IgG for NEED8 (1:3000 dilution) and goat anti-mouse IgG for YBX2 (1:2000 dilution) at room temperature (28 °C) for 1 h, followed by three washes (5 min each) in TBS-T. Before detection with immunocomplex, the membrane was washed with TBS and then, immunocomplex was detected by using the ECL-HRP substrate (SuperSignal West Femto, Thermo Scientific, USA). The membrane was incubated with a substrate for 5 min in the dark and the signal was captured by the chemi-documentation system (GBOX, iChemiXR, Syngene, UK). The bands were analyzed in ImageJ software (https://imagej.nih.gov/ij/). The β-actin (42 kDa) was used as normalizing control to obtain the relative expression of SPINK2, NEDD8 and YBX2 proteins in sperm.
Statistical analysis
The sperm functional parameters were arcsine transformed before being subjected to statistical analysis. The level of significance (p < 0.05) was determined using the Student t test. The values are expressed as mean ± SEM.
Results
Sperm functional parameters
The sperm concentration and total semen production capacity were significantly higher in good as compared with poor semen producers (Fig. 1a). In the neat semen samples, the sperm positive for acrosome integrity, high mitochondrial membrane potential, and normal chromatin distribution were significantly (p < 0.05) higher in good as compared with poor quality samples (Fig. 1b).
In the post-thaw semen samples, the percentages of progressive and type-A (Fig. 2a), curvilinear velocity and average path velocity (Fig. 2b) and linearity (Fig. 2c) were significantly (p < 0.05) higher in good than poor quality samples. The percentage of straightness did not differ significantly between groups (Fig. 2c). Though the percentages of sperm having high mitochondrial membrane potential and normal chromatin distribution did not differ significantly between groups, the percentage of acrosome intact sperm was significantly (p < 0.05) higher in good as compared with poor quality semen samples (Fig. 2d).
Sperm functional parameter analyses in frozen-thawed good and poor semen samples. The sperm functional parameters such as (a): progressive motility (PM), fast progressive motility (FP) and Type A; (b): straight-line velocity (VSL), curvilinear velocity (VCL) and average path velocity (VAP); (c): linearity and straightness; (d):plasmalemma integrity (live), acrosomal integrity (AP), mitochondrial membrane potential (MMP) and chromatin distribution (CD) were assessed
Mass spectrometry analysis and protein identification
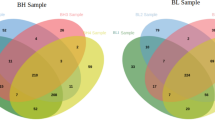
In total, 1365 proteins were identified from buffalo sperm, of which good and poor quality sperm had 935 and 888 proteins, respectively (Fig. 3). Out of 477 unique proteins in good quality sperm (Table 1), 13, 464 and 63 proteins were found to be known, predicted and LOC numbers, respectively (Supplementary data set 1). Correspondingly, out of 430 unique proteins in poor quality sperm (Table 2), 23, 407 and 51 proteins were found to be known, predicted and LOC numbers, respectively (Supplementary data set 2). The number of uncharacterized proteins was 20 and 9 in good and poor quality semen, respectively.
Processes, functions and pathways
Total sperm proteome
GO analysis of the whole proteome of buffalo sperm revealed that the proteins associated with sexual reproduction (PEBP1, FETUB and ACR; enrichment FDR 8.44E−19), generation of precursor metabolites and energy (MDH1, CS, and PFKP; enrichment FDR 1.03E−18), multi-organism reproductive process (PEBP1, FETUB and ACR; enrichment FDR 2.04E−16), etc. were observed to be the enriched biological processes (Fig. 4). The molecular functions of these proteins were nucleoside phosphate binding (HSPA5, PFKP and RHOA; enrichment FDR 7.32E−19), apart from cell adhesion molecule binding (VCL, HSPA5 and PKM; enrichment FDR 1.05E−05) and microtubule motor activity (DYNLL1, DNAH7 and DNAI1; enrichment FDR 3.33E−05). The proteins were enriched to be localized in the extracellular exosome (LAP3, AK2 and RALA; enrichment FDR 3.77E−38), sperm part (SPA17, ACRBP and SPACA1; enrichment FDR 1.52E−27), cilium (PROM1, CATSPERG and CEP164; enrichment FDR 2.41E−25), etc. The KEGG pathways associated with sperm proteins were metabolic pathways such as carbon metabolism pathway (CS, DLD and DLST; enrichment FDR 7.93E−29), glycolysis (GALM, ENO1 and FBP1; enrichment FDR 6.35E−17) and insulin signaling pathways (HK1, HK2 and PPP1CC; enrichment FDR 0.001), apart from thermogenesis (COX5B, ACSL6 and NDUFA8; enrichment FDR 0.003), oocyte meiosis (YWHAQ, SMC1B and PRKACA; enrichment FDR 0.007; Fig. 5), vascular smooth muscle contraction (NPPC, PPP1CC and PRKACA; p = 0.009; Fig. 6), etc. The reactome pathways also identified various metabolic pathways such as cilium assembly (AKAP9, ALMS1 and CCT2; enrichment FDR 4.74E−08), innate immune system (ABCA13, ACTB and AGA; enrichment FDR 4.45E−07), immune system (ABCA13, ALDOA and APOB; enrichment FDR 3.58E−05), cellular response to external stimuli (ATG4, CAPZA3 and CAPZB; enrichment FDR 5.15E−05), etc. as enriched in the sperm proteins.
Top 10 gene ontological functions and pathways enriched in the sperm proteins of buffalo. The majority of the proteins were associated with the reproductive process, metabolic function and located in the extracellular exosome. Among the pathways, carbon metabolism was enriched in the sperm proteins of buffalo
The oocyte meiosis pathway is enriched in buffalo sperm proteins. Sperm proteins, such as CALM and PP2A, may take part in the oocyte meiosis pathway. (The observed proteins in the sperm were highlighted in the red oval box; the pathways are re-created based on the KEGG database, Kanehisa Laboratories)
Smooth muscle contraction pathway is enriched in buffalo sperm proteins. Proteins such as NPPC (CNP), RHOA and MLCP present in the sperm may regulate smooth muscle (uterus and oviduct) contraction in order to reach the sperm to the site of the oocyte. (The observed proteins in the sperm are highlighted in the red oval box; the pathways are re-created based on the KEGG database, Kanehisa Laboratories)
It is also important to mention that the sperm carries proteins associated with Huntington’s disease (COX5B, DNAH2 and DNAH8; p = 1.52E−05), Alzheimer’s disease (NCSTN, GAPDH and HSD17B10; p = 3.91E−05) and Parkinson’s disease (PARK7, HTRA2 and NDUFA8; p = 5.88E−05).
Unique protein analyses
The study revealed that mesenchyme migration (ACTA2, ACTA1 and ACTC1; p = 1.246E−07), organelle organization (MTA2, RHOA and TIMM4; p = 5.917E−05), tissue migration (RHOA, ATP5F1A and MACF1; p = 0.0013), skeletal muscle satellite cell migration (RHOA and RHOC; p = 0.0013), etc. were the unique enriched biological process in the good quality sperm (Table 3).
The good quality semen was enriched with the proteins involved in fluid shear stress (GSTM3, HSP90AA1 and RHOA; p = 0.01), oxidative phosphorylation (NDUFA8, ATP5F1A and ATP5F1D; p = 0.02), carbon metabolism (ENO1, HIBCH and IDH3A; p = 0.023), vascular smooth muscle contraction (RHOA, ACTA2 and ACTG2; p = 0.023) and thermogenesis (NDUFA8, ATP5F1A and ATP5F1D; p = 0.03) pathways (Table 3). Importantly, the poor quality semen was enriched with proteins responsible for stress and the catabolic process. In the catabolic process, beta-oxidation of octanoyl-CoA to hexanoyl-CoA (p = 6.08E−08) was significantly upregulated (Table 3). The pathways such as valine, leucine, and isoleucine degradation (HSD17B10, HADHA, HADH, ACADM; p = 3.80E−05) and glycolysis (GALM, FBP1, PDHA1, PDHA2, PDHB; p = 6.9E−06) were enriched in the poor quality semen (Table 3). In the good quality semen, the proteins responsible for the mitochondrial membrane organization, cilia development, microtubule-based process, antigen processing and thermogenesis were also enriched (Supplementary Table 1), whereas in the poor quality semen samples, such processes were not observed (Supplementary Table 2).
Differentially expressed proteins
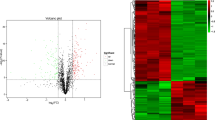
By employing the protein quantification methodology, a total of 497 proteins were differentially expressed (Supplementary Table 3). In good quality sperm, 136 proteins were upregulated and 361 proteins were downregulated. Among them, seven proteins were significantly (p < 0.05) upregulated and 33 proteins were significantly (p < 0.05) downregulated (Table 4). The significantly upregulated proteins were functionally associated with spermatogenesis (SPINK2), motility (NIN), capacitation (PPP1CB) and other metabolic processes (ACAT1). While the proteins in good quality semen samples were associated with spermatogenesis and sperm function, the proteins involved in infertility (NCOA2), stress (HSP70) and glycolysis (SORDX1) were significantly downregulated (Table 4).
Functional analysis of differentially expressed proteins
The upregulated proteins of the good quality semen were involved in the generation of precursor metabolites and energy (SDHA, ATP5F1 and ACAT1; p = 0.0008), oxidation–reduction process (MDH2, QSOX1 and VAT1; p = 0.008), fusion of sperm to egg plasma membrane involved in single fertilization (EQTN, IZUMO1 and LYZL6; p = 0.02) and plasma membrane fusion (EQTN, IZUMO1 and LYZL6; p = 0.04). Oxidoreductase activity (AKR1B1, SDHA and QSOX1; p = 0.02) and adenylate kinase activity (AK1 and AK3; p = 0.014) were the enriched molecular functions. The pathways such as calcium-dependent and PKA activations (PRKAR2A and PRKAR1B; p = 0.019), fertilization (CATSPERG and IZUMO1; p = 0.025) and post-translational protein phosphorylation (ALB, SPP1 and QSOX1; p = 0.03) were enriched. The glycolysis (ALDOA, ENO1 and ENO2; p = 9.05E−15), lysine catabolism pathways (ALDH7A1, DLD and DLST; p = 7.86E−05) and fatty acid beta-oxidation (ACADM, ACOP13 and DDI; p = 4.4E−05) were downregulated in these semen samples.
The poor quality semen samples were enriched with proteins involved in cellular component biogenesis (TEX11, DNALI1 and SRR) and the catabolic process. The protein–protein interaction study suggested that the proteins involved in the catabolic process were involved in valine, leucine and isoleucine degradation (1.54E−06), lysine degradation (5.80E−05), fatty acid degradation (3.82E−05) and tryptophan metabolism (3.82E−05). These proteins were identified to have protein domains such as the ubiquitin-like domain (8.11E−06), 3-hydroxyacyl-CoA dehydrogenase and NAD binding domain (7.22E−05).
Validation with western blotting
Proteome analysis revealed that the proteins namely SPINK2 (10.34-fold) and NEDD8 (2.06-fold) were significantly (p < 0.05) abundant in good quality semen (Fig. 7). SPINK2 and NEDD8 were upregulated (SPINK2 = 2.17-fold, NEDD8 = 1.13) and YBX2 was downregulated (0.41-fold) in good quality semen (Fig. 7).
Discussion
In the current study, buffalo sperm proteome was deciphered using LC–MS/MS coupled with bioinformatics. The whole sperm proteome analysis revealed that the sperm contains 1365 proteins. The existence of 477 and 430 unique proteins in good and poor quality semen, respectively, reflects the inherent differences in sperm functions that may influence the fertilizing capacity of the sperm and early embryonic development.
The sperm of both groups were enriched with proteins associated with spermatogenesis, fertilization and other reproductive processes apart from metabolic activities. The proteins involved in the biosynthesis of amino acids, glucagon, insulin signaling pathway and fatty acid degradation pathway were enriched in sperm indicating the essential role of energy metabolism for sperm kinematics and other functions. Sperm is a metabolically active cell requiring high energy for gaining motility to travel in the female reproductive tract and sequential events of fertilization especially penetration through cumulus cells and zona pellucida (Ruiz-Pesini et al. 2007). Glucagon-like peptide 1 receptor is expressed in human sperm and has a role in regulating sperm motility and cholesterol efflux (Parker et al.2020). Insulin and leptin also improved sperm motility and acrosome reaction in mammals (Lampiao and Du Plessis 2008). The sperm motility, acrosomal reaction and sperm–oocyte fusion depend on the fatty acid profile (Collodel et al. 2020). Fatty acid degradation pathway occurs through an oxidation cycle and yields lower metabolites to generate energy for the sperm cellular function. However, further studies on the beneficial effects of different types of fatty acids and their metabolic pathways are warranted for assessing the type of fatty acid that undergoes denaturation during sperm metabolism (Collodel et al. 2020).
The sperm were also enriched to have proteins responsible for carbon metabolism, oocyte meiosis and vascular smooth muscle contraction. Carbon metabolic pathway aids in the transcriptional regulation via methylation. The role of the DNA methylation process in spermatogenesis is well known. Thus, carbon metabolism aids in the spermatogenesis and any mutations in the enzymes involved in the pathways are reported to cause DNA damage and affect normal fertility (Singh and Jaiswal 2013). In an earlier study from our group, we observed that the transcripts responsible for oocyte meiosis were also present in bovine sperm (Selvaraju et al. 2017). Though the sperm motility is important for the sperm fertilization process, maintenance and stimulation of sperm motility and ability to reach the site of fertilization are favored by contraction of vascular smooth muscles in the uterus and fallopian tube (Wanggren et al.2008). In the present study, we observed that the proteins, such as NPPC present in the sperm, have the property of stimulating sperm motility and vascular smooth muscle contraction, which may facilitate the sperm to reach the site of fertilization. In mice, NPPC protein was also abundantly expressed in oviducts and attracts the sperm towards oocyte for fertilization (Kong et al. 2017). The sperm were also enriched with disease-associated proteins such as PARK7. PARK7 is a stress-responsive protein that protects the cells against ROS and mitochondrial damage (Sun et al. 2014) and as reviewed by Agarwal et al. (2020), a decrease in this protein expression is associated with oligozoospermia in humans. Reactome pathways also identified various metabolic pathways including the proteins responsible for immune regulation. A recent review (Archana et al. 2019) suggested that the proteins in semen and sperm provide immune tolerant mechanisms for protecting the sperm and developing an embryo from immunological reactions in the female reproductive tract. The molecular function indicates that the proteins are mainly involved in DNA or RNA binding. The proteins that bind to the DNA are transcription factors that can either act to repress gene activity or regulate biological functions. Thus, the majority of the sperm proteins were found to be involved in transcription and translation-related functions.
The localization of sperm proteins was mainly observed to be in extracellular exosomes. Exosomes, the nano-sized membrane vesicles, carry a cargo of proteins, lipids, microRNAs and mRNA. They are involved in intracellular communication. Exosomes transfer proteins to the sperm during their epididymal transit, help sperm in acquiring the fertilizing ability and protect it against the oxidative stress. Proteins associated with prostasomes were reported to regulate capacitation, thus avoiding the premature induction of acrosome reaction (Baskaran et al. 2020).
In the present study, mesenchyme migration was enriched in the unique proteins of good quality semen. Mesenchyme migration is an essential process during early embryonic development, wherein it migrates from the blastocyst to the endometrial layer for placental attachment. The presence of proteins associated with mesenchymal migration in sperm suggests that these proteins might be involved in early embryonic development (Hay 2005). Though the contribution of sperm protein on early embryo development has been reported, so far, no previous reports have suggested that sperm proteins are involved in mesenchymal migration during embryonic development.
In the good quality semen, fluid shear stress, vascular smooth muscle contraction, thermogenesis and amino acid synthesis pathways were uniquely present. The fluid shear force can either positively or negatively affect sperm function. But, in the present study, the presence of fluid shear force proteins observed in the good quality semen suggests a positive influence on semen quality. The protein involved in fluid shear force, GSTM3, is secreted from the seminal vesicle (Westfalewicz et al. 2017) and the presence of GSTM proteins in the sperm head the reported to facilitate sperm–zona binding (Hemachand et al. 2002) and may improve fertility.
Importantly, biosynthesis of amino acids was enriched in good quality semen, whereas valine, leucine and isoleucine degradation pathways were enriched in the poor quality semen samples. These essential amino acids promote protein synthesis, signaling pathways and metabolism of glucose. The degradation of these amino acids may lead to oxidative stress and poor semen motility (Bahadorani et al. 2019; Huang et al. 2019). The expression of proteins in the amino acid degradation pathway might also be attributed to the spontaneous acrosome reaction in sperm resulting in poor fertilizing ability (Moos et al. 1993).
The upregulated differentially expressed proteins of the good quality semen were involved in sperm metabolic activity such as the generation of precursor metabolites and the energy oxidation–reduction process, apart from the fusion of sperm to egg plasma membrane involved in single fertilization, wherein equatorin and IZUMO1 proteins are involved. The equatorin protein present in the sperm head equatorial region is involved in sperm–oocyte binding (Toshimori 1998). IZUMO1 is present in the acrosome membrane and translocated to the plasma membrane after the acrosome reaction (Satouh et al. 2012) and the sperm from the IZUMO1 knockout males had a fusion defect with the vitelline membrane (Inoue et al. 2005). Besides, the proteins related to oxidoreductase activity and pathways such as PKA activation and post-translational protein phosphorylation were enriched indicating the importance of these proteins for sperm metabolism and competence in counteracting the stress. The post-translational protein phosphorylations are essential for the capacitation process apart from controlling sperm motility, cytoskeleton assembly, ionic current regulation and receptor regulation (Visconti and Kopf 1998). One of the proteins involved in post-translational protein phosphorylations, the SPP1, stimulates cell–cell adhesion, increases cell–extracellular matrix communication, promotes migration of immune cells and decreases cell death by reducing reactive oxygen species with other roles in the male and female reproductive systems. In porcine, SPP1 actively participates in sperm–zona interactions and is reported to be preventing polyspermy. Studies also reported that SPP1 is involved in adhesion of early germ cells to the basement membrane and epididymal maturation of the sperm (Hao et al. 2006).
In bull sperm, the oxidative phosphorylation pathway is predominant for energy production and high glycolytic activity may reduce intracellular pH and hinder capacitation (review: Storey 2008). Both the pathways can be used by the sperm to produce energy (Krzyzosiak et al. 1999; Magdanz et al. 2019) but different species may utilize various energy sources depending on the condition. In humans, the glycolytic pathway has been the predominant source for the generation of ATP to support the energy requirement for progressive motility and capacitation (Hereng et al. 2011). The aberrant expression of sperm-specific glycolytic enzymes is associated with immature and asthenozoospermic samples (Liu et al. 2019). The predominant glycolytic pathway in poor quality semen tends to indicate that the sperm were under stress and may hamper capacitation. Lysine is shown to increase the sperm concentrations (Nizza et al. 2010); its degradation of which in the poor quality semen may have a negative effect.
Quantitative proteomics analysis revealed that seven proteins (SPINK2, ACAT1, FAHD2A, PPP1CB, LPCAT2B, NEDD8 and NIN) were significantly upregulated and 33 proteins (YBX2, PACRG, VCP, ATP5F1D, SGTA, PGP, SORD, etc.) were significantly downregulated. In the present study, one of the significant abundant proteins, SPINK2, was validated with western blotting. SPINK2 protein is highly essential during different stages of spermatogenesis. The SPINK2 knockout male mice are infertile due to disrupted testicular integrity and increased germ cell apoptosis (Lee et al. 2011). SPINK2 plays a critical role to neutralize protease during the morphogenesis of sperm and formation of acrosomal structure (Jalkanen et al. 2006). In the present study also, the percentage of sperm with acrosome intactness was significantly higher in neat and frozen-thawed samples of good quality semen. Heterologous and homologous mutations of SPINK2 in humans cause oligozoospermia and azoospermia, respectively (Kherraf et al. 2017). In chicken, the expression of SPINK2 in seminal plasma was positively correlated with fertility (Thelie et al. 2019). SPINK2 also acts as a regulatory protein that blocks the action of a serine protease and prevents spontaneous capacitation and acrosome reaction (Ou et al. 2012). These studies along with the present study suggest that SPINK2 protein might regulate buffalo sperm quality by maintaining acrosome integrity.
Another abundant protein validated in the present study, NEDD8, has been reported to be essential for mitosis, cell cycle progression and morphogenesis (Tateishi et al. 2001). The functional role of NEDD8 has not been reported in sperm. However, the NEDD8-related protein, Cullin-associated neddylation/NEDD8-dissociated protein1 (CAND1), is associated with low sperm motility in humans (Amaral et al. 2014). Through the process of neddylation, it modifies non-cullin protein to regulate protein activity, stability and localization (Yu et al. 2019). The positive association between the expression levels of NEDD8 and sperm motility in this study suggests that NEDD8 regulates buffalo bull fertility through promoting sperm progressive motility, which was significantly higher in neat and frozen-thawed samples of good quality semen. However, the exact role of NEDD8 in buffalo sperm function needs to be elucidated. The other abundantly expressed proteins in good quality sperm namely, ACAT1, PPP1CB, lysophosphatidylcholine acyltransferase 2B-like and NIN were reported to regulate various sperm functions importantly, fatty acid metabolism (Wakazono et al. 1995), sperm capacitation (Rotman et al. 2010), lipid metabolism (Moessinger et al. 2011) and locomotion (Bouckson-Castaing et al. 1996), respectively.
YBX2, an RNA binding protein, was significantly downregulated in good quality buffalo sperm. YBX2 showed stage-specific expression during spermatogenesis, i.e., highly expressed from pachytene spermatocyte to the elongating spermatid phase in adult mice (Kleene 2016). Due to the termination of RNA synthesis during mid-spermiogenesis, YBX2 regulates the stabilization, storage and transient suppression of mRNA (PRM1, PRM2, SMCP) translation as these proteins are essential during the final phases of spermatogenesis (Hecht 1998; Giorgini et al. 2001). However, the occurrence of YBX2 protein in matured sperm has not been reported in any species. In our study, YBX2 was upregulated in the poor quality semen indicating that genes, such as PRM1 and PRM2, required for the chromatin condensation may be translationally repressed during spermiogenesis. This may result in the improper translation of protamine transcripts and lead to the defects in the chromatin distribution as observed in the poor quality semen samples of the present study.
We observed downregulation of HSP proteins (HSPA8, HSP90, HSP70) in good quality sperm. HSP proteins are induced to express during stimuli and help to withstand the stress (Baena et al. 2018) and improve sperm survival through membrane repair and protecting mechanism (Moein-Vaziri et al. 2014; Weber et al. 2020). The downregulation of HSP proteins in good quality sperm suggests that these animals are less prone to stress conditions, whereas subfertile/poor population of sperm is susceptible to stress and manifested as upregulation of these proteins for sperm survival and repair mechanisms (Erata et al. 2008). Similar to the present study, other proteins such as FH (Martinez-Heredia et al. 2008), ELSPBP1 (D’Amours et al. 2012), ARMC3 (D’Amours et al. 2019) and NCO2A (Qin et al. 2014) downregulated in good quality sperm were earlier reported to be associated with decreased sperm quality.
Taken together, in the upregulated proteins of good quality semen, the biological processes such as metabolic activity, mitochondrial membrane organization, cilia development and function, fusion of sperm to egg plasma membrane (EQTN, IZUMO1 and LYZL6; p = 0.02) and adenylate kinase activity (AK1 and AK3; p = 0.014) were enriched along with the pathways associated with oxidative phosphorylation and calcium-dependent events including PKA activation (PRKAR2A and PRKAR1B; p = 0.019) and post-translational protein phosphorylation (ALB, SPP1 and QSOX1; p = 0.03). These enriched functions are indicative of the presence of metabolically active, acrosome intact sperm essential for traveling through the female reproductive tract and can undergo acrosome reaction at a suitable microenvironment. In addition, a significantly higher percentage of progressive forward motile and acrosome intact sperm observed in the neat and frozen-thawed good quality semen samples suggests that these attributes are likely associated with the differentially expressed proteins of good quality semen.
No single test has been developed and also no single protein can be used for assessing semen quality and bull fertility. Since the fertilization is a multifaceted process, many proteins regulate the sperm function and earlier study from our lab also indicates that the ratio of different proteins influences sperm fertility (Somashekar et al. 2017). In-depth studies on these differentially expressed proteins may reveal their regulatory role in sperm fertilization competence.
Conclusion
The sperm proteome profile deciphered the molecular functions of sperm during fertilization and early embryonic development. The proteins regulating membrane integrity were enriched in the good quality semen, whereas the amino acid degradation pathways were enriched in the poor quality semen samples. These differentially expressed proteins will be useful for predicting the presence of functionally competent sperm in the ejaculate for reproductive success.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Change history
24 February 2021
A Correction to this paper has been published: https://doi.org/10.1007/s00441-021-03417-8
References
Agarwal A, Mulgund A, Hamada A, Chyatte MR (2015) A unique view on male infertility around the globe. Reprod Biol Endocrinol 13:37. https://doi.org/10.1186/s12958-015-0032-1
Agarwal A, Panner Selvam MK, Baskaran S (2020) Proteomic analyses of human sperm cells: understanding the role of proteins and molecular pathways affecting male reproductive health. Int J Mol Sci 21:1621. https://doi.org/10.3390/ijms21051621
Amaral A, Paiva C, Attardo Parrinello C, Estanyol JM, Ballesca JL, Ramalho-Santos J, Oliva R (2014) Identification of proteins involved in human sperm motility using high-throughput differential proteomics. J Proteome Res 13:5670–5684. https://doi.org/10.1021/pr500652y
Amini-Bavil-Olyaee S, Choi YJ, Lee JH, Shi M, Huang IC, Farzan M, Jung JU (2013) The antiviral effector IFITM3 disrupts intracellular cholesterol homeostasis to block viral entry. Cell Host Microbe 13:452–464. https://doi.org/10.1016/j.chom.2013.03.006
Archana SS, Selvaraju S, Binsila BK, Arangasamy A, Krawetz SA (2019) Immune regulatory molecules as modifiers of semen and fertility: a review. Mol Reprod Dev 86:1485–1504. https://doi.org/10.1002/mrd.23263
Baena MM, Tizioto PC, Meirelles SLC, Regitano LCDA, de Almeida LC (2018) HSF1 and HSPA6 as functional candidate genes associated with heat tolerance in Angus cattle. Rev Bras Zootec 47. https://doi.org/10.1590/rbz4720160390
Bahadorani M, Tavalaee M, Abedpoor N, Ghaedi K, Nazem MN, Nasr-Esfahani MH (2019) Effects of branched-chain amino acid supplementation and/or aerobic exercise on mouse sperm quality and testosterone production. Andrologia 51:e13183. https://doi.org/10.1111/and.13183
Bansal SK, Gupta N, Sankhwar SN, Rajender S (2015) Differential genes expression between fertile and infertile spermatozoa revealed by transcriptome analysis. PLoS ONE 10. https://doi.org/10.1371/journal.pone.0127007
Baskaran S, Panner Selvam MK, Agarwal A (2020) Exosomes of male reproduction. Adv Clin Chem 95:149–163. https://doi.org/10.1016/bs.acc.2019.08.004
Bhakat M, Mohanty TK, Raina VS, Gupta AK, Khan HM, Mahapatra RK, Sarkar M (2011) Effect of age and season on semen quality parameters in Sahiwal bulls. Trop Anim Health Prod 43:1161–1168. https://doi.org/10.1007/s11250-011-9817-1
Bouckson-Castaing V, Moudjou M, Ferguson DJP, Mucklow S, Belkaid Y, Milon G, Crocker PR (1996) Molecular characterisation of ninein, a new coiled-coil protein of the centrosome. J cell Sci 109:179–190
Cao J, Xu H, Zhao H, Gong W, Dunaway-Mariano D (2009) The mechanisms of human hotdog-fold thioesterase 2 (h0054HEM2) substrate recognition and catalysis illuminated by a structure and function based analysis. Biochemistry 48:1293–1304. https://doi.org/10.1021/bi801879z
Card CJ, Anderson EJ, Zamberlan S, Krieger KE, Kaproth M, Sartini BL (2013) Cryopreserved bovine spermatozoal transcript profile as revealed by high-throughput ribonucleic acid sequencing. Biol Reprod 88:49–50. https://doi.org/10.1095/biolreprod.112.103788
Codognoto VM, Yamada PH, Schmith RA, de Ruediger FR, Scott C, de Faria LP, de Brochine S, Paula Freitas-Dell’Aqua C, de Souza Oba FFE (2018) Functional insights into the role of seminal plasma proteins on sperm motility of buffalo. Anim Reprod Sci 195:251–258. https://doi.org/10.1016/j.anireprosci.2018.06.002
Collodel G, Castellini C, Chung-Yung Lee J, Signorini C (2020) Relevance of fatty acids to sperm maturation and quality. Oxid Med Cell Longev. https://doi.org/10.1155/2020/7038124
Crisa A, Marchitelli C, Pariset L, Contarini G, Signorelli F, Napolitano F, Catillo G, Valentini A, Moioli B (2010) Exploring polymorphisms and effects of candidate genes on milk fat quality in dairy sheep. J Dairy Sci 93:3834–3845. https://doi.org/10.3168/jds.2009-3014
D’Amours O, Bordeleau LJ, Frenette G, Blondin P, Leclerc P, Sullivan R (2012) Binder of sperm 1 and epididymal sperm binding protein 1 are associated with different bull sperm subpopulations. Reproduction 143:759–771. https://doi.org/10.1530/REP-11-0392
D’Amours O, Calvo E, Bourassa S, Vincent P, Blondin P, Sullivan R (2019) Proteomic markers of low and high fertility bovine spermatozoa separated by Percoll gradient. Mol Reprod Dev 86:999–1012. https://doi.org/10.1002/mrd.23174
Danshina PV, Geyer CB, Dai Q, Goulding EH, Willis WD, Kitto GB, McCarrey JR, Eddy EM, O’Brien DA (2010) Phosphoglycerate kinase 2 (PGK2) is essential for sperm function and male fertility in mice. Biol Reprod 82:136–145. https://doi.org/10.1095/biolreprod.109.079699
de Mateo S, Castillo J, Estanyol JM, Ballesca JL, Oliva R (2011) Proteomic characterization of the human sperm nucleus. Proteomics 11:2714–2726. https://doi.org/10.1002/pmic.201000799
Erata GO, Toker NK, Durlanık O, Kadıoglu A, Aktan G, Toker GA (2008) The role of heat shock protein 70 (Hsp 70) in male infertility: is it a line of defense against sperm DNA fragmentation? Fertil Steril 90:322–327
Filippou PS, Karagiannis GS, Constantinidou A (2020) Midkine (MDK) growth factor: a key player in cancer progression and a promising therapeutic target. Oncogene 39:2040–2054. https://doi.org/10.1038/s41388-019-1124-8
Fujihara Y, Oji A, Kojima-Kita K, Larasati T, Ikawa M (2018) Co-expression of sperm membrane proteins CMTM2A and CMTM2B is essential for ADAM3 localization and male fertility in mice. J Cell Sci 131. https://doi.org/10.1242/jcs.221481
Fu Q, Pan L, Huang D, Wang Z, Hou Z, Zhang M (2019) Proteomic profiles of buffalo spermatozoa and seminal plasma. Theriogenology 134:74–82. https://doi.org/10.1016/j.theriogenology.2019.05.013
Giorgini F, Davies HG, Braun RE (2001) MSY2 and MSY4 Bind a conserved sequence in the 3′ untranslated region of protamine 1 mRNA in vitro and in vivo. Mol Cell Biol 21:7010–7019. https://doi.org/10.1128/mcb.21.20.7010-7019.2001
Gmachl M, Sagan S, Ketter S, Kreil G (1993) The human sperm protein PH-20 has hyaluronidase activity. FEBS Lett 336:545–548. https://doi.org/10.1016/0014-5793(93)80873-S
Goodarzi MO, Xu N, Cui J, Guo X, Chen YI, Azziz R (2008) Small glutamine-rich tetratricopeptide repeat-containing protein alpha (SGTA), a candidate gene for polycystic ovary syndrome. Hum Reprod 23:1214–1219. https://doi.org/10.1093/humrep/den065
Hao Y, Mathialagan N, Walters E, Mao J, Lai L, Becker D, Li W, Critser J, Prather RS (2006) Osteopontin reduces polyspermy during in vitro fertilization of porcine oocytes. Biol Reprod 75:726–733. https://doi.org/10.1095/biolreprod.106.052589
Hay ED (2005) The mesenchymal cell, its role in the embryo and the remarkable signaling mechanisms that create it. Dev Dyn 233:706–720. https://doi.org/10.1002/dvdy.20345
Hecht NB (1998) Molecular mechanisms of male germ cell differentiation. Bio Essays 20:555–561. https://doi.org/10.1002/(SICI)1521-1878(199807)
Hemachand T, Gopalakrishnan B, Salunke DM, Totey SM, Shaha C (2002) Sperm plasma-membrane-associated glutathione S-transferases as gamete recognition molecules. J Cell Sci 115:2053–2065. https://doi.org/10.1093/humrep/des452
Hereng TH, Elgstoen KBP, Cederkvist FH, Eide L, Jahnsen T, Skalhegg BS, Rosendal KR (2011) Exogenous pyruvate accelerates glycolysis and promotes capacitation in human spermatozoa. Hum Reprod 26:3249–3263. https://doi.org/10.1093/humrep/der317
Hou Z, Fu Q, Huang Y, Zhang P, Chen F, Li M, Xu Z, Yao S, Chen D, Zhang M (2019) Comparative proteomic identification buffalo spermatozoa during in vitro capacitation. Theriogenology 126:303–309. https://doi.org/10.1016/j.theriogenology.2018.12.025
Huang Q, Liu L, Wu Y, Wang X, Luo L, Nan B, Zhang J, Tian M, Shen H (2019) Seminal plasma metabolites mediate the associations of multiple environmental pollutants with semen quality in Chinese men. Environ Int 132:105066. https://doi.org/10.1016/j.envint.2019.105066
Inoue N, Ikawa M, Isotani A, Okabe M (2005) The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature 434:234–238. https://doi.org/10.1038/nature03362
Jalkanen J, Kotimaki M, Huhtaniemi I, Poutanen M (2006) Novel epididymal protease inhibitors with Kazal or WAP family domain. Biochem Biophys Res Commun 349:245–254. https://doi.org/10.1016/j.bbrc.2006.08.023
Jiang H, Rao KS, Yee VC, Kraus JP (2005) Characterization of four variant forms of human propionyl-CoA carboxylase expressed in Escherichia coli. J Biol Chem 280:27719–27727. https://doi.org/10.1074/jbc.M413281200
Jodar M, Sendler E, Krawetz SA (2016) The protein and transcript profiles of human semen. Cell Tissue Res 363:85–96. https://doi.org/10.1007/s00441-015-2237-1
Kherraf Z, Christou‐Kent M, Karaouzene T, Amiri‐Yekta A, Martinez G, Vargas AS, Lambert E, Borel C, Dorphin B, Aknin‐Seifer I, Mitchell MJ (2017) SPINK 2 deficiency causes infertility by inducing sperm defects in heterozygotes and azoospermia in homozygotes. EMBO Mol Med 9:1132–1149. https://doi.org/10.15252/emmm.201607461
Kleene KC (2016) Position-dependent interactions of Y-box protein 2 (YBX2) with mRNA enable mRNA storage in round spermatids by repressing mRNA translation and blocking translation-dependent mRNA decay. Mol Reprod Dev 83:190–207. https://doi.org/10.1002/mrd.22616
Kong N, Xu X, Zhang Y, Wang Y, Hao X, Zhao Y, Qiao J, Xia G, Zhang M (2017) Natriuretic peptide type C induces sperm attraction for fertilization in mouse. Sci Rep 7:1–12. https://doi.org/10.1038/srep39711
Kowalik D, Haller F, Adamski J, Moeller G (2009) In search for function of two human orphan SDR enzymes: hydroxysteroid dehydrogenase like 2 (HSDL2) and short-chain dehydrogenase/reductase-orphan (SDR-O). J Steroid Biochem Mol Biol 117:117–124. https://doi.org/10.1016/j.jsbmb.2009.08.001
Krzyzosiak J, Molan P, Vishwanath R (1999) Measurements of bovine sperm velocities under true anaerobic and aerobic conditions. Anim Reprod Sci 55:163–173. https://doi.org/10.1016/S0378-4320(99)00016-0
Lampiao F, du Plessis SS (2008) Insulin and leptin enhance human sperm motility, acrosome reaction and nitric oxide production. Asian J Androl 10:799–807. https://doi.org/10.1111/j.1745-7262.2008.00421
Lee B, Park I, Jin S, Choi H, Kwon JT, Kim J, Jeong J, Cho BN, Eddy EM, Cho C (2011) Impaired spermatogenesis and fertility in mice carrying a mutation in the Spink2 gene expressed predominantly in testes. J Biol Chem 286:29108–29117. https://doi.org/10.1074/jbc.M111.244905
Li X, Zhang N, Ye HY, Song PP, Chang W, Chen L, Wang Z, Zhang L, Wang NN (2019) HYOU1 promotes cell growth and metastasis via activating PI3K/AKT signaling in epithelial ovarian cancer and predicts poor prognosis. Eur Rev Med Pharmaco 23:4126–4135. https://doi.org/10.26355/eurrev_201901_17914
Liu X, Li Q, Wang W, Liu F (2019) Aberrant expression of sperm-specific glycolytic enzymes are associated with poor sperm quality. Mol Med Rep 19:2471–2478. https://doi.org/10.3892/mmr.2019.9926
Liu X, Huang W, Li C, Li P, Yuan J, Li X, Qiu XB, Ma Q, Cao C (2006) Interaction between c-Abl and Arg tyrosine kinases and proteasome subunit PSMA7 regulates proteasome degradation. Mol Cell 22:317–327. https://doi.org/10.1016/j.molcel.2006.04.007
Lorenzetti D, Bishop CE, Justice MJ (2004) Deletion of the Parkin coregulated gene causes male sterility in the quakingviable mouse mutant. Proc Natl Acad Sci 101:8402–8407. https://doi.org/10.1073/pnas.0401832101
Aslam M, Kumaresan MK, Yadav A, Mohanty S, Datta TK (2019) Comparative proteomic analysis of high- and low-fertile buffalo bull spermatozoa for identification of fertility-associated proteins. Reprod Domest Anim 54:786–794. https://doi.org/10.1111/rda.13426
Magdanz V, Boryshpolets S, Ridzewski C, Eckel B, Reinhardt K (2019) The motility-based swim-up technique separates bull sperm based on differences in metabolic rates and tail length. PLoS ONE 14. https://doi.org/10.1371/journal.pone.0223576
Martinez-Heredia J, de Mateo S, Vidal-Taboada JM, Ballesca JL, Oliva R (2008) Identification of proteomic differences in asthenozoospermic sperm samples. Hum Reprod 23:783–791. https://doi.org/10.1093/humrep/den024
Martinez G, Kherraf ZE, Zouari R, Fourati Ben Mustapha S, Saut A, Pernet-Gallay K, Bertrand A, Bidart M, Hograindleur JP, Amiri-Yekta A, Kharouf M (2018) Whole-exome sequencing identifies mutations in FSIP2 as a recurrent cause of multiple morphological abnormalities of the sperm flagella. Hum Reprod 33:1973–1984. https://doi.org/10.1093/humrep/dey264
Matsumoto M, Fujimoto H (1990) Cloning of a hsp70-related gene expressed in mouse spermatids. Biochem Biophys Res Commun 166:43–49. https://doi.org/10.1016/0006-291X(90)91909-C
Moein-Vaziri N, Phillips I, Smith S, Alminana C, Maside C, Gil MA, Roca J, Martinez EA, Holt WV, Pockley AG, Fazeli A (2014) Heat-shock protein A8 restores sperm membrane integrity by increasing plasma membrane fluidity. Reproduction 147:719–732. https://doi.org/10.1530/REP-13-0631
Moessinger C, Kuerschner L, Spandl J, Shevchenko A, Thiele C (2011) Human lysophosphatidylcholine acyltransferases 1 and 2 are located in lipid droplets where they catalyze the formation of phosphatidylcholine. J Biol Chem 286:21330–21339. https://doi.org/10.1074/jbc.M110.202424
Moos J, Peknicova J, Tesarik J (1993) Protein—protein interactions controlling acrosin release and solubilization during the boar sperm acrosome reaction. Biol Reprod 49:408–415. https://doi.org/10.1095/biolreprod49.2.408
Naaby-Hansen S, Herr JC (2010) Heat shock proteins on the human sperm surface. J Reprod Immunol 84:32–40. https://doi.org/10.1016/j.jri.2009.09.006
Nizza A, Dimeo C, Taranto S (2010) Effect of lysine and methionine on libido and semen characteristics of bucks. World Rabbit Sci 8:181–184. https://doi.org/10.4995/wrs.2000.437
Olahova M, Yoon WH, Thompson K, Jangam S, Fernandez L, Davidson JM, Kyle JE, Grove ME, Fisk DG, Kohler JN, Holmes M (2018) Biallelic mutations in ATP5F1D, which encodes a subunit of ATP synthase, cause a metabolic disorder. Am J Hum Genet 102:494–504. https://doi.org/10.1016/j.ajhg.2018.01.020
Ou CM, Tang JB, Huang MS, Sudhakar Gandhi PS, Geetha S, Li SH, Chen YH (2012) The mode of reproductive-derived Spink (serine protease inhibitor Kazal-type) action in the modulation of mammalian sperm activity. Int J Androl 35:52–62. https://doi.org/10.1111/j.1365-2605.2011.01159.x
Parker V, Robertson D, Wang T, Hornigold DC, Petrone M, Cooper AT, Posch MG, Heise T, Plum-Moerschel L, Schlichthaar H, Klaus B (2020) Efficacy, safety, and mechanistic insights of cotadutide, a dual receptor glucagon-like peptide-1 and glucagon agonist. J Clin Endocrinol Metab 105:803–820
Pasek RC, Malarkey E, Berbari NF, Sharma N, Kesterson RA, Tres LL, Kierszenbaum AL, Yoder BK (2016) Coiled-coil domain containing 42 (Ccdc42) is necessary for proper sperm development and male fertility in the mouse. Dev Biol 412:208–218. https://doi.org/10.1016/j.ydbio.2016.01.042
Pini T, Rickard JP, Leahy T, Crossett B, Druart X, de Graaf SP (2018) Cryopreservation and egg yolk medium alter the proteome of ram spermatozoa. J Proteomics 181:73–82. https://doi.org/10.1016/j.jprot.2018.04.001
Pinto TMF, Moreira RF, Matos MNC, Soares VV, Aguiar MV, Aragao PD, Alves Filho JG, Moreno FB, Monteiro-Moreira AC, Costa CR, Lima Filho JL (2019) Evaluation of the proteomic profiles of ejaculated spermatozoa from Saanen bucks (Capra hircus). Anim Reprod 16:902–913. https://doi.org/10.21451/1984-3143-AR2019-0001
Qin J, Lee HJ, Wu SP, Lin SC, Lanz RB, Creighton CJ, DeMayo FJ, Tsai SY, Tsai MJ (2014) Androgen deprivation–induced NCoA2 promotes metastatic and castration-resistant prostate cancer. J Clin Invest 124:5013–5026. https://doi.org/10.1172/JCI76412
Rezende FM, Dietsch GO, Penagaricano F (2018) Genetic dissection of bull fertility in US Jersey dairy cattle. Anim Genet 49:393–402. https://doi.org/10.1111/age.12710
Roca J, Perez-Patino C, Barranco I, Padilla LC, Martinez EA, Rodriguez-Martinez H, Parrilla I (2020) Proteomics in fresh and preserved pig semen: recent achievements and future challenges. Theriogenology 150:41–47. https://doi.org/10.1016/j.theriogenology.2020.01.066
Rotman T, Etkovitz N, Spiegel A, Rubinstein S, Breitbart H (2010) Protein kinase A and protein kinase C (a)/PPP1CC2 play opposing roles in the regulation of phosphatidylinositol 3-kinase activation in bovine sperm. Reproduction 140:43–56. https://doi.org/10.1530/REP-09-0314
Ruiz-Pesini E, Diez-Sanchez C, Lopez-Perez MJ, Enriquez JA (2007) The role of the mitochondrion in sperm function: is there a place for oxidative phosphorylation or is this a purely glycolytic process? Curr Top Dev Biol 77:3–19. https://doi.org/10.1016/S0070-2153(06)77001-6
Satouh Y, Inoue N, Ikawa M, Okabe M (2012) Visualization of the moment of mouse sperm-egg fusion and dynamic localization of IZUMO1. J Cell Sci 125:4985–4990. https://doi.org/10.1242/jcs.100867
Selvaraju S, Parthipan S, Somashekar L, Kolte AP, Binsila BK, Arangasamy A, Ravindra JP, (2017) Occurrence and functional significance of the transcriptome in bovine (Bos taurus) spermatozoa. Scientific Reports 7 (1)
Selvaraju S, Krishnan BB, Archana SS, Ravindra JP (2016a) IGF1 stabilizes sperm membrane proteins to reduce cryoinjury and maintain post-thaw sperm motility in buffalo (Bubalus bubalis) spermatozoa. Cryobiology 73:55–62. https://doi.org/10.1016/j.cryobiol.2016.05.012
Selvaraju S, Ravindra JP, Ghosh J, Gupta PS, Suresh KP (2008) Evaluation of sperm functional attributes in relation to in vitro sperm-zona pellucida binding ability and cleavage rate in assessing frozen thawed buffalo (Bubalus bubalis) semen quality. Anim Reprod Sci 106:311–321. https://doi.org/10.1016/j.anireprosci.2007.05.005
Selvaraju S, Somashekar L, Krishnan BB, Parthipan S, Pushparani G, Arangasamy A, Rajendran D, Ravindra JP (2016b) Relationship between seminal plasma tuberoinfundibular peptide of 39 residues and sperm functional attributes in buffalo (Bubalus bubalis). Reprod Fertil Dev 28:1622. https://doi.org/10.1071/RD15008
Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M (2007) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1:2856–2860. https://doi.org/10.1038/nprot.2006.468
Singh K, Jaiswal D (2013) One-carbon metabolism, spermatogenesis, and male infertility. Reprod Sci 20:622–630. https://doi.org/10.1177/1933719112459232
Somashekar L, Selvaraju S, Parthipan S, Patil SK, Binsila BK, Venkataswamy MM, Karthik Bhat S, Ravindra JP (2017) Comparative sperm protein profiling in bulls differing in fertility and identification of phosphatidylethanolamine-binding protein 4, a potential fertility marker. Andrology 5:1032–1051. https://doi.org/10.1111/andr.12404
Stillwell EE, Zhou J, Joshi HC (2004) Human ninein is a centrosomal autoantigen recognized by CREST patient sera and plays a regulatory role in microtubule nucleation. Cell Cycle 3:921–928. https://doi.org/10.4161/cc.3.7.947
Storey BT (2008) Mammalian sperm metabolism: oxygen and sugar, friend and foe. Int J Dev Biol 52:427–437. https://doi.org/10.1387/ijdb.072522bs
Sun Y, Zhang W, Zhao X, Yuan RP, Jiang H, Pu XP (2014) PARK7 protein translocating into spermatozoa mitochondria in Chinese asthenozoospermia. Reproduction 143:249–257. https://doi.org/10.1530/REP-14-0222
Tateishi K, Omata M, Tanaka K, Chiba T (2001) The NEDD8 system is essential for cell cycle progression and morphogenetic pathway in mice. J Cell Biol 155:571–579. https://doi.org/10.1083/jcb.200104035
Thelie A, Rehault-Godbert S, Poirier J, Govoroun M, Fouchécourt S, Blesbois E (2019) The seminal acrosin-inhibitor ClTI1/SPINK2 is a fertility-associated marker in the chicken. Mol Reprod Dev 86:762–775. https://doi.org/10.1002/mrd.23153
Toshimori K (1998) Maturation of mammalian spermatozoa: modifications of the acrosome and plasma membrane leading to fertilization. Cell Tissue Res 293:177–187
van Koningsbruggen S, Straasheijm KR, Sterrenburg E, de Graaf N, Dauwerse HG, Frants RR, van der Maarel SM (2007) FRG1P-mediated aggregation of proteins involved in pre-mRNA processing. Chromosoma 116:53–64. https://doi.org/10.1007/s00412-006-0083-3
Verma A, Rajput S, Kumar S, De S, Chakravarty AK, Kumar R, Datta TK (2015) Differential histone modification status of spermatozoa in relation to fertility of buffalo bulls. J Cell Biochem 116:743–753. https://doi.org/10.1002/jcb.25029
Visconti PE, Kopf GS (1998) Regulation of protein phosphorylation during sperm capacitation1. Biol Reprod 59:1–6. https://doi.org/10.1095/biolreprod59.1.1
Wakazono A, Fukao T, Yamaguchi S, Hori T, Orii T, Lambert M, Mitchell GA, Lee GW, Hashimoto T (1995) Molecular, biochemical, and clinical characterization of mitochondrial acetoacetyl-coenzyme A thiolase deficiency in two further patients. Hum Mutat 5:34–42. https://doi.org/10.1002/humu.1380050105
Wanggren K, Stavreus-Evers A, Olsson C, Andersson E, Gemzell-Danielsson K (2008) Regulation of muscular contractions in the human fallopian tube through prostaglandins and progestagens. Human Reprod 23(2359):2368. https://doi.org/10.1093/humrep/den260
Weber A, Argenti LE, de Souza APB, Santi L, Beys-da-Silva WO, Yates JR, Bustamante-Filho IC (2020) Ready for the journey: a comparative proteome profiling of porcine cauda epididymal fluid and spermatozoa. Cell Tissue Res 379:389–405. https://doi.org/10.1007/s00441-019-03080-0
Westfalewicz B, Dietrich MA, Mostek A, Partyka A, Bielas W, Nizanski W, Ciereszko A (2017) Identification and functional analysis of bull (Bos taurus) cauda epididymal fluid proteome. J Dairy Sci 100:6707–6719. https://doi.org/10.3168/jds.2016-12526
Yu G, Liu X, Zhang D, Wang J, Ouyang G, Chen Z, Xiao W (2019) Zebrafish Nedd8 facilitates ovarian development and the maintenance of female secondary sexual characteristics via suppression of androgen receptor activity. SSRN Electron J. https://doi.org/10.2139/ssrn.3323376
Zhu W, Zhang Y, Ren C, huan C, Cheng X, Chen JH, Ge ZY, Sun ZP, Zhuo X, Sun FF, Jia XJ, Zhang Z (2020) Identification of proteomic markers for ram spermatozoa motility using a tandem mass tag (TMT) approach. J Proteomics 210:103438. https://doi.org/10.1016/j.jprot.2019.103438
Zimmerman S, Sutovsky P (2009) The sperm proteasome during sperm capacitation and fertilization. J Reprod Immunol 83:19–25. https://doi.org/10.1016/j.jri.2009.07.006
Acknowledgments
The study was supported by ICAR-All India Coordinated Research Project on “Nutritional and physiological interventions for enhancing reproductive performance in animals.” The authors sincerely acknowledge the support of the Director, ICAR-NIANP, Bangalore, and Coordinator AICRP for conducting research at ICAR-NIANP. The authors sincerely thank Dr. J.P Ravindra, Former Head, Animal Physiology Division, ICAR-NIANP for the technical inputs and critical reviewing of the manuscript. Dr. S. Selvaraju is supported by ICAR-National Fellow Project, ICAR, Ministry of Agriculture, Government of India.
Author information
Authors and Affiliations
Contributions
BBK, SS and NSKG: designed the experiment; ASS, RL and SD: performed experiments; RA: data interpretation; BBK, SS, ASS and NSKG: wrote the paper; RB: critically discussed the data; BBK, SS, NSKG and DTP: critically discussed the data, carried out data interpretation and prepared manuscript. All authors reviewed the manuscript.
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Binsila, B.K., Archana, S.S., Ramya, L. et al. Elucidating the processes and pathways enriched in buffalo sperm proteome in regulating semen quality. Cell Tissue Res 383, 881–903 (2021). https://doi.org/10.1007/s00441-020-03303-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00441-020-03303-9