Abstract
Runx1 transcription factor is a key developmental regulator. However, little is known about the effects of Runx1 on embryo implantation and decidualization. The aim of this study is to examine the expression and regulation of Runx1 in mouse uterus during the peri-implantation period. There was no evident Runx1 mRNA signal on days 1–4 of pregnancy. On day 5 of pregnancy, Runx1 mRNA was mainly localized in the subluminal stroma surrounding the implanting blastocyst. A similar result was observed in the estrogen-activated implantation uterus. Simultaneously, a high level of Runx1 mRNA expression was detected on days 6–8 of pregnancy and under artificial decidualization. 8-Br-cAMP could induce the expression of Runx1 mRNA in the uterine stromal cells. Moreover, the induction was obviously blocked by PKA inhibitor H89. Inhibition of Runx1 with specific siRNA could decrease the proliferation of stromal cells and expression of decidual markers Prl8a2 and Prl3c1 in the uterine stromal cells. Further study found that inhibition of Runx1 could also suppress the expression of Cox-2, mPGES-1 and Mmp2 genes in uterine stromal cells. Estrogen and progesterone could induce the expression of Runx1 mRNA in ovariectomized mouse uterus and uterine stromal cells. Taken together, these data suggest that Runx1 may play an important role during mouse decidualization.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The runt-related transcription factor (Runx) family, also known as acute myeloid leukaemia (Aml), core-binding factor-α (Cbfα) and polyoma enhancer-binding protein-2α (Pebp2α), is a crucial transcription factor that is involved in cellular proliferation, differentiation and apoptosis (Blyth et al. 2005; Kilbey et al. 2010; Chuang et al. 2013). In mammals, there are three Runx family members: Runx1 (Aml1/Cbfα2/Pebp2αB), Runx2 (Aml3/Cbfα1/Pebp2αA) and Runx3 (Aml3/Cbfα1/Pebp2αC). They share a highly conserved runt domain that is responsible for DNA binding and heterodimerization with core-binding factor β (Cbfβ) (Blyth et al. 2005; Chimge and Frenkel 2013; Ozaki et al. 2013). All Runx proteins bind to the specific DNA consensus sequences (ACCACA) to either activate or repress gene transcription by forming heterodimers with Cbfβ, which can enhance the DNA-binding activity of Runx and stabilize the heterodimeric complex by preventing ubiquitin-dependent degradation (Blyth et al. 2005; Kitoh et al. 2009; Klunker et al. 2009; Kim et al. 2014; Wu et al. 2014).
Despite their structural similarity, the evidence from gene knockout mice showed that Runx members possessed distinct physiological functions during mammalian development (Blyth et al. 2005). Runx1-deficient mice displayed a major defect in hematopoiesis while Runx2 knockout resulted in lack of osteogenesis (Okuda et al. 1996; Komori et al. 1997; Otto et al. 1997). Runx3 has been shown to play a role in thymogenesis, neurogenesis and gastrointestinal development (Levanon and Groner 2004; Chimge and Frenkel 2013; Kim et al. 2014). Subsequent studies revealed that Runx genes were closely related to female reproduction. In Runx3 knockout mouse ovaries, the numbers of primary, preantral and antral follicles were significantly reduced and corpora lutea were not observed (Sakuma et al. 2008; Blyth et al. 2010). Our previous study demonstrated that Runx3 might be essential for mouse embryo implantation and decidualization (Bai et al. 2013). Runx1 and Runx2 were expressed in Mullerian ducts, which were the rudimentary structure that ultimately formed the uterine tubes, uterus, cervix and upper part of the vagina (Blyth et al. 2010). Further studies found that Runx1 was also documented to be highly expressed in cumulus–oocyte complexes and granulosa cells of periovulatory follicles (Jo and Curry 2006; Liu et al. 2009). However, little is known about the expression and regulation of Runx1 in mouse uterus. According to our (unpublished) microarray data, Runx1 was highly expressed in day 8 decidua and deciduoma under artificial decidualization compared with the untreated uterine horn. Thus, this study was undertaken to investigate the expression and regulation of the Runx1 gene in mouse uterus during the peri-implantation period.
Materials and methods
Animal
Matured mice (Kunming White strain) were caged in a controlled environment with a cycle of 14 L:10 D. All animal procedures were approved by the Institutional Animal Care and Use Committee of Jilin University. To confirm reproducibility of results, at least three mice per group were used in each stage or treatment in this study.
Pregnancy and pseudopregnancy
Adult female mice were mated with fertile or vasectomized males of the same strain to induce pregnancy or pseudopregnancy by cocaging, respectively (day 1 = day of vaginal plug). On days 1–4, pregnancy was confirmed by recovering embryos from the oviducts or uterus. The implantation sites on day 5 were identified by intravenous injection of 0.1 ml of 1 % Chicago blue (Sigma, St. Louis, MO, USA).
Delayed implantation and activation
To induce delayed implantation, pregnant mice were ovariectomized under ether anesthesia at 0830–0900 hours on day 4 of pregnancy. Progesterone (1 mg/mouse; Sigma) was injected subcutaneously to maintain delayed implantation from days 5 to 7. Estrogen (25 ng/mouse; Sigma) was given to progesterone-primed delayed-implantation mice to activate blastocyst implantation. The mice were sacrificed to collect uteri 24 h after estrogen treatment. The implantation sites were identified by intravenous injection of Chicago blue solution. Delayed implantation was confirmed by flushing the blastocysts from the uterus.
Artificial induced decidualization
Artificial decidualization was induced by intraluminally infusing 25 μl sesame oil into one uterine horn on day 4 of pseudopregnancy, while the contralateral uninjected horn served as a control. The uteri were collected on day 8 of pseudopregnancy. Decidualization was confirmed by weighing the uterine horn and by histological examination of uterine sections.
Steroid hormonal treatments
Mature female mice were ovariectomized and, after 2 weeks, given a single sc injection of estrogen (100 ng/mouse) or progesterone (2 mg/mouse). Uteri were then collected at 1, 3, 6, 12 and 24 h after steroid treatment. All steroids were dissolved in sesame oil and injected subcutaneously. Controls received the vehicle only (0.1 ml/mouse).
Uterine stromal cells from day 4 of pregnancy were isolated and cultured as previously described (Tian et al. 2013). Cultured stromal cells were also treated with 100 nM of progesterone or 0.1 nM of estrogen, respectively. Then cells were collected at 1, 3, 6, 12 and 24 h for further quantitative analysis by real-time PCR. All steroids were dissolved in ethanol. Controls received the vehicle only.
In situ hybridization
Total RNAs from the mouse uteri were reverse-transcribed and amplified with Runx1 primers. Runx1 forward primer 5′- CCTTGAACCACTCCACTGCC and reverse primer 5′- GACGGCAGAGTAG GGAACTG were designed according to the Mus musculus runt-related transcription factor 1 gene (Genbank accession number: NM_001111021). The amplified fragment (268 bp) of Runx1 was cloned into pGEM-T plasmid (pGEM-T Vector System 1; Promega, Madison, WI, USA) and verified by sequencing. Runx1-containing plasmid was amplified with the primers for T7 and SP6 to prepare templates for labeling. Digoxigenin (DIG)-labeled antisense and sense cRNA probes were transcribed in vitro using a DIG RNA labeling kit (Roche Diagnostics, Mannheim, Germany).
Frozen sections (10 μm) were mounted on 3-aminopropyltriethoxy-silane (Sigma)-coated slides and fixed in 4 % paraformaldehyde solution in PBS. Hybridization was performed as described previously (Tian et al. 2013). Sections were counterstained with 1 % methyl green in 0.12 M glacial acetic acid. The sense probe was also hybridized and served as a negative control. There was no detectable signal from sense probes.
Real-time PCR
Total RNAs from mouse uteri or cultured cells were isolated using TRIPURE reagent according to the manufacturer’s instructions (Roche) and reverse-transcribed into cDNA using M-MLV reverse-transcriptase (Promega). Reverse transcriptase was performed at 42 °C for 60 min with 2 μg total RNA in 25 μl volume. For real-time PCR, cDNA was amplified using FS Universal SYBR Green Real Master (Roche) on the BIO-RAD CFX96TM Real Time Detection System. The conditions used for real-time PCR were as follows: 95 °C for 3 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. All reactions were run in triplicate. The results were analyzed using CFX Manager Software. After analysis using the 2-ΔΔCt method, data were normalized to Gapdh expression. Primer sequences for real-time PCR are listed in Table 1.
RNA interference
The small-interfering RNA (siRNA) duplexes for targeting Runx1 as well as a scrambled sequence (control siRNA duplex, negative control) were synthesized by GenePharma. The sequences were shown as follows: Runx1 siRNA 1 sense: 5′- CCGCCGCUUCACGCCGCCUUCTT, antisense: 5′-GAAGGCGGCGUGAAGCGGCGGTT; nonspecific scrambled siRNA (negative control) sense: 5′- UUCUCCGAACGUGUCACGUTT, antisense: 5′- ACGUGACACGUUCGGAGAATT. Transfections for siRNA were performed according to the Lipofectamine 2000 protocol.
Cell proliferation
Proliferation assays were performed using MTS reagent (Promega) according to the manufacturer’s directions. Uterine stromal cells were seeded at a density of 1 × 105/well in 96-well plates and cultured in the DMEM/F12 medium containing 2 % heat-inactivated FBS. After transfection with Runx1 siRNA, the stromal cells were cultured for 48 h. Finally, 20 μl MTS reagent was added to each well and incubated for 4 h. Absorbance was measured at 490 nm using a 96-well plate reader. Every experiment was performed in triplicate.
Statistics
All the experiments were independently repeated at least three times. The significance of difference was analyzed by one-way ANOVA or Independent-Samples t Test using the SPSS software program (SPS, Chicago, IL, USA). The differences were considered significant at P < 0.05.
Results
Runx1 mRNA expression during early pregnancy
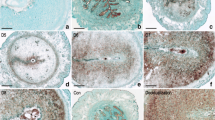
In situ hybridization was performed to examine the spatial distribution of Runx1 mRNA in mouse uterus. From days 1 to 4 of pregnancy, there was no obvious Runx1 mRNA signal in the uteri (Fig. 1b). On day 5 of pregnancy when the blastocyst implanted, a specific Runx1 mRNA signal was detected in the subluminal stroma surrounding the implanting blastocyst at implantation site (Fig. 1c). On days 6–8 of pregnancy, Runx1 mRNA was expressed in decidualized cells and its expression was increased with the development of decidua (Fig. 1d–f). In addition, Runx1 mRNA signal was also detected in the embryos on day 8 of pregnancy by in situ hybridization (Fig. 1f). To quantify Runx1 mRNA expression, real-time PCR was performed. The result showed that Runx1 mRNA expression gradually increased from days 3 to 8 of pregnancy and reached a peak on day 8 (Fig. 2).
In situ hybridization of Runx1 expression in mouse uteri during early pregnancy on days 4 (b), 5 (c), 6 (d), 7 (e) and 8 (f). No hybridization signals were seen in the mouse uterus on day 8 of pregnancy when a DIG-labeled Runx1 sense probe was used to replace the antisense probe as a negative control (a). D stands for day of pregnancy. Asterisks indicate embryo. Bar 60 μm
Runx1 mRNA expression during pseudopregnancy
By in situ hybridization, no evident Runx1 mRNA signal was observed in the uteri from days 1 to 5 of pseudopregnancy (Fig. 3a, b). However, real-time PCR results showed that a significantly higher level of Runx1 expression was detected on day 2 of pseudopregnancy, although Runx1 expression was seen from days 1 to 5 (Fig. 4a).
In situ hybridization of Runx1 expression in mouse uteri. a Day 3 of pseudopregnancy (PD3). b Day 5 of pseudopregnancy (PD5). c Delayed implantation (Delay). d Activation of delayed implantation by estrogen (Activation). e Uninjected uterine horn of control (Control). f Uterine horn under artificial decidualization (Decidualization)
Real-time PCR analysis of Runx1 expression in mouse uteri. a Real-time PCR analysis of Runx1 expression in mouse uterus on days 1–5 of pseudopregnancy. b Real-time PCR analysis of Runx1 expression in mouse uterus during delayed implantation and activation. c Real-time PCR analysis of Runx1 expression under artificial decidualization
Runx1 mRNA expression during delayed implantation and activation
In the progesterone-primed delayed implantation uterus, Runx1 was located in the luminal and glandular epithelium, whereas no expression was observed in the stromal cells (Fig. 3c). After delayed implantation was terminated by estrogen treatment and the blastocyst implanted, Runx1 mRNA signal was detected in the subluminal stromal cells surrounding the implanting blastocyst (Fig. 3d). Real-time PCR analysis showed that there was no significant difference in Runx1 mRNA expression between the activated and delayed implantation uterus (Fig. 4b).
Runx1 mRNA expression under artificial decidualization
Runx1 mRNA signal was obviously detected in the decidualized cells when decidualization was induced in the uterine horn by injecting sesame oil into the uterine lumen, whereas it was barely found in the uninjected control uterus (Fig. 3e, f). Likewise, a significantly higher level of Runx1 expression was observed in the decidualized uterus compared with the control uterus by real-time PCR (Fig. 4c).
Regulatory effects of cAMP and H89 on Runx1 expression
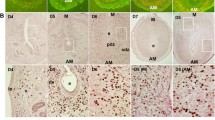
It has been demonstrated that initiation of the decidual process is dependent on increased intracellular cAMP levels and sustained activation of the protein kinase A (PKA) pathway (Gellersen and Brosens 2003). Here, we tested the effects of cAMP analog 8-bromoadenosine-cAMP (8-Br-cAMP, 500 μM) and PKA inhibitor H89 (10 μM) on Runx1 expression in cultured uterine stromal cells. The results showed that 8-Br-cAMP could significantly induce the expression of Runx1 at 3 h (Fig. 5a). As expected, the induction effect of 8-Br-cAMP on Runx1 expression was obviously blocked by PKA inhibitor H89 (Fig. 5b).
Steroid hormonal regulation of Runx1 expression
To clarify the role of steroid hormones in Runx1 expression, ovariectomized mice were given a single inection of oil (control), estrogen, or progesterone and then Runx1 mRNA expression was examined in the uteri by in situ hybridization and real-time PCR. In situ hybridization results revealed that neither estrogen nor progesterone affected the expression of Runx1 (data not shown). Real-time PCR results showed that elevated expression levels of Runx1 were observed in the uteri of ovariectomized mice with a time-dependent increase after injection of progesterone (Fig. 6b). Similarly, Runx1 expression was significantly enhanced in the uteri of ovariectomized mice after injection of estrogen and reached the highest level at 3 h (Fig. 6a). In the in vitro cultured stromal cells, progesterone treatment resulted in an increase of the Runx1 mRNA level at 12 and 24 h (Fig. 6d). Likewise, estrogen could also induce the expression of Runx1 mRNA at 3 h (Fig. 6c).
Hormonal regulation of Runx1 expression. a Real-time PCR analysis of Runx1 expression in ovariectomized mouse uterus after estrogen treatments for 1, 3, 6, 12 and 24 h. b Runx1 expression in ovariectomized mouse uterus after injection of progesterone. c Runx1 expression in uterine stromal cells after estrogen treatments for 1, 3, 6, 12 and 24 h. d Runx1 expression in uterine stromal cells after progesterone treatments for 1, 3, 6, 12 and 24 h
Effects of Runx1 on decidualization
Because Runx1 was highly expressed in the decidualized cells, we hypothesized that Runx1 might be essential for decidualization. Stromal cell proliferation was the first step of decidualization. To determine the effects of Runx1 on stromal cell proliferation, we treated the uterine stromal cells with specific Runx1 siRNA, which efficiently suppressed the expression level of Runx1 mRNA (Fig. 7a). After transfection with Runx1 siRNA, proliferation activity of stromal cells was dramatically inhibited (Fig. 7b). To further study the function of Runx1 on decidualization, we also examined the effects of Runx1 on the expression of prolactin family 8, subfamily a, member 2 (Prl8a2) and prolactin family 3, subfamily c, member 1 (Prl3c1), which were two well-known markers for decidualization in mice. The results demonstrated that Runx1 knockdown could notably inhibit the expression of Prl8a2 and Prl3c1 genes in the uterine stromal cells (Fig. 7c, d).
Effects of Runx1 on decidualization. a Effects of Runx1 siRNA on Runx1 mRNA expression in uterine stromal cells. After transfection with control siRNA or Runx1 siRNA, Runx1 mRNA expression was determined by real-time PCR. b Effects of Runx1 siRNA on the proliferation of uterine stromal cells. Stromal cells after transfection Runx1 siRNA were analyzed by MTS assay. c Effects of Runx1 siRNA on Prl8a2 expression. d Effects of Runx1 siRNA on Prl3c1 expression
Regulation of Runx1 on Cox-2, mPGES-1, Mmp2 and Mmp9 expression
Because decidualization was involved in the uterine extracellular matrix remodeling and angiogenesis (McConaha et al. 2011), we investigated the effects of Runx1 on the expression of cyclooxygenase-2 (Cox-2), microsomal prostaglandin E synthase 1 (mPGES-1), matrix metallopeptidase 2 (Mmp2) and Mmp9. The results showed that inhibition of Runx1 with specific Runx1 siRNA could suppress the expression of Cox-2, mPGES-1 and Mmp2 genes in the cultured stromal cells, whereas had no obvious change on the expression of the Mmp9 gene (Fig. 8a–d).
Effects of Runx1 on Cox-2, mPGES-1, Mmp2 and Mmp9 expression. a Effects of Runx1 siRNA on Cox-2 expression. After transfection with control siRNA or Runx1 siRNA, Cox-2 mRNA expression was determined by real-time PCR. b Effects of Runx1 siRNA on mPGES-1 expression. c Effects of Runx1 siRNA on Mmp2 expression. d Effects of Runx1 siRNA on Mmp9 expression
Discussion
Runx1 transcription factor is a key regulator involved in normal development. However, the implication of Runx1 in the process of embryo implantation and decidualization is currently unknown. The present study aimed to investigate the expression and regulation of Runx1 in mouse uterus during the peri-implantation period. Our results showed that Runx1 mRNA was expressed in the subluminal stromal cells surrounding the implanting blastocyst on day 5 of pregnancy but not in the uterus on day 5 of pseudopregnancy. A similar expression pattern was also observed in the estrogen-activated implantation sites rather than in the progesterone-primed delayed implantation uterus. These results suggest that Runx1 may be important for mouse embryo implantation and is influenced by the active blastocyst. However, the majority of Runx1-deficient embryos were viable at embryonic day 11.5 (E11.5), while very few Runx1-deficient embryos were found alive at E12.5 (Okuda et al. 1996). In the meantime, uterine-specific deletion of the Runx1 gene in mice led to embryonic lethality on days 10–12 of pregnancy due to severe hemorrhage and embryo resorption (Athilakshmi et al. 2011). These results imply that Runx1 may be dispensable for mouse embryo implantation.
In response to implantation, the uterine stromal cells surrounding the implantation embryo initiate the extensive proliferation and subsequent differentiation into polyploidy decidual cells (Zhang et al. 2013). Once decidualization is impaired, it will induce embryonic lethality and spontaneous abortion, thereby resulting in pregnancy failure (Shao et al. 2013). The present results found that Runx1 mRNA was highly expressed in the decidualized cells on days 6–8 of pregnancy and under artificial decidualization. Previous studies have evidenced that injection of sesame oil resulted in an increase in uterine cAMP level, which could induce and regulate the decidualization of uterine stromal cells (Rankin et al. 1977; Gellersen and Brosens 2003). In the uterine stromal cells, cAMP analog 8-Br-cAMP induced the expression of Runx1. As a ubiquitous second messenger molecule, cAMP can activate the PKA signal transduction pathway (Gellersen and Brosens 2003). Thus, PKA inhibitor H89 suppressed the induction effect of 8-Br-cAMP on Runx1 expression in the stromal cells. Taken together, these results suggest that Runx1 may be crucial for decidualization. Runx1 knockdown could remarkably decrease the proliferation of stromal cells and expression of decidual markers Prl8a2 and Prl3c1 in the uterine stromal cells, which verified the effects of Runx1 on mouse decidualization. Further analysis revealed that compromised decidualization was also observed in Runx1 conditional knockout mice (Athilakshmi et al. 2011).
Angiogenesis was crucial for decidualization due to the fact that administration of AGM-1470 (a nonspecific angiogenesis inhibitor) to pregnant mice caused an impaired decidualization (Klauber et al. 1997). Cumulative evidence showed that Cox-2 was a key regulator of uterine angiogenesis during decidualization (Dey et al. 2004). In Cox-2-deficient mice, one cause of decidualization failure was the deregulated vascular events (Dey et al. 2004). The present results evidenced that Runx1 might modulate the expression of Cox-2 in the uterine stromal cells. Further studies found that there were two consensus Runx1 binding motifs in the Cox-2 promoter region (Liu et al. 2009). Moreover, mutation of the Runx1 consensus sequence significantly reduced the agonist-stimulated luciferase activity of Cox-2 promoter reporter constructs in preovulatory granulose cells, whereas overexpression of Runx1 increased the agonist-stimulated Cox-2 promoter activity (Liu et al. 2009). Meanwhile, in the uterine stromal cells, Runx1 was also capable of regulating the expression of mPGES-1, which was an inducible enzyme downstream of Cox-2 in prostaglandin E2 biosynthesis and involved in uterine angiogenesis (Numao et al. 2011). Collectively, these results indicate that Runx1 may direct uterine angiogenesis by affecting the expression of Cox-2 and mPGES-1 genes during decidualization. Indeed, several key observations have supported a role of Runx1 in angiogenesis including the finding that mice in which the Runx1 gene has been disrupted die in utero with vascular abnormalities (Okuda et al. 1996; Takakura et al. 2000; Sun et al. 2004; Athilakshmi et al. 2011). Additionally, extracellular matrix (ECM) degradation also occurs in the uterus during decidualization (Dey et al. 2004). Mmps were thought to be key mediators for ECM degradation, because inhibition of total Mmp activity could significantly reduce the length and size of the decidua (Dey et al. 2004; Chen et al. 2009). However, it was unclear whether Runx1 could mediate the expression of Mmp2 and Mmp9 in the uterus, although Mmp2 and Mmp9 were also expressed in the decidualized cells (Bany et al. 2000). The present data showed that inhibition of Runx1 with specific siRNA downregulated the expression of Mmp2 but had no effects on the expression of Mmp9, demonstrating that Runx1 might regulate the uterine ECM degradation through influencing the expression of Mmp2 during decidualization.
It is well established that estrogen and progesterone are essential for embryo implantation and decidualization (Dey et al. 2004; Zhang et al. 2013). Estrogen can induce the expression of Runx1 in the MCF-7 cells and uterine epithelial cells (Giroux et al. 2008; Wall et al. 2013). Similar results were observed in ovariectomized mouse uterus and uterine stromal cells. In estrogen receptor β (ERβ)-deficient mice, estrogen could still stimulate the expression of Runx1 (Giroux et al. 2008), indicating that the stimulation action of estrogen on Runx1 expression could be dependent on ERα rather than ERβ. Emerging evidence has illustrated that Runx1 might mediate the recruitment of ERα to enhancer sites of target genes (Stender et al. 2010). Likewise, progesterone could upregulate the expression of Runx1 mRNA in ovariectomized mouse uterus and uterine stromal cells. In conclusion, Runx1 may play an important role during mouse decidualization.
References
Athilakshmi K, Shanmugasundaram N, Li QX, DeMayo FJ, Lydon JP, Bagchi MK, Bagchi IC (2011) Runx1 functions downstream of BMP2 to regulate uterine stromal differentiation and blood vessel formation at the maternal-fetal interface. Biol Reprod 85(Meeting Abstracts):180
Bai ZK, Guo B, Tian XC, Li DD, Wang ST, Cao H, Wang QY, Yue ZP (2013) Expression and regulation of Runx3 in mouse uterus during the peri-implantation period. J Mol Histol 44:519–526
Bany BM, Harvey MB, Schultz GA (2000) Expression of matrix metalloproteinases 2 and 9 in the mouse uterus during implantation and oil-induced decidualization. J Reprod Fertil 120:125–134
Blyth K, Cameron ER, Neil JC (2005) The RUNX genes: gain or loss of function in cancer. Nat Rev Cancer 5:376–387
Blyth K, Vaillant F, Jenkins A, McDonald L, Pringle MA, Huser C, Stein T, Neil J, Cameron ER (2010) Runx2 in normal tissues and cancer cells: A developing story. Blood Cells Mol Dis 45:117–123
Chen L, Belton RJ Jr, Nowak RA (2009) Basigin-mediated gene expression changes in mouse uterine stromal cells during implantation. Endocrinology 150:966–976
Chimge NO, Frenkel B (2013) The RUNX family in breast cancer: relationships with estrogen signaling. Oncogene 32:2121–2130
Chuang LS, Ito K, Ito Y (2013) RUNX family: Regulation and diversification of roles through interacting proteins. Int J Cancer 132:1260–1271
Dey SK, Lim H, Das SK, Reese J, Paria BC, Daikoku T, Wang H (2004) Molecular cues to implantation. Endocr Rev 25:341–373
Gellersen B, Brosens J (2003) Cyclic AMP and progesterone receptor cross-talk in human endometrium: a decidualizing affair. J Endocrinol 178:357–372
Giroux V, Lemay F, Bernatchez G, Robitaille Y, Carrier JC (2008) Estrogen receptor beta deficiency enhances small intestinal tumorigenesis in ApcMin/+ mice. Int J Cancer 123:303–311
Jo M, Curry TE Jr (2006) Luteinizing hormone-induced RUNX1 regulates the expression of genes in granulosa cells of rat periovulatory follicles. Mol Endocrinol 20:2156–2172
Kilbey A, Terry A, Jenkins A, Borland G, Zhang Q, Wakelam MJ, Cameron ER, Neil JC (2010) Runx regulation of sphingolipid metabolism and survival signaling. Cancer Res 70:5860–5869
Kim JH, Jang JW, Lee YS, Lee JW, Chi XZ, Li YH, Kim MK, Kim DM, Choi BS, Kim J, Kim HM, van Wijnen A, Park I, Bae SC (2014) RUNX family members are covalently modified and regulated by PIAS1-mediated sumoylation. Oncogenesis 3:e101
Kitoh A, Ono M, Naoe Y, Ohkura N, Yamaguchi T, Yaguchi H, Kitabayashi I, Tsukada T, Nomura T, Miyachi Y, Taniuchi I, Sakaguchi S (2009) Indispensable role of the Runx1-Cbfbeta transcription complex for in vivo-suppressive function of FoxP3+ regulatory T cells. Immunity 31:609–620
Klauber N, Rohan RM, Flynn E, D'Amato RJ (1997) Critical components of the female reproductive pathway are suppressed by the angiogenesis inhibitor AGM-1470. Nat Med 3:443–446
Klunker S, Chong MM, Mantel PY, Palomares O, Bassin C, Ziegler M, Rückert B, Meiler F, Akdis M, Littman DR, Akdis CA (2009) Transcription factors RUNX1 and RUNX3 in the induction and suppressive function of Foxp3+ inducible regulatory T cells. J Exp Med 206:2701–2715
Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T (1997) Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89:755–764
Levanon D, Groner Y (2004) Structure and regulated expression of mammalian RUNX genes. Oncogene 23:4211–4219
Liu J, Park ES, Jo M (2009) Runt-related transcription factor 1 regulates luteinized hormone-induced prostaglandin-endoperoxide synthase 2 expression in rat periovulatory granulosa cells. Endocrinology 150:3291–3300
McConaha ME, Eckstrum K, An J, Steinle JJ, Bany BM (2011) Microarray assessment of the influence of the conceptus on gene expression in the mouse uterus during decidualization. Reproduction 141:511–527
Numao A, Hosono K, Suzuki T, Hayashi I, Uematsu S, Akira S, Ogino Y, Kawauchi H, Unno N, Majima M (2011) The inducible prostaglandin E synthase mPGES-1 regulates growth of endometrial tissues and angiogenesis in a mouse implantation model. Biomed Pharmacother 65:77–84
Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR (1996) AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84:321–330
Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ (1997) Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89:765–771
Ozaki T, Nakagawara A, Nagase H (2013) RUNX Family Participates in the Regulation of p53-Dependent DNA Damage Response. Int J Genomics 2013:271347
Rankin JC, Ledford BE, Baggett B (1977) Early involvement of cyclic nucleotides in the artificially stimulated decidual cell reaction in the mouse uterus. Biol Reprod 17:549–554
Sakuma A, Fukamachi H, Ito K, Ito Y, Takeuchi S, Takahashi S (2008) Loss of Runx3 affects ovulation and estrogen-induced endometrial cell proliferation in female mice. Mol Reprod Dev 75:1653–1661
Shao J, Li MQ, Meng YH, Chang KK, Wang Y, Zhang L, Li DJ (2013) Estrogen promotes the growth of decidual stromal cells in human early pregnancy. Mol Hum Reprod 19:655–664
Stender JD, Kim K, Charn TH, Komm B, Chang KC, Kraus WL, Benner C, Glass CK, Katzenellenbogen BS (2010) Genome-wide analysis of estrogen receptor alpha DNA binding and tethering mechanisms identifies Runx1 as a novel tethering factor in receptor-mediated transcriptional activation. Mol Cell Biol 30:3943–3955
Sun L, Vitolo MI, Qiao M, Anglin IE, Passaniti A (2004) Regulation of TGFbeta1-mediated growth inhibition and apoptosis by RUNX2 isoforms in endothelial cells. Oncogene 23:4722–4734
Takakura N, Watanabe T, Suenobu S, Yamada Y, Noda T, Ito Y, Satake M, Suda T (2000) A role for hematopoietic stem cells in promoting angiogenesis. Cell 102:199–209
Tian XC, Wang QY, Li DD, Wang ST, Yang ZQ, Guo B, Yue ZP (2013) Differential expression and regulation of Cryab in mouse uterus during preimplantation period. Reproduction 145:577–585
Wall EH, Hewitt SC, Liu L, del Rio R, Case LK, Lin CY, Korach KS, Teuscher C (2013) Genetic control of estrogen-regulated transcriptional and cellular responses in mouse uterus. FASEB J 27:1874–1886
Wu M, Li C, Zhu G, Wang Y, Jules J, Lu Y, McConnell M, Wang YJ, Shao JZ, Li YP, Chen W (2014) Deletion of core-binding factor β (Cbfβ) in mesenchymal progenitor cells provides new insights into Cbfβ/Runxs complex function in cartilage and bone development. Bone 65:49–59
Zhang S, Lin H, Kong S, Wang S, Wang H, Wang H, Armant DR (2013) Physiological and molecular determinants of embryo implantation. Mol Aspects Med 34:939–980
Acknowledgments
This work was financially supported by the Special Funds for Scientific Research on Public Causes (201303119) and the National Natural Science Foundation of China (31302048, 31372390 and 31472158).
Author information
Authors and Affiliations
Corresponding author
Additional information
Zhi-Kun Bai and Dang-Dang Li contributed equally to this work.
Rights and permissions
About this article
Cite this article
Bai, ZK., Li, DD., Guo, CH. et al. Differential expression and regulation of Runx1 in mouse uterus during the peri-implantation period. Cell Tissue Res 362, 231–240 (2015). https://doi.org/10.1007/s00441-015-2174-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00441-015-2174-z