Abstract
In light sheet-based fluorescence microscopy (LSFM), only the focal plane is illuminated by a laser light sheet. Hence, only the fluorophores within a thin volume of the specimen are excited. This reduces photo-bleaching and photo-toxic effects by several orders of magnitude compared with any other form of microscopy. Therefore, LSFM (aka single/selective-plane illumination microscopy [SPIM] or digitally scanned light sheet microscopy [DSLM]) is the technique of choice for the three-dimensional imaging of live or fixed and of small or large three-dimensional specimens. The parallel recording of millions of pixels with modern cameras provides an extremely fast acquisition speed. Recent developments address the penetration depth, the resolution and the recording speed of LSFM. The impact of LSFM on research areas such as three-dimensional cell cultures, neurosciences, plant biology and developmental biology is increasing at a rapid pace. The development of high-throughput LSFM is the next leap forward, allowing the application of LSFM in toxicology and drug discovery screening.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Principles of light sheet-based fluorescence microscopy
Light sheet-based fluorescence microscopy (LSFM) is revolutionizing the live imaging of three-dimensional (3D) cell cultures and of both live and fixed whole-mount tissue specimens. Zsigmondy and Siedentopf are the acknowledged inventors of the first light sheet-based microscope, the “slit-ultramicroscope” (Mappes et al. 2012). In the 1902 ultramicroscope, the diffraction-limited light sheet was obtained by focusing the sunlight passing an adjustable horizontal slit aperture (Siedentopf and Zsigmondy 1902). Zsigmondy employed the ultramicroscope to answer fundamental questions in colloids physics. This work earned him the 1925 Nobel Prize in chemistry. However, Zsigmondy could not exploit an important property of modern light sheet-based microscopy, namely the possibility of producing images with a greatly increased depth of field by translating the specimen along the detection optical axis. This principle was applied, for the first time, in the “deep-field microscope”, a low-magnification and low-numerical-aperture (N.A.) scanning light sheet microscope that was developed and patented in the 1960s (McLachlan Jr 1964, 1968), which was suitable for the imaging millimeter-sized non-biological specimens. The orthogonal-plane fluorescence optical sectioning macroscope (OPFOS; Voie et al. 1993; Voie and Spelman 1995; Voie 2002) and the confocal theta microscope (Stelzer and Lindek 1994) are the closest forerunners of modern LSFM. The OPFOS macroscope is the first system employing a cylindrical lens to produce a light sheet and to combine fluorescence contrast with light sheet macroscopy. The confocal theta microscope anticipates several theoretical and technical solutions that appear in most of the modern LSFM set-ups, such as water-dipping high N.A. objective lenses (e.g., Carl Zeiss 40×, N.A. 0.75) for both illumination and detection, multiview microscopy by independent observation of the specimen from several sides and a glass capillary serving the mounting and positioning of the specimen.
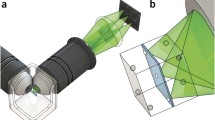
Modern LSFM takes advantage of a modern charge-coupled device (CCD) and complementary metal oxide semiconductor (CMOS) cameras, which allow massive and parallel acquisition of millions of pixels, extremely fast recording, excellent dynamic range, high-quality imaging and high sensitivity. The principle and rationale of LSFM are summarized in Fig. 1. The light sheet is generated by employing a cylindrical lens to form a static light sheet in a single/selective plane illumination microscope (SPIM; Huisken et al. 2004) or a rapidly scanned, diffraction-limited laser line in a digitally scanned laser light sheet-based fluorescence microscope (DSLM; Keller et al. 2008a, b; Tomer et al. 2012). The light sheet of a DSLM is not only generated dynamically but in contrast to the static light sheet of a SPIM, it is incoherent and, therefore, less affected by scatter. Thus, specimen-induced imaging artifacts are reduced. Structured illumination is easily implemented and, in combination with simple imaging processing algorithms, strongly improves the image quality (Breuninger et al. 2007; Neil et al. 1997). Current set-ups provide live imaging of extremely large specimens such as cellular spheroids, whole brains and complete plants at cellular resolution (Ahrens et al. 2013; Lucas et al. 2013; Maizel et al. 2011; Rosquete et al. 2013; Sena et al. 2011).
Rationale of light sheet-based fluorescence microscopy (LSFM). The laser light sheet illumination and wide-field detection of the fluorescence signal at 90° provides optical sectioning (top right). The combination of the point-spread functions (PSF) of the excitation and detection systems (top left) provides a PSF that has an axial extent smaller than that in a confocal microscope equipped with low-numerical-aperture (N.A.) or mid-N.A. objective lenses. The use of a high performance and a fast charge-coupled device (CCD) and complementary metal oxide semiconductor (CMOS) cameras provides an extremely versatile microscope that is well suited for the long-term imaging of live three-dimensional systems. The applications range from single molecule biophysics and disperse systems to three-dimensional cultures and developmental biology (FCS fluorescence correlation spectroscopy, FLIM fluorescence lifetime imaging, FRET Förster resonance energy transfer, FRAP fluorescence recovery after photobleaching, SIM structured illumination microscopy, PALM photo-activated localization microscopy, STORM stochastic optical reconstruction microscopy)
The most recent advances further improve the performance of LSFM in terms of acquisition speed and image quality. New technical variations, which improve specific features of LSFM, are published and demonstrated on an almost monthly basis. Among these are the multidirectional SPIM (mSPIM) and four-lens SPIM (MuVi-SPIM, SiMView), which boost the recording speed of in toto developmental biology studies (Huisken 2012; Krzic et al. 2012; Tomer et al. 2012; Weber and Huisken 2012) and LSFM based on multiple interfering Bessel beams (Chen et al. 2014). Bessel beams (Chen et al. 2014; Fahrbach et al. 2013b; Fahrbach and Rohrbach 2010, 2012; Gao et al. 2012, 2014; Olarte et al. 2012; Planchon et al. 2011) allow one to generate thinner light sheets at the expense of higher illumination angles and higher laser power. Incoherent extended focusing (Dean and Fiolka 2014) and two-photon scanned SPIM (Fahrbach et al. 2013a; Cella Zanacchi et al. 2013; Mahou et al. 2014; Truong et al. 2011) improve the penetration depth but increase the thickness of the light sheet and expose the specimen to extremely high laser powers. Recently, confocal slit detection was combined with scanned light sheet microscopy. Both scanned Gaussian beams (Baumgart and Kubitscheck 2012; Silvestri et al. 2012) and Bessel beams (Fahrbach and Rohrbach 2012; Zhang et al. 2014) were employed in these studies for light sheet excitation. Confocal slit detection significantly improves the image contrast by spatially filtering the blurring attributable to scattered light and to the side lobes of the Bessel beams.
Several versions of LSFM are available commercially. The Lightsheet Z.1 (Carl Zeiss, www.zeiss.com) is based on the original SPIM design (Huisken et al. 2004) and features double-sided illumination and light sheet pivoting that supports an even illumination and a reduction of the stripe artifacts (Huisken and Stainier 2007). A macroscope LSFM based on the design by Voie (2002) and Dodt et al. (2007) is available (http://lavisionbiotec.com/). Finally, a system based on an inverted SPIM (iSPIM) and dual-view inverted SPIM (diSPIM) design (Y. Wu et al. 2011, 2013; J. Wu et al. 2013) is offered as a kit (http://www.asiimaging.com/products/light-sheet-microscopy/). All systems provide LSFM that is adapted to different applications, allowing biologists to take advantage of LSFM and to address biological questions that could not be considered until now.
The increasing popularity of LSFM requires software packages able to process a huge amount of generated data. An important image processing step in light sheet microscopy is the fusion of 3D image stacks recorded at different angles by multiview imaging. Several efficient multiview reconstruction algorithms have been published to date (Preibisch et al. 2010, 2014; Temerinac-Ott et al. 2012).
Combination of LSFM with quantitative single-molecule detection techniques: FCS, FLIM, FRET, localization, STED and Raman microscopy
LSFM boosts the signal-to-noise ratio and allows shorter exposure times and faster acquisition rates. Therefore, it is well suited for the analysis of single molecules in cells. Single molecule imaging and the tracking of fluorescent proteins in live cells with LSFM have been demonstrated and achieve a temporal resolution up to 10 ms (Gebhardt et al. 2013; Ritter et al. 2010). A further intriguing application of LSFM in living specimens is fluorescence correlation spectroscopy (FCS), which requires two necessary conditions: fast acquisition and small observation volume. Modern cameras can meet the first condition and a dynamic range of more than 10 bits. The thin and diffraction-limited light sheet provides a small observation volume extended over the whole field of view. This enables massively parallel FCS measurements. Parallel measurement is essential to generate the data required for analyzing the diffusion in spatially non-homogeneous environments. Recently, the dynamics of fluorescent nano-beads injected into the blood stream of living zebrafish embryos at 48 h post-fertilization has been observed. Interestingly, the flow speed of the beads, which was extracted from the auto-correlation functions (ACFs), were consistent with the movements of single beads, which were directly estimated from the images of time series. This means that the heartbeat rate can be obtained directly from the peaks of the ACF measured close to the heart (Wohland et al. 2010). FCS at the protein level in a living cell is also feasible, e.g., two-dimensional (2D) FCS measurement of the nuclear localization signal (NLS) green fluorescent protein (GFP) in isolated wing imaginal discs of Drosophila melanogaster larvae has been demonstrated (Capoulade et al. 2011). In the same study, protein interactions such as the α isoform of heterochromatin protein 1 (HP1α) with chromatin in 3 T3 cells were investigated. Recently, the technique was also extended to two-color fluorescence cross-correlation spectroscopy (Krieger et al. 2014). In short, the application of LSFM in FCS enables parallel measurement per time interval per sample; this is feasible in heterogeneous 3D specimens and in vivo. More than 100,000 FCS curves can be collected in a single day. Open-source software provides calculations of spatial and temporal auto- and cross-correlations and of the differences in cross-correlation functions (Sankaran et al. 2010). Different 2D array detectors for parallel FCS measurement based on LSFM have also been compared (Singh et al. 2013).
Long-term recording in fluorescence lifetime imaging (FLIM) has been achieved with LSFM (Greger et al. 2011). The combination of Förster resonance energy transfer (FRET) and LSFM has been demonstrated in order to show that it is an ideal approach for monitoring Ca2+ signaling in vivo (Costa et al. 2013). It is also feasible to combine LSFM with super-resolution techniques such as localization and stimulated emission depletion (STED) techniques (Cella Zanacchi et al. 2011). Finally, Raman imaging also benefits from light sheet illumination with improved axial resolution and acquisition speed. The wide-field Raman images of polystyrene beads at a diameter of 20 μm have been acquired within less than 2 min instead of within an hour with conventional confocal Raman spectroscopy (Barman et al. 2010). Raman imaging of a living young fish (Quintet line) has also been achieved (Oshima et al. 2012).
LSFM for 3D cell cultures
Scientists working on drug discovery, toxicology, stem cells and regenerative medicine increasingly employ 3D cell cultures in order to obtain physiologically relevant data (Pampaloni et al. 2007, 2009; Pampaloni and Stelzer 2010). LSFM is optimally suited for the imaging of 3D cell cultures. The high acquisition speed and good penetration depth of LSFM allow the fast recording of particularly large specimens, such as multicellular spheroids (Verveer et al. 2007) and cells embedded in surrogates of the extracellular matrix, e.g., collagen or Matrigel (Pampaloni et al. 2014a). These favorable imaging properties facilitate the prolonged live imaging (up to several days) of 3D cultures (Pampaloni et al. 2014a). Technologists have developed specific mounting protocols for the observation of 3D cultures with LSFM. The easiest approach consists in embedding the specimen in low-melting agarose. This method is particularly well-suited for the multi-view imaging of live and fixed multicellular spheroids (Swoger et al. 2014a; Pampaloni et al. 2013; Verveer et al. 2007; Fig. 2a). However, the embedding of material in full agarose cylinders mechanically constrains the specimen and potentially biases its biological response. In order to overcome this drawback, tiny transparent cylindrical wells (aka beaker) cast with inert hydrogels such as agarose or gelrite (aka phytagel) were employed for the time-lapse observation of multicellular spheroids (Fig. 2b; Lorenzo et al. 2011; (Pampaloni et al. 2015) and of MDCK-cell cysts growing in 3D collagen (Pampaloni et al. 2014a; Fig. 2c, and see below). In the “hydrogel-beaker” method, one cellular spheroid (or a cell suspension) is gently pipetted in the hydrogel beaker. The spheroid or the cells quickly sediment at the bottom (Fig. 2b). Thus, the spheroid forms in the beaker exactly as in the “liquid overlay” method (Friedrich et al. 2009). The mitosis dynamics in large (diameter about 400 μm) live HCT116 human colon carcinoma spheroids growing in hydrogel beakers has been observed over a period of hours by LSFM (Lorenzo et al. 2011). The hydrogel beakers have a cylindrical surface. This can introduce aberration in the images. In order to reduce aberrations, squared cross-section agarose beakers have been produced by using 3D-printed templates (Fig. 3a–c; Desmaison et al. 2013; www.ip3d.fr/IP3D/SPIM/SPIM.html). The squared beakers contain multiple chambers that facilitate the observation of multiple spheroids during one time-lapse experiment in parallel. In general, mounting 3D cell cultures within hydrogels such as agarose (both by embedding or within “agarose beakers”) has the advantage of favoring the diffusion of nutrients and soluble factors to and from the cells. However, several experiments require the confinement of the cells within a completely isolated environment in order to avoid any exchange with the external environment. This is the case in drug screening studies, in which the cells are exposed to a well-defined concentration of drugs, growth factors, cytokines, etc. For these applications, different mounting approaches are necessary. Recently, a microfluidic chamber suitable for LSFM-imaging was achieved by soft lithography with inert and bio-compatible silicone rubber (polydimethysiloxane, PDMS). Single spheroids were cultured in nine cubical micro-wells, with a volume of 200 × 200 × 250 μm3 each. Both the highly transparent planar walls and the geometrical arrangement of the PDMS micro-wells allowed imaging with LSFM (Fig. 3d, e; Patra et al. 2014). Another approach for isolating specimens is by placing them in glass capillaries. However, imaging through cylindrical capillaries is not suitable because of strong optical aberration. The answer to this problem is to employ tiny square-cross section glass capillaries (Pampaloni et al. 2014b; Bruns et al. 2012, 2014). These caplillaries allow the specimen to be confined within a volume of about 20 μl and avoid any mass or gas exchange with the outside environment (Bruns et al. 2012). The squared cross-section allows an aberration-free recording of multiple views of the specimen from four rotation angles (Pampaloni et al. 2014b). If more angles are required, an octagonal cross-section can be employed (Pampaloni et al. 2014b).
Specimen preparation for live imaging of three-dimensional cell cultures with LSFM. a Full embedding of pre-formed multi-cellular spheroids in low-melting agarose in a glass capillary and extrusion of the agarose cylindrical slab for LSFM imaging. a’ Positioning of the mounted spheroid in a medium-filled LSFM chamber. a’’ Enlarged view of the embedded spheroid in front of a water-dipping detection objective lens. Bars 1 mm. Modified from Swoger et al. (2014a). b One multicellular spheroid located at the conical bottom of an “agarose beaker”. Both the agarose beaker and the cellular spheroid are visible under dark field illumination. Image recorded with a stereomicroscope at 25× magnification. b’ Enlarged view of the spheroid at the bottom of the beaker. Phase contrast image recorded with a 20× objective. Bar 300 μm. b’’ Same spheroid, fluorescence image. Marker: H2B-GFP. Modified from Pampaloni et al. (2015). c From left to right, agarose beaker containing MDCK cell cysts embedded in collagen type I gel and three views at increasing magnifications (c’, c’’, c’’’). A single MDCK cell embedded in the gel within the beaker grows over several days forming polarized epithelial hollow spheres (cysts). The morphology of single cysts is visible in c’’’. Bar 1 mm. Modified from Pampaloni et al. (2014a)
Specimen preparation for live imaging of cellular spheroids. a Square-cross section hydrogel beaker containing four separate chambers for the observation of multiple spheroids. Modified from http://www.ip3d.fr/IP3D/SPIM/SPIM.html. b LSFM holder for the square cross-section hydrogel beaker. Modified from http://www.ip3d.fr/IP3D/SPIM/SPIM.html. c Possible geometry of excitation and detection in the square cross-section hydrogel beaker. Each chamber is imaged sequentially. d Microfluidic polydimethylsiloxane (PDMS) microwells suitable for the observation of cellular spheroids with LSFM. e Mounting of the PDMS microwells on the light sheet microscope employed by Patra et al. (2014), whose detection optics consist in an inverted microscope (CCD charge-coupled device). d, e Modified from Patra et al. (2014)
In order to perform long-term time lapse imaging of live specimens, an optimized environmental control system fully integrated with LSFM is required. Tissue-culture LSFM (TC-LSFM; Pampaloni et al. 2014a, 2015) was employed to monitor the development of MDCK cell cysts in a 3D collagen gel. The cell-gel mixture when still in the liquid stage was poured into agarose beakers and allowed to polymerize in the incubator (Fig. 2c). Up to sixty embedded MDCK cells were observed in parallel for up to 5 days. The details of the set-up are shown in Fig. 4. In a further study (Pampaloni et al. 2015), the aggregation, compaction and growth of HeparRG liver cell spheroids was imaged over a period of 150 h (6.25 days) at intervals of 15 min. In total, 600 3D stacks with two channels (transmitted light and fluorescence from the H2B-GFP nuclear marker) were recorded. Each 3D stack contained 135 single frames spaced 2.6 μm along the z-axis (81,000 frames in total over 150 h). The analysis of the time-lapse data showed that the progressive aggregation and compaction of the HepaRG cells lasted for approximately 60 h. From 60 h onwards, spheroid compaction occurred, as inferred from the volume reduction of the spheroid. The mitotic events during the spheroid formation were counted, showing that cell proliferation occurred in correspondence to the outer cell layers.
Tissue-culture LSFM (TC-LSFM) allows the long-term imaging of live three-dimensional cell cultures. a The perfusion chamber, which is made of inert polyoxymethylene (POM, Delrin), features an embedded heater with active temperature control, an inlet and outlet for media perfusion and an airtight flexible cover that allows the “xyz” translation of the specimen and isolates the internal environment from bacterial contamination. b Photograph of the TC-LSFM perfusion chamber. c Overview of the entire perfusion system: CO2/air mixer, gas exchanger box to saturate the flowing medium with the gas mixture of desired composition, supply and waste bottles, peristaltic pump regulating the flow and TC-LSFM chamber. d Development of MDCK cysts over 70 h recorded in three-dimensions by TC-LSFM. Modified from Pampaloni et al. (2014a)
LSFM for plant biology
In contrast to animals, the majority of plant organs develop post-embryonically. Thus, in many instances, organ formation (e.g., the development of lateral roots) occurs within a pre-existing tissue in plants (Maizel et al. 2011). This implies that whole plant specimens, which are usually large, must be imaged in order to observe organ morphogenesis. Hence, the imaging of organ formation in a living plant with a conventional microscope is challenging. In contrast, the imaging of a living plant by LSFM is relatively straightforward. This is because of the long penetration depth and the possibility of mounting the specimen in three-dimensions under close-to-natural conditions. In a study performed with a SPIM, a vertically growing Arabidopsis thaliana root was imaged at cellular resolution every 6 to 15 min over several days. The cell nuclei were tracked and the cell divisions were identified (Sena et al. 2011). In further work, the growth of the lateral roots of Arabidopsis thaliana was observed with the more advanced mDSLM system (Maizel et al. 2011). The plant root was mounted in a phytagel slab, whereas the leaves were in the air. The lateral root was able to grow entirely outside the phytagel. Thus, the light sheet did not pass any substrate during the imaging, ensuring the best image quality. A solar-spectrum light source was used to illuminate the leaves. Multi-color 3D imaging of the living plants was achieved at organ, cellular and subcellular scales over long observation periods with the minimal damage of light to the sample (Maizel et al. 2011). With this approach, many quantitative descriptions of growth processes can be addressed in plants by investigating the interplay between the patterns of cell division, organ shape and surrounding tissues. Dynamic 3D analyses of the morphogenesis of lateral root primordia (LRP) were performed with mDSLM (Lucas et al. 2013) and revealed two important new aspects of LRP morphogenesis. First, atypical radial and tangential cell divisions establish the radial symmetry of the new primordium by forming a ring that confines the growth of the rapidly proliferating cells at the apex. Second, stereotypical patterns of cell division are not essential for correct primordium morphogenesis. Rather, the mechanical properties of the tissue overlying a new primordium are determinant for morphogenesis.
The dynamics of pericycle and endodermis before and during the first asymmetric cell division in LRP formation in Arabidopsis thaliana have also been quantified. The LRP must grow through overlying endodermal cell layers and previous observations with confocal microscopy led to the prediction that the endodermal cells lose volume, change shape and relinquish their tight junctions (i.e., forming holes) to allow the emerging LRP to pass through these layers. In this new study, however, these phenomena were investigated live in a living plant by LSFM. With live imaging, Vermeer et al. (2014) observed, for the first time, that the break in the endodermis and the growth of the pericycle do not occur simultaneously; instead, the endodermis cells shrink first and the primordium cells swell later.
LSFM for developmental biology
The first applications of modern LSFM were the observations of developing model organisms such as medakafish, zebrafish and fruit fly. Since then, further model organisms have been investigated, including Caenorhabditis elegans and the red flour beetle Tribolium castaneum (Strobl and Stelzer 2014). Substantial technical advancement has been made towards real-time in toto imaging of developing embryos with LSFM. The image quality and the recording speed have been significantly increased by using dual-sided illumination and detection, faster and higher-resolving cameras (e.g., scientific CMOS cameras) and improved control electronics (Vladimirov et al. 2014; Ahrens et al. 2013). The imaging depth has been improved with self-reconstructing beams (such as the Bessel and Airy beam; Vettenburg et al. 2014) in combination with multi-photon excitation (Truong et al. 2011; Pantazis and Supatto 2014). Several strategies to improve the rejection of the background signal have been suggested, including structured illumination (Breuninger et al. 2007) and confocal slit detection (Pantazis and Supatto 2014). Several reviews illustrate the contribution of LSFM to advancements in developmental biology (Huisken and Stainier 2009; Tomer et al. 2011; Weber and Huisken 2011). The main challenge is currently to improve the computational efficiency along the whole imaging pipeline, rather than the technical refinement of the microscope hardware (optics, electronics). Indeed, in the case of in toto live embryo imaging, the amount of raw data easily approaches the terabyte range. One key advancement will be to perform as many image processing tasks as possible in real time during live imaging, such as cell segmentation and lineaging (Keller 2013). Alternatively, a radial maximum intensity of the raw data is computed in real time to achieve a 240-fold reduction in data rate, as originally suggested by Keller et al. (2008a, 2008b) for the analysis of microtubule asters and by Schmid et al. (2013) for the analysis of zebrafish embryo development.
LSFM for the neurosciences
The high recording speed of LSFM, combined with its sub-micrometer resolution and low photo-bleaching, provides the means to perform whole-brain neuroanatomy of fixed specimens and whole-brain functional imaging in live animals at cellular and sub-cellular resolution. This addresses essential questions in the neurosciences, such as the finding of the neuronal connections between and within the various regions of the nervous system (i.e., a comprehensive wiring diagram of the brain) and the correlation of patterns of neuronal activity with behavior and sensorial stimulation (Kasthuri and Lichtman 2007). Among other light microscopy techniques, LSFM is contributing to map the connectivity of the mouse brain, the so-called “mesoscopic connectome” project (Oh et al. 2014), whose aim is to build a comprehensive neuroanatomical online atlas of the mouse brain (Osten and Margrie 2013). The combination of fluorescent labeling of long-range synaptic connections with a modified rabies virus, optical clearing (see also the following paragraph) and LSFM imaging has produced a high-resolution 3D map of the neurons connecting the ventral forebrain with the bulbar interneurons (Niedworok et al. 2012). Protocols for the clearing and the LSFM imaging of the mouse brain have been published (Ludovico Silvestri et al. 2013). LSFM is the tool of choice for in vivo functional studies of the brain in model animals. Turaga and Holy (2012) recorded the simultaneous chemosensory response of many thousands of neurons in explants of the mouse vomeronasal organ stimulated with various pheromones. In a further study by the same authors, a large-scale functional mapping of the vomeronasal glomerular layer was carried out by LSFM (Hammen et al. 2014). The mouse brain is opaque and too large for live in toto observation. However, in small and transparent model organisms such as larval zebrafish, the activity of the entire brain can be monitored live. For example, the simultaneous activity of almost all neurons, about 105, was recorded in transgenic animals expressing the fluorescent calcium indicator GCaMP5G, a marker of neuronal activity (Ahrens et al. 2013). DSLM with simultaneous multiview imaging (Tomer et al. 2012) was employed to achieve the required high recording speed. The system records a volume of 800 × 600 × 200 μm3 composed of 41 slices, spaced 5 μm apart, in 1.3 s, i.e., a sampling frequency of 0.8 Hz. Large anatomical regions showing a correlated neuronal activity (functional circuits) were mapped and two functional oscillatory circuits consistently present in the larval brain were found. One circuit was located in the hindbrain behind the cerebellum, whereas the other was part of the lateral inferior olive. In a further study, LSFM-based whole-brain functional imaging was combined with visual inputs to induce fictive swimming in a paralyzed larval zebrafish. The original dual-side laser sheet illumination was modified in order to avoid a bias in the data attributable to the stimulation of the retina by the light sheet. With this new method, the neuronal activity in the live zebrafish larva could be observed in real-time during visuo-motor behavior (Vladimirov et al. 2014).
Three-dimensional models of human nervous tissue (aka neurospheres or central nervous system [CNS] organoids) based on, for example, differentiated iPSC (induced pluripotent stem cells; Lancaster et al. 2013) or progenitor cells are available (Brito et al. 2012). These tissue models are increasingly important for drug and toxicity screening. LSFM is emerging as an important imaging tool for analyzing both the morphology and dynamic processes in CNS organoids (Gualda et al. 2014) and in organoids mimicking various other tissues (Pampaloni et al. 2013; Swoger et al. 2014a, b).
Outlook: high-throughput LSFM and “microtome-free histology”
The technical improvements of LSFM have focused on increasing the recording speed for high-end imaging of developing embryos or of the beating heart in zebrafish embryos (Tomer et al. 2012; Mickoleit et al. 2014; Krzic et al. 2012). Nevertheless, one obviously unsolved limitation of LSFM is the low throughput, which limits its potential for screening a large number of specimens. Multi-specimen imaging has been successfully performed on collagen-embedded MDCK cells by TC-LSFM (Pampaloni et al. 2014a). However, in the case of large samples, such as large spheroids or embryos, usually only one single specimen per experiment can be realistically recorded. A suggested system able to increase the throughput of LSFM is similar to that of a flow cytometer, in which the samples (e.g., cellular spheroids) are passed through a square cross-section glass capillary (Bruns et al. 2012; J. Wu et al. 2013; Y. Wu et al. 2013). A completely different approach to increasing the throughput of LSFM-imaging consists in using a “farm” of LSFM set-ups working in parallel. The open-access project “OpenSPIM” (http://openspim.org) provides a blueprint for building a compact, relatively inexpensive LSFM set-up that could be suitable for building a SPIM farm (Pitrone et al. 2013; Fig. 5). However, the gold standard for high-throughput imaging is a substrate such as 96- or 384-well plates. Multi-well plates are ubiquitous in cellular assays. A huge choice of laboratory equipment used in high-throughput assays works in combination with micro-well plates, ranging from multiple-tip pipettes to fully automated pipetting robots. Thus, a truly practicable high-throughput screening LSFM should be compatible with the geometry of multi-well plates and with standard motorized microscope stages. Indeed, a module adding LSFM functionality to an inverted microscope has been developed (so-called iSPIM/diSPIM; Y. Wu et al. 2011, 2013; J. Wu et al. 2013) and is also available commercially (http://www.asiimaging.com/products/light-sheet-microscopy/). Whereas iSPIM/diSPIM can image specimens in Petri dishes and in similar planar substrates, it is not suitable for multi-wells because of geometrical constraints. Clearly, new ideas are needed in order to combine LSFM imaging with current high-throughput screening technology, e.g., oblique plane microscopy (OPM) might be feasible (Abstract book p 64, 1st LightSheet Fluorescence Microscopy International Conference, 25–26 Sept. 2014, http://www.lsfm2014.com/). A recent technical innovation assessed this issue and a high-throughput LSFM could soon be a reality (F. Pampaloni, E.H.K. Stelzer, patent application submitted).
Towards high-throughput LSFM. a OpenSPIM, a compact, relatively low-cost, open-access LSFM from Pitrone et al. (2013). b An “OpenSPIM farm”, an ensemble of LSFM working in parallel as a medium-throughput imaging solution in developmental biology from http://openspim.org/File:2I_1D_OpenSPIM_farm_02.jpg
A further field in which LSFM is going to play an important role is microtome-free histology (Dobosz et al. 2014). The idea is to employ optical clearing methods, in combination with 3D fluorescence microscopy, in order to perform high-resolution optical sectioning of tumor or organ biopsies (Fig. 6). This approach, once clinically validated, could replace classical histologal methods. Optical clearing works by immersing fixed specimens in solutions matching the refractive index of the specimen. The medium penetrates the tissue minimizing the differences in the refractive index. Thus, whereas specimens become transparent, the scattering of light is strongly reduced. Since the re-discovery of the classical Spalteholz clearing method in combination with LSFM (Dodt et al. 2007; Keller and Dodt 2012), several new index-matching clearing solutions have been described (Ertürk et al. 2012; Spence et al. 2014; Susaki et al. 2014; Tomer et al. 2014; Yang et al. 2014). We anticipate that the high recording speed of LSFM combined with optical clearing will open completely new perspectives in this emerging alternative method to classical histology.
“Microtome-free histology”. a Optically cleared KPL-4 xenograft tumor imaged with LSFM recording the autofluorescence signal. b Upon optical slicing, i.e., optical clearing and collection of the complete three-dimensional stack of images in an LSFM, the same specimen was embedded in paraffin, mechanically sliced with a microtome and stained with hematoxylin and eosin. Comparison of a, b shows a perfect correlation between optical and mechanical slices. Necrotic areas (white arrows) and solid tumor areas (yellow arrows) are clearly distinguishable in both images. c Volume rendering of an optically cleared KPL-4 xenograft tumor imaged with LSFM, showing blood vessels. d Same specimen as in c showing the penetration of Trastuzumab-Alexa 750 inside the tumor. Bars 250 μm
Concluding remarks
LSFM is the culmination of developments in biology, chemistry, physics and technology of the past century, e.g., the development of fluorophores, cameras and the laser. Without any or each of them, in particular without the laser, LSFM will not work. Consequently, LSFM is not merely a newly developed technique but it truly shifts the paradigm of modern fluorescence microscopy. This is not only because it can be applied to almost all kinds of specimens, e.g., the whole embryo or the whole plant, thick tissue such as brain, a tissue-like spheroid, a single cell and a single molecule or particle but also because it can be combined with almost any kind of imaging technique, e.g., FCS, FLIM, FRET, Raman imaging, structured illumination, super-resolution localization and STED. The main motivation behind the development of LSFM is the avoidance of the waste of fluorophores during the imaging process. LSFM provides for low photo-bleaching and low photo-toxicity, true optical sectioning, long-term and in vivo observations, three dimensionality and live imaging, all being available at the same time without any compromise. More techniques are currently developed around LSFM, e.g., sample preparation methodologies, high-volume image processing and data analyses and the storage and/or transmission of huge datasets. Thus, we can reasonably say that LSFM is not only opening a new field in microscopy but is also pushing development in bioinformatics, biophysics and drug development.
References
Ahrens MB, Orger MB, Robson DN, Li JM, Keller PJ (2013) Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat Methods 10:413–420. doi:10.1038/nmeth.2434
Barman I, Tan KM, Singh GP (2010) Optical sectioning using single-plane-illumination Raman imaging. J Raman Spectrosc 41:1099–1101. doi:10.1002/jrs.2785
Baumgart E, Kubitscheck U (2012) Scanned light sheet microscopy with confocal slit detection. Opt Express 20:21805–21814. doi:10.1364/OE.20.021805
Breuninger T, Greger K, Stelzer EHK (2007) Lateral modulation boosts image quality in single plane illumination fluorescence microscopy. Opt Lett 32:1938. doi:10.1364/OL.32.001938
Brito C, Simão D, Costa I, Malpique R, Pereira CI, Fernandes P, Alves PM (2012) 3D cultures of human neural progenitor cells: dopaminergic differentiation and genetic modification. [corrected]. Methods 56:452–460. doi:10.1016/j.ymeth.2012.03.005
Bruns T, Schickinger S, Wittig R, Schneckenburger H (2012) Preparation strategy and illumination of three-dimensional cell cultures in light sheet-based fluorescence microscopy. J Biomed Opt 17:101518. doi:10.1117/1.JBO.17.10.101518
Bruns T, Schickinger S, Schneckenburger H (2014) Single plane illumination module and micro-capillary approach for a wide-field microscope. J Vis Exp 90:e51993. doi:10.3791/51993
Capoulade J, Wachsmuth M, Hufnagel L, Knop M (2011) Quantitative fluorescence imaging of protein diffusion and interaction in living cells. Nat Biotechnol 29:835–839. doi:10.1038/nbt.1928
Cella Zanacchi F, Lavagnino Z, Perrone Donnorso M, Del Bue A, Furia L, Faretta M, Diaspro A (2011) Live-cell 3D super-resolution imaging in thick biological samples. Nat Methods 8:1047–1049. doi:10.1038/nmeth.1744
Cella Zanacchi F, Lavagnino Z, Faretta M, Furia L, Diaspro A (2013) Light-sheet confined super-resolution using two-photon photoactivation. PLoS One 8:e67667. doi:10.1371/journal.pone.0067667
Chen B-C, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, Betzig E (2014) Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science 346:1257998. doi:10.1126/science.1257998
Costa A, Candeo A, Fieramonti L, Valentini G, Bassi A (2013) Calcium dynamics in root cells of Arabidopsis thaliana visualized with selective plane illumination microscopy. PLoS One 8:e75646. doi:10.1371/journal.pone.0075646
Dean KM, Fiolka R (2014) Uniform and scalable light-sheets generated by extended focusing. Opt Express 22:26141. doi:10.1364/OE.22.026141
Desmaison A, Lorenzo C, Rouquette J, Ducommun B, Lobjois V (2013) A versatile sample holder for single plane illumination microscopy. J Microsc 251:128–132. doi:10.1111/jmi.12051
Dobosz M, Ntziachristos V, Scheuer W, Strobel S (2014) Multispectral fluorescence ultramicroscopy: three-dimensional visualization and automatic quantification of tumor morphology, drug penetration, and antiangiogenic treatment response. Neoplasia 16:1–W7. doi:10.1593/neo.131848
Dodt H-U, Leischner U, Schierloh A, Jährling N, Mauch CP, Deininger K, Becker K (2007) Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat Methods 4:331–336. doi:10.1038/nmeth1036
Ertürk A, Becker K, Jährling N, Mauch CP, Hojer CD, Egen JG, Dodt H-U (2012) Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat Protoc 7:1983–1995. doi:10.1038/nprot.2012.119
Fahrbach FO, Rohrbach A (2010) A line scanned light-sheet microscope with phase shaped self-reconstructing beams. Opt Express 18:24229–24244. doi:10.1364/OE.18.024229
Fahrbach FO, Rohrbach A (2012) Propagation stability of self-reconstructing Bessel beams enables contrast-enhanced imaging in thick media. Nat Commun 3:632. doi:10.1038/ncomms1646
Fahrbach FO, Gurchenkov V, Alessandri K, Nassoy P, Rohrbach A (2013a) Light-sheet microscopy in thick media using scanned Bessel beams and two-photon fluorescence excitation. Opt Express 21:13824–13839. doi:10.1364/OE.21.013824
Fahrbach FO, Gurchenkov V, Alessandri K, Nassoy P, Rohrbach A (2013b) Self-reconstructing sectioned Bessel beams offer submicron optical sectioning for large fields of view in light-sheet microscopy. Opt Express 21:11425–11440. doi:10.1364/OE.21.011425
Friedrich J, Seidel C, Ebner R, Kunz-Schughart LA (2009) Spheroid-based drug screen: considerations and practical approach. Nat Protoc 4:309–324. doi:10.1038/nprot.2008.226
Gao L, Shao L, Higgins CD, Poulton JS, Peifer M, Davidson MW, Betzig E (2012) Noninvasive imaging beyond the diffraction limit of 3D dynamics in thickly fluorescent specimens. Cell 151:1370–1385. doi:10.1016/j.cell.2012.10.008
Gao L, Shao L, Chen B-C, Betzig E (2014) 3D live fluorescence imaging of cellular dynamics using Bessel beam plane illumination microscopy. Nat Protoc 9:1083–1101. doi:10.1038/nprot.2014.087
Gebhardt JCM, Suter DM, Roy R, Zhao ZW, Chapman AR, Basu S, Xie XS (2013) Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nat Methods 10:421–426. doi:10.1038/nmeth.2411
Greger K, Neetz MJ, Reynaud EG, Stelzer EHK (2011) Three-dimensional fluorescence lifetime imaging with a single plane illumination microscope provides an improved signal to noise ratio. Opt Express 19:20743–20750. doi:10.1364/OE.19.020743
Gualda EJ, Simão D, Pinto C, Alves PM, Brito C (2014) Imaging of human differentiated 3D neural aggregates using light sheet fluorescence microscopy. Front Cell Neurosci 8:221. doi:10.3389/fncel.2014.00221
Hammen GF, Turaga D, Holy TE, Meeks JP (2014) Functional organization of glomerular maps in the mouse accessory olfactory bulb. Nat Neurosci 17:953–961. doi:10.1038/nn.3738
Huisken J (2012) Slicing embryos gently with laser light sheets. Bioessays 34:406–411. doi:10.1002/bies.201100120
Huisken J, Stainier DYR (2007) Even fluorescence excitation by multidirectional selective plane illumination microscopy (mSPIM). Opt Lett 32:2608. doi:10.1364/OL.32.002608
Huisken J, Stainier DYR (2009) Selective plane illumination microscopy techniques in developmental biology. Development 136:1963–1975. doi:10.1242/dev.022426
Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EHK (2004) Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305:1007–1009. doi:10.1126/science.1100035
Kasthuri N, Lichtman JW (2007) The rise of the “projectome”. Nat Methods 4:307–308. doi:10.1038/nmeth0407-307
Keller PJ (2013) Imaging morphogenesis: technological advances and biological insights. Science 340:1234168. doi:10.1126/science.1234168
Keller PJ, Dodt H-U (2012) Light sheet microscopy of living or cleared specimens. Curr Opin Neurobiol 22:138–143. doi:10.1016/j.conb.2011.08.003
Keller PJ, Pampaloni F, Lattanzi G, Stelzer EHK (2008a) Three-dimensional microtubule behavior in Xenopus egg extracts reveals four dynamic states and state-dependent elastic properties. Biophys J 95:1474–1486. doi:10.1529/biophysj.107.128223
Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EHK (2008b) Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science 322:1065–1069. doi:10.1126/science.1162493
Krieger JW, Singh AP, Garbe CS, Wohland T, Langowski J (2014) Dual-color fluorescence cross-correlation spectroscopy on a single plane illumination microscope (SPIM-FCCS). Opt Express 22:2358–2375. doi:10.1364/OE.22.002358
Krzic U, Gunther S, Saunders TE, Streichan SJ, Hufnagel L (2012) Multiview light-sheet microscope for rapid in toto imaging. Nat Methods 9:730–733. doi:10.1038/nmeth.2064
Lancaster MA, Renner M, Martin C-A, Wenzel D, Bicknell LS, Hurles ME, Knoblich JA (2013) Cerebral organoids model human brain development and microcephaly. Nature 501:373–379. doi:10.1038/nature12517
Lorenzo C, Frongia C, Jorand R, Fehrenbach J, Weiss P, Maandhui A, Lobjois V (2011) Live cell division dynamics monitoring in 3D large spheroid tumor models using light sheet microscopy. Cell Div 6:22. doi:10.1186/1747-1028-6-22
Lucas M, Kenobi K, von Wangenheim D, Voβ U, Swarup K, De Smet I, Bennett MJ (2013) Lateral root morphogenesis is dependent on the mechanical properties of the overlaying tissues. Proc Natl Acad Sci U S A 110:5229–5234. doi:10.1073/pnas.1210807110
Mahou P, Vermot J, Beaurepaire E, Supatto W (2014) Multicolor two-photon light-sheet microscopy. Nat Methods 11:600–601. doi:10.1038/nmeth.2963
Maizel A, von Wangenheim D, Federici F, Haseloff J, Stelzer EHK (2011) High-resolution live imaging of plant growth in near physiological bright conditions using light sheet fluorescence microscopy. Plant J 68:377–385. doi:10.1111/j.1365-313X.2011.04692.x
Mappes T, Jahr N, Csaki A, Vogler N, Popp J, Fritzsche W (2012) The invention of immersion ultramicroscopy in 1912—the birth of nanotechnology? Angew Chem 51:11208–11212. doi:10.1002/anie.201204688
McLachlan D Jr (1964) Extreme focal depth in microscopy. Appl Opt 3:1009. doi:10.1364/AO.3.001009
McLachlan D Jr (1968) Microscope. US. Retrieved from https://www.google.com/patents/US3398634?dq=D.+McLachlan+j.+microscope&hl=en&sa=X&ei=na8HVMh9wbI8_uWBoAI&ved=0CCIQ6AEwAA
Mickoleit M, Schmid B, Weber M, Fahrbach FO, Hombach S, Reischauer S, Huisken J (2014) High-resolution reconstruction of the beating zebrafish heart. Nat Methods 11:919-922. doi:10.1038/nmeth.3037
Neil MAA, Juskaitis R, Wilson T (1997) Method of obtaining optical sectioning by using structured light in a conventional microscope. Opt Lett 22:1905. doi:10.1364/OL.22.001905
Niedworok CJ, Schwarz I, Ledderose J, Giese G, Conzelmann K-K, Schwarz MK (2012) Charting monosynaptic connectivity maps by two-color light-sheet fluorescence microscopy. Cell Rep 2:1375–1386. doi:10.1016/j.celrep.2012.10.008
Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, Zeng H (2014) A mesoscale connectome of the mouse brain. Nature 508:207–214. doi:10.1038/nature13186
Olarte OE, Licea-Rodriguez J, Palero JA, Gualda EJ, Artigas D, Mayer J, Loza-Alvarez P (2012) Image formation by linear and nonlinear digital scanned light-sheet fluorescence microscopy with Gaussian and Bessel beam profiles. Biomed Opt Express 3:1492–1505. doi:10.1364/BOE.3.001492
Oshima Y, Sato H, Kajiura-Kobayashi H, Kimura T, Naruse K, Nonaka S (2012) Light sheet-excited spontaneous Raman imaging of a living fish by optical sectioning in a wide field Raman microscope. Opt Express 20:16195. doi:10.1364/OE.20.016195
Osten P, Margrie TW (2013) Mapping brain circuitry with a light microscope. Nat Methods 10:515–523. doi:10.1038/nmeth.2477
Pampaloni F, Stelzer E (2010) Three-dimensional cell cultures in toxicology. Biotechnol Genet Eng Rev 26:117–38. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/21415878
Pampaloni F, Reynaud EG, Stelzer EHK (2007) The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol 8:839–845. doi:10.1038/nrm2236
Pampaloni F, Stelzer EHK, Masotti A (2009) Three-dimensional tissue models for drug discovery and toxicology. Recent Patents Biotechnol 3:103–117. doi:10.2174/187220809788700201
Pampaloni F, Ansari N, Stelzer EHK (2013) High-resolution deep imaging of live cellular spheroids with light-sheet-based fluorescence microscopy. Cell Tissue Res 352:161–177. doi:10.1007/s00441-013-1589-7
Pampaloni F, Berge U, Marmaras A, Horvath P, Kroschewski R, Stelzer EHK (2014a) Tissue-culture light sheet fluorescence microscopy (TC-LSFM) allows long-term imaging of three-dimensional cell cultures under controlled conditions. Integr Biol (Camb) 6:988-998. doi:10.1039/c4ib00121d
Pampaloni F, Stelzer EHK, Mattheyer C (2014b) Kapillarzelle, anordnung und verfahren zur aufnahme, zur positionierung und zur untersuchung einer mikroskopischen probe. Retrieved from https://www.google.com/patents/WO2014033320A1?cl=de&dq=francesco+pampaloni&hl=en&sa=X&ei=_yIUVMjOHcHMyAPRq4GwDQ&ved=0CDsQ6AEwBA
Pampaloni F, Richa R, Ansari N, Stelzer EHK (2015) Live spheroid formation recorded with light sheet-based fluorescence microscopy. Methods Mol Biol 1251:43-57. doi: 10.1007/978-1-4939-2080-8_3
Pantazis P, Supatto W (2014) Advances in whole-embryo imaging: a quantitative transition is underway. Nat Rev Mol Cell Biol 15:327–339. doi:10.1038/nrm3786
Patra B, Peng Y-S, Peng C-C, Liao W-H, Chen Y-A, Lin K-H, Lee C-H (2014) Migration and vascular lumen formation of endothelial cells in cancer cell spheroids of various sizes. Biomicrofluidics 8:052109. doi:10.1063/1.4895568
Pitrone PG, Schindelin J, Stuyvenberg L, Preibisch S, Weber M, Eliceiri KW, Tomancak P (2013) OpenSPIM: an open-access light-sheet microscopy platform. Nat Methods 10:598–599. doi:10.1038/nmeth.2507
Planchon TA, Gao L, Milkie DE, Davidson MW, Galbraith JA, Galbraith CG, Betzig E (2011) Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat Methods 8:417–423. doi:10.1038/nmeth.1586
Preibisch S, Saalfeld S, Schindelin J, Tomancak P (2010) Software for bead-based registration of selective plane illumination microscopy data. Nat Methods 7:418–419. doi:10.1038/nmeth0610-418
Preibisch S, Amat F, Stamataki E, Sarov M, Singer RH, Myers E, Tomancak P (2014) Efficient Bayesian-based multiview deconvolution. Nat Methods 11:645–648. doi:10.1038/nmeth.2929
Ritter JG, Veith R, Veenendaal A, Siebrasse JP, Kubitscheck U (2010) Light sheet microscopy for single molecule tracking in living tissue. PLoS One 5:e11639. doi:10.1371/journal.pone.0011639
Rosquete MR, von Wangenheim D, Marhavý P, Barbez E, Stelzer EHK, Benková E, Kleine-Vehn J (2013) An auxin transport mechanism restricts positive orthogravitropism in lateral roots. Curr Biol 23:817–822. doi:10.1016/j.cub.2013.03.064
Sankaran J, Shi X, Ho LY, Stelzer EHK, Wohland T (2010) ImFCS: a software for imaging FCS data analysis and visualization. Opt Express 18:25468–25481. doi:10.1364/OE.18.025468
Schmid B, Shah G, Scherf N, Weber M, Thierbach K, Campos CP, Huisken J (2013) High-speed panoramic light-sheet microscopy reveals global endodermal cell dynamics. Nat Commun 4:2207. doi:10.1038/ncomms3207
Sena G, Frentz Z, Birnbaum KD, Leibler S (2011) Quantitation of cellular dynamics in growing Arabidopsis roots with light sheet microscopy. PLoS One 6:e21303. doi:10.1371/journal.pone.0021303
Siedentopf H, Zsigmondy R (1902) Über Sichtbarmachung und Größenbestimmung ultramikoskopischer Teilchen, mit besonderer Anwendung auf Goldrubingläser. Ann Phys 315:1–39. doi:10.1002/andp.19023150102
Silvestri L, Bria A, Sacconi L, Iannello G, Pavone FS (2012) Confocal light sheet microscopy: micron-scale neuroanatomy of the entire mouse brain. Opt Express 20:20582–20598. doi:10.1364/OE.20.020582
Silvestri L, Bria A, Costantini I, Sacconi L, Peng H, Iannello G, Pavone FS (2013) Micron-scale resolution optical tomography of entire mouse brains with confocal light sheet microscopy. J Vis Exp 80:e50696. doi:10.3791/50696
Singh AP, Krieger JW, Buchholz J, Charbon E, Langowski J, Wohland T (2013) The performance of 2D array detectors for light sheet based fluorescence correlation spectroscopy. Opt Express 21:8652–8668. doi:10.1364/OE.21.008652
Spence RD, Kurth F, Itoh N, Mongerson CRL, Wailes SH, Peng MS, MacKenzie-Graham AJ (2014) Bringing CLARITY to gray matter atrophy. Neuroimage 101:625-632. doi:10.1016/j.neuroimage.2014.07.017
Stelzer EHK, Lindek S (1994) Fundamental reduction of the observation volume in far-field light microscopy by detection orthogonal to the illumination axis: confocal theta microscopy. Opt Commun 111:536–547. doi:10.1016/0030-4018(94)90533-9
Strobl F, Stelzer EHK (2014) Non-invasive long-term fluorescence live imaging of Tribolium castaneum embryos. Development 141:2331–2338. doi:10.1242/dev.108795
Susaki EA, Tainaka K, Perrin D, Kishino F, Tawara T, Watanabe TM, Ueda HR (2014) Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell 157:726–739. doi:10.1016/j.cell.2014.03.042
Swoger J, Pampaloni F, Stelzer EHK (2014a) Imaging cellular spheroids with a single (selective) plane illumination microscope. Cold Spring Harb Protoc 2014:106–113. doi:10.1101/pdb.prot080176
Swoger J, Pampaloni F, Stelzer EHK (2014b) Light-sheet-based fluorescence microscopy for three-dimensional imaging of biological samples. Cold Spring Harb Protoc 2014:1–8. doi:10.1101/pdb.top080168
Temerinac-Ott M, Ronneberger O, Ochs P, Driever W, Brox T, Burkhardt H (2012) Multiview deblurring for 3-D images from light-sheet-based fluorescence microscopy. IEEE Trans Image Process 21:1863–1873. doi:10.1109/TIP.2011.2181528
Tomer R, Khairy K, Keller PJ (2011) Shedding light on the system: studying embryonic development with light sheet microscopy. Curr Opin Genet Dev 21:558–565. doi:10.1016/j.gde.2011.07.003
Tomer R, Khairy K, Amat F, Keller PJ (2012) Quantitative high-speed imaging of entire developing embryos with simultaneous multiview light-sheet microscopy. Nat Methods 9:755–763. doi:10.1038/nmeth.2062
Tomer R, Ye L, Hsueh B, Deisseroth K (2014) Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat Protoc 9:1682–1697. doi:10.1038/nprot.2014.123
Truong TV, Supatto W, Koos DS, Choi JM, Fraser SE (2011) Deep and fast live imaging with two-photon scanned light-sheet microscopy. Nat Methods 8:757–760. doi:10.1038/nmeth.1652
Turaga D, Holy TE (2012) Organization of vomeronasal sensory coding revealed by fast volumetric calcium imaging. J Neurosci 32:1612–1621. doi:10.1523/JNEUROSCI. 5339-11.2012
Vermeer JEM, von Wangenheim D, Barberon M, Lee Y, Stelzer EHK, Maizel A, Geldner N (2014) A spatial accommodation by neighboring cells is required for organ initiation in Arabidopsis. Science 343:178–183. doi:10.1126/science.1245871
Verveer PJ, Swoger J, Pampaloni F, Greger K, Marcello M, Stelzer EHK (2007) High-resolution three-dimensional imaging of large specimens with light sheet-based microscopy. Nat Methods 4:311–313. doi:10.1038/NMETH1017
Vettenburg T, Dalgarno HIC, Nylk J, Coll-Lladó C, Ferrier DEK, Čižmár T, Dholakia K (2014) Light-sheet microscopy using an Airy beam. Nat Methods 11:541–544. doi:10.1038/nmeth.2922
Vladimirov N, Mu Y, Kawashima T, Bennett DV, Yang C-T, Looger LL, Ahrens MB (2014) Light-sheet functional imaging in fictively behaving zebrafish. Nat Methods 11:883-884. doi:10.1038/nmeth.3040
Voie AH (2002) Imaging the intact guinea pig tympanic bulla by orthogonal-plane fluorescence optical sectioning microscopy. Hear Res 171:119–128. doi:10.1016/S0378-5955(02)00493-8
Voie AH, Spelman FA (1995) Three-dimensional reconstruction of the cochlea from two-dimensional images of optical sections. Comput Med Imaging Graph 19:377–384. doi:10.1016/0895-6111(95)00034-8
Voie AH, Burns DH, Spelman FA (1993) Orthogonal-plane fluorescence optical sectioning: three-dimensional imaging of macroscopic biological specimens. J Microsc 170:229–236. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/8371260
Weber M, Huisken J (2011) Light sheet microscopy for real-time developmental biology. Curr Opin Genet Dev 21:566–572. doi:10.1016/j.gde.2011.09.009
Weber M, Huisken J (2012) Omnidirectional microscopy. Nat Methods 9:656–657. doi:10.1038/nmeth.2022
Wohland T, Shi X, Sankaran J, Stelzer EHK (2010) Single plane illumination fluorescence correlation spectroscopy (SPIM-FCS) probes inhomogeneous three-dimensional environments. Opt Express 18:10627–10641. doi:10.1364/OE.18.010627
Wu J, Li J, Chan RKY (2013) A light sheet based high throughput 3D-imaging flow cytometer for phytoplankton analysis. Opt Express 21:14474–14480. doi:10.1364/OE.21.014474
Wu Y, Ghitani A, Christensen R, Santella A, Du Z, Rondeau G, Shroff H (2011) Inverted selective plane illumination microscopy (iSPIM) enables coupled cell identity lineaging and neurodevelopmental imaging in Caenorhabditis elegans. Proc Natl Acad Sci U S A 108:17708–17713. doi:10.1073/pnas.1108494108
Wu Y, Wawrzusin P, Senseney J, Fischer RS, Christensen R, Santella A, Shroff H (2013) Spatially isotropic four-dimensional imaging with dual-view plane illumination microscopy. Nat Biotechnol 31:1032–1038. doi:10.1038/nbt.2713
Yang B, Treweek JB, Kulkarni RP, Deverman BE, Chen C-K, Lubeck E, Gradinaru V (2014) Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 158:945–958. doi:10.1016/j.cell.2014.07.017
Zhang P, Phipps ME, Goodwin PM, Werner JH (2014) Confocal line scanning of a Bessel beam for fast 3D imaging. Opt Lett 39:3682–3685. doi:10.1364/OL.39.003682
Acknowledgments
We thank Nariman Ansari for many discussions on 3D cell culture, Daniel von Wangenheim for the comments on LSFM applications in plant research and Christian Mattheyer for his contribution to optical clearing.
Author information
Authors and Affiliations
Corresponding author
Additional information
The research of the authors was funded by the Cluster of Excellence for Macromolecular Complexes (CEF-MC, EXC-115) granted to the Goethe Universität Frankfurt am Main by the Deutsche Forschungsgemeinschaft (DFG) and by the German Ministry for Education and Research (BMBF, Forschungsschwerpunkt Biophotonik IV, project ProMEBS) and additionally by the National Science Council (NSC100-2917-I-564-030) in Taiwan, R.O.C. and EMBO (ASTF 404–2012).
Rights and permissions
About this article
Cite this article
Pampaloni, F., Chang, BJ. & Stelzer, E.H.K. Light sheet-based fluorescence microscopy (LSFM) for the quantitative imaging of cells and tissues. Cell Tissue Res 360, 129–141 (2015). https://doi.org/10.1007/s00441-015-2144-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00441-015-2144-5