Abstract
The central nervous system-specific serotonin receptor 2C (5HT2C) controls key physiological functions, such as food intake, anxiety, and motoneuron activity. Its deregulation is involved in depression, suicidal behavior, and spasticity, making it the target for antipsychotic drugs, appetite controlling substances, and possibly anti-spasm agents. Through alternative pre-mRNA splicing and RNA editing, the 5HT2C gene generates at least 33 mRNA isoforms encoding 25 proteins. The 5HT2C is a G-protein coupled receptor that signals through phospholipase C, influencing the expression of immediate/early genes like c-fos. Most 5HT2C isoforms show constitutive activity, i.e., signal without ligand binding. The constitutive activity of 5HT2C is decreased by pre-mRNA editing as well as alternative pre-mRNA splicing, which generates a truncated isoform that switches off 5HT2C receptor activity through heterodimerization; showing that RNA processing regulates the constitutive activity of the 5HT2C system. RNA processing events influencing the constitutive activity target exon Vb that forms a stable double stranded RNA structure with its downstream intron. This structure can be targeted by small molecules and oligonucleotides that change exon Vb alternative splicing and influence 5HT2C signaling in mouse models, leading to a reduction in food intake. Thus, the 5HT2C system is a candidate for RNA therapy in multiple models of CNS disorders.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Overview
Serotonin is an important neurotransmitter, both in the central nervous system and the periphery. There are 14 distinct receptors for serotonin, of which 13 couple to G-proteins and one is a ligand gated ion channel. Depending on the subtype, the receptors couple to G-alphai, G-alphaq11, or G-alphas and with one exception (5HT5B) are expressed in the brain, where they regulate various functions, such as appetite, mood, and sleep cycles making them important pharmaceutical drug targets (McCorvy and Roth 2015).
A well-studied receptor subtype is the serotonin receptor 2C (5HT2C), a seven transmembrane, G-protein coupled receptor that is involved in regulating key brain functions including food intake, anxiety, stress response, and sleep (Heisler et al. 2007; Mongeau et al. 2010; Monti 2011; Tecott et al. 1995). A deregulation of 5HT2C receptors is seen in depression (Martin et al. 2014), schizophrenia (Castensson et al. 2003, 2005), suicidal behavior (Gurevich et al. 2002a), and spinal cord injury (Murray et al. 2010) in humans as well as in mouse models of obesity (Schellekens et al. 2012). The understanding of 5HT2C’s role in appetite control resulted in the development of Lorcarserin, an approved drug for the short-term treatment of obesity, which enhances the activity of the 5HT2C receptor (Greenway et al. 2016). A large number of antipsychotic drugs used to treat depression, anxiety, and schizophrenic disorders interact with the 5HT2C (Chagraoui et al. 2016; Martin et al. 2014), which contributes to the drug’s efficacy, but frequently causes weight gain as a major side effect (Shams and Muller 2014). Based upon its constitutive expression in motoneurons after spinal cord injury (Murray et al. 2010), targeting the 5HT2C in the spinal cord system may hold great potential for combating uncontrolled muscle spasms.
Processing of 5HT2C pre-mRNA
The activity of the 5HT2C system is controlled to a large degree by pre-mRNA processing. The 5HT2C pre-mRNA undergoes pre-mRNA splicing, which generates a truncated receptor (5HT2C_tr) and pre-mRNA editing, generating 24 full-length receptor protein isoforms (5HT2C_Fl). Most of the full-length receptors are constitutively active, i.e., they signal without the serotonin ligand. This constitutive activity is decreased by the changes in amino acids caused by editing (Berg et al. 2001). In contrast to the well-studied 5HT2C editing, the importance of the truncated 5HT2C_tr receptor generated by alternative splicing has emerged only recently. The truncated receptor isoform is located intracellularly and decreases receptor signaling through heterodimerization with the full-length receptor (Martin et al. 2013; Zhang et al. 2016). Thus, alternative splicing and RNA editing regulate the constitutive activity of the 5HT2C system. The ratio between truncated and full-length receptors can be manipulated through oligonucleotides, allowing selective modulation of 5HT2C receptor activity. Herein, we review the splicing and editing regulation of the 5HT2C, their influence on the encoded proteins, and highlight diseases/pathologies that could be targeted by modulating the pre-mRNA processing of the 5HT2C. It is important to note that, prior to 1992, initial pharmacological characterization of the 5HT2C receptor classified it as 5HT1C instead of the current 5HT2C nomenclature (Humphrey et al. 1993).
Gene structure
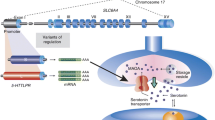
The 5HT2C gene is located on chromosome X and spans at least 326 kb. Intron V is unusually large, spanning at least 326 kb. The gene contains six exons and hosts four miRNAs in intron II. The first three exons are expressed in all cell types tested, but exons IV–VI are neuron-specific, suggesting a termination signal in intron III that is silenced in neurons. 5HT2C proteins are thus only expressed in neurons, as the start codon is located in exon III. In contrast, the four miRNAs are ubiquitously expressed, though their targets are unknown (Zhang et al. 2013), (Fig. 1a). The neuronal pre-mRNA containing exons I–VI undergoes both RNA editing and alternative pre-mRNA splicing of exon Vb. Exon V contains a proximal and distal splice site defining alternative exon Vb. Alternative splicing generates two classes of RNAs: RNA1 lacking exon Vb and RNA2 including exon Vb (Fig. 1b). The core of the splicing regulation is a pre-mRNA structure formed by exon Vb and intron V (Fig. 1c) that was determined in vitro using SHAPE (Selective 2′-hydroxyl acylation analyzed by primer extension) analysis (Shen et al. 2013). This RNA structure contains the two regulated splice sites and the five Adenosine → Inosine (A → I) editing sites.
Gene structure and regulation of the 5HT2C pre-mRNA. a Gene structure of the serotonin receptor 2C. PS, DS: proximal and distal splice sites. The start codon in exon III is indicated by a circle, the two stop codons in exon VI by open and filled squares. The blue shaded area forms the dsRNA structure shown in C. The location of the miRNAs in intron II is indicated. b The exon structure of RNA1 and RNA2 generated through alternative splicing of exon Vb is indicated. c dsRNA structure formed by exon Vb and Intron V. a–e Sites changed by deamination. The binding site for SNORD115 is boxed in orange. Blue nucleotides depict the five adenosine residues (a–e) that can be deaminated to inosines. The first two nucleotides of the proximal and distal 5′ splice sites (gu) are highlighted in yellow and red. d Site of regulation in the ds RNA structure. PLC phosphor lipase C, IP6 hexakisphosphate, HTP helix-threading peptide binding to the central loop (Schirle et al. 2010), PP pyrvinium pamoate (Shen et al. 2013); oligo#5: binding site for splice site changing oligo, indicated by a yellow line (Zhang et al. 2016). The SNORD116 binding site is indicated by an orange box, and adenosines shown in blue indicate the editing sites
Pre-mRNA editing
The A → I editing is catalyzed by ADARs (adenosine deaminase acting on RNA), enzymes that require double stranded RNA as a substrate, which strongly suggests that a dsRNA structure likely similar to the one determined in vitro also forms in vivo. Since Intron V is part of this dsRNA structure, the ADAR substrate is most likely the pre-mRNA. Transfection studies showed that use of the proximal splice site is the default splicing pathway in cultured cells (Flomen et al. 2004; Kishore and Stamm 2006) resulting in predominant exon Vb skipping. However, when the RNA editing sites are mutated from adenine to guanine, where guanine resembles the deamination product inosine, a strong activation of the distal splice site and inclusion of exon Vb is observed in non-neuronal cells upon transfection of A → G mutant constructs (Flomen et al. 2004). Thus, RNA editing promotes exon Vb inclusion, possibly by changing the pre-mRNA secondary structure of exon Vb, as the resulting U:I base pairs are energetically weaker than the original U:A base pairs, which is especially significant when the U:I pairs are clustered, as in sites A, B, E, and C (Serra et al. 2004). RNA editing changes the sequence of the RNA at five positions, leading to 25 = 32 mRNA isoforms. Collectively, these RNA isoforms including exon Vb are referred to as RNA2 (Fig. 1b). Since inosine is read as a guanosine by the ribosome, the editing changes the amino acid composition at three sites, resulting in 3 × 4 × 2 = 24 different proteins encoded by RNA2 (see also Fig. 2e).
Proteins generated from the 5HT2C gene. a Protein sequence of human 5HT2C_tr. The transmembrane spanning helices are indicated. The black amino acid marks the C23S polymorphism that is associated with suicide. b Protein sequence of human 5HT2C_FL receptor. The blue marked amino acids SSV at the C-terminus are a PDZ-ligand (Becamel et al. 2004). The amino acids changed through RNA editing are shown in red, green, and blue and enlarged in e. c Localization of 5HT2C_tr, which is only associates with internal membranes, likely the endoplasmic reticulum (ER), indicated as an orange oval. d Localization of the 5HT2C_FL receptors that shuttle between internal membranes, likely endoplasmic reticulum (ER, orange) and the plasma membrane. e Effect of RNA editing on amino acids encoded by 5HT2C and effect on receptor activity. The amino acids changed by editing are highlighted in b. The effect of RNA editing on signaling is shown on the right
Alternative splicing
Exon Vb not only undergoes pre-mRNA editing, but it is also alternatively spliced. The RNA generated by exon Vb skipping is called RNA1 (Fig. 1b). Skipping of exon Vb causes a frameshift and usage of an early stop codon in exon VI generates a truncated protein (5HT2C_tr) that contains only three transmembrane domains. Since the stop codon is located in the last exon, the mRNA escapes nonsense-mediated decay both RNA1 and the truncated receptor protein can be detected in brain, the latter using an antiserum against the 5HT2C_tr-specific C-terminus (Zhang et al. 2016). Exon Vb shares an 18 nucleotide perfect base complementarity with SNORD115, a neuron-specific C/D box snoRNA. C/D box snoRNAs (SNORDs) are a highly expressed class of non-coding RNAs (60–300 nt in length) that accumulate in the nucleolus. About half of the known SNORDs work in rRNA processing, either by guiding the methylase fibrillarin to rRNA or by guiding the cleavage of pre-rRNAs. SNORD115 forms non-methylating ribonuclear protein complexes containing hnRNPs but lacking the methylase fibrillarin. Thus, upon binding to exon Vb, SNORD115 acting in a non-methylating SNORD complex promotes exon Vb inclusion (Falaleeva et al. 2017; Kishore and Stamm 2006). SNORD115 is expressed in most brain regions, but is excluded from the choroid plexus, a brain structure lining the ventricles that generates cerebrospinal fluid (Kishore and Stamm 2006). Similarly, exon Vb is predominantly included in all brain areas, but is not used in the choroid plexus, supporting the physiological role of SNORD115 in promoting exon Vb inclusion (Kishore and Stamm 2006). Thus, due to pre-mRNA splicing and editing, a total of 25 proteins (24 full length and one truncated isoform) and 33 mRNAs and four miRNAs are generated from the 5HT2C gene.
Regulation of RNA editing
The dsRNA region formed by exon Vb and intron V is the substrate for ADAR (adenosine deaminase acting on RNA) enzymes, proteins that bind to dsRNA, and deaminate adenosine residues (Deffit and Hundley 2016), resulting in changing adenosine residues to inosine (A → I editing). The comparison of cDNA to genomic DNA showed five editing sites in exon Vb, termed A, B, E, C, and D (the E site is also referred to as C’ site). Sites A, B, and D are highly edited compared to sites C and E (Hartner et al. 2004). The usage of the editing sites is coordinated, for example, the editing states of site A and B depend on each other (Carmel et al. 2012). The analysis of ADAR mouse knock-outs suggests that sites A and B are predominantly edited by ADAR1, the D and C sites by ADAR2, and the E site by both enzymes (Hartner et al. 2004). In vitro editing experiments showed that the loop opposite the SNORD115 binding site is needed for the correct placement of ADAR and thus for the recognition of the editing sites (Fukuda et al. 2015). This loop also binds to synthetic cyclic peptides, known as helix-threading peptides (HTP) (Schirle et al. 2010), suggesting that it is part of a defined three-dimensional structure (Fig. 1d). ADAR2 stably binds Inositol hexakisphosphate (IP6) in the enzyme core and IP6 is required for ADAR2’s activity (Macbeth et al. 2005). 5HT2C couples to a heterotrimeric G-protein containing Gq/11 that generates phospholipase C and IP3, which can be converted to IP6. It is thus possible that feedback between 5HT2C activity and 5HT2C editing occurs (Schmauss et al. 2010).
Regulation of alternative splicing
Similar to 5HT2C editing, the regulation of alternative splicing is based on the dsRNA structure. The RNA elements in exon Vb regulation were determined by transfection experiments. In these experiments, RNA editing was mimicked by A → G substitutions, as the guanosine resembles Inosine base pairing. A → G base changes of single sites slightly promoted exon Vb inclusion with site E having the strongest effect. However, substitutions at two or more sites strongly activated exon Vb inclusion. Deletion analysis further showed that the 3′ stem (Fig. 1d) is important in suppressing the distal splice site, as its removal leads to a strong activation (Flomen et al. 2004).
Transfection experiments that tested numerous splicing factors for their effect on 5HT2C did not have a strong effect on exon Vb inclusion (Kishore and Stamm 2006). However, a C/D box snoRNA (SNORD) has been identified that binds to the regulated dsRNA structure. This RNA, SNORD115, previously named HBII-52, is a major activator for exon Vb. To gain insight into the underlying mechanism, SNORD115 was further characterized. In humans, there are 47 highly similar SNORD115 copies. Similar to other C/D box snoRNAs, SNORD115 is localized by an intron, each SNORD115 copy surrounded by two exons.
SNORD-RNPs are generated in a multistep assembly pathway, where SNORDs form a SNORD-RNP containing NHP2L1 (15.5k, SNU13), NOP56, NOP58, and fibrillarin. In this complex, the SNORD binds to a target RNA, allowing for methylation 5 nt upstream of the D box by the methylase fibrillarin (Falaleeva et al. 2017).
SNORD115 does not follow this classical model, which was determined upon analysis of the RNAs made from SNORD115-expressing units, composed of two exons flanking an intron containing SNORD115. RNase protection analysis showed that these units expressed shorter SNORD115-RNAs, termed psnoRNAs for processed snoRNAs (Kishore et al. 2010; Shen et al. 2011). Such shorter SNORDs have been described for SNORD116 and SNORD113—114 families—as well as SNORDs—50, 19, 32B, 123, 111, 72 93, 23, and 85 (Jorjani et al. 2016; Shen et al. 2011). Furthermore, SNORD115 associates mainly with hnRNPs and forms variable protein complexes in sucrose gradients (Kishore et al. 2010; Soeno et al. 2010). Thus, SNORD115 acts not in a traditional snoRNP containing fibrillarin, NOP56, NOP58, and NHP2L1, but rather as an RNA stabilized by hnRNPs.
Since exon Vb is part of a stable dsRNA structure, a screen for substances that activate this exon was performed, giving further insight into the splicing regulation (Shen et al. 2013). The screen identified pyrvinium pamoate (PP), a drug that binds to double stranded nucleic acids, with a greater propensity to dsRNA. SHAPE and CD spectrum analysis showed that pyrvinium pamoate binds directly to the regulated RNA region and changes its structure. Without pyrvinium pamoate, the distal splice site is in a predominantly double stranded conformation that blocks U1 binding. Pyrvinium pamoate opens up this structure, and U1 binding increases, which leads to selection of the distal splice site and inclusion of exon Vb (Shen et al. 2013).
It is thus likely that SNORD115 causes a structural change of the regulated dsRNA region, which activates the distal splice site. The SNORD115 binding site partially overlaps with the ADAR binding sites, suggesting that both molecules compete for exon Vb binding in the 5HT2C-pre-mRNA.
Proteins made from the 5HT2C gene
The 5HT2C pre-mRNA generates two classes of proteins: a truncated receptor 5HT2C_tr encoded by RNA1 and the full-length receptors, 5HT2C_Fl encoded by RNA1 (Fig. 2a, b). The full-length receptors can either be non-edited (5HT2C_FL_INI), having the amino acids INI at the editing sensitive sites, or be edited to form one of 23 edited receptors, 5HT2C_Fl_ed (Fig. 2e).
Full-length receptor
5HT2C_Fl are seven transmembrane receptors that couple to heterotrimeric G-proteins containing Gq/11 and activate the phospholipase C beta (PLCbeta) signaling pathway. PLCbeta cleaves phosphatidylinositol 4,5-bisphosphate to inositol trisphosphate (IP3), which increases intracellular calcium levels, as well as diacylglycerol, which activates protein kinase C (PKC). PKC induces immediate/early genes and also phosphorylates ion channels. In addition, 5HT2C signals through Gq/13, which activates phospholipase A2 (PLA2), which releases arachidonic acid and activates cGMP via nitric oxide (Berg et al. 1998). The site of G-protein coupling is the intracellular second transmembrane domain, where three amino acids are changed by RNA editing. The amino acid changes caused by RNA editing decrease the affinity of the heterotrimeric G-proteins, which is lowered by RNA editing (Burns et al. 1997). The active receptor is a likely a dimer, but higher order structures can be detected (Herrick-Davis 2013). The full-length receptors shuttle between intracellular membranes and the cell surface and these trafficking properties are altered by editing as well (Burns et al. 1997; Herrick-Davis et al. 1999; Wang et al. 2000) (Fig. 2d).
All 5HT2C full-length receptors contain a PDZ-ligand motif in their C-terminus that binds to at least seven different PDZ domain containing proteins. The 5HT2C_FL receptor shares 75% sequence identity with the 5HT2A receptor and both receptors are pharmacologically similar. However, both receptors bind to different PDZ domain containing proteins, with SAP102 specific for the 5HT2C (Becamel et al. 2004).
An important property of the 5HT2C full-length receptors is their constitutive activity, i.e., the activity without ligand. The receptors fluctuate between an inactive and an active conformation with ligands stabilizing the active conformation, increasing the overall activity. The non-edited 5HT2C_Fl_INI shows the highest constitutive activity, which is decreased by editing, with 5HT2C_FL_VGV showing no constitutive activity (Niswender et al. 1999), (Fig. 2e). Thus, RNA editing has two effects: it increases the amount of full-length receptors due to activation of exon Vb and it decreases their constitutive activity.
Truncated receptor
The truncated receptor 5HT2C_tr generated through skipping of exon Vb contains only the first three transmembrane domains that are identical to the full-length receptors. Due to the frameshift caused by exon Vb skipping, it lacks the second intracellular loop (Fig. 2a) and differs from the full-length receptors in its C-terminus. Whereas the first three transmembrane domains are evolutionary highly conserved, there are large species differences in the C-termini. For example, in mouse, the C-terminus contains 19 amino acids, but it is 96 amino acids long in humans. Due to the lack of the second transmembrane domain, 5HT2C_tr cannot couple to a G-protein and does not activate phospholipase C (Martin et al. 2013). 5HT2C_tr is localized in internal membranes, likely the endoplasmic reticulum (ER), and is not detectable at the plasma membrane (Martin et al. 2013; Zhang et al. 2016) (Fig. 2c). The 5HT2C_tr protein is expressed in the brain and can be extracted from internal membranes, supporting the idea that the results from transfection studies are physiologically relevant. In contrast to the full-length receptor, the truncated receptor does not contain a PDZ-ligand sequence, which could contribute to the differences in membrane association and localization between the full-length and truncated receptors (Zhang et al. 2016).
(Hetero)dimerization of serotonin receptors
Similar to other seven transmembrane receptors, the 5HT2C_FL forms dimers (Herrick-Davis et al. 2004) at the plasma membrane. These dimers originate during 5HT2C’s synthesis in the endoplasmic reticulum (Herrick-Davis et al. 2006). Disulfide trapping experiments, based on the rhodopsin structure, showed that 5HT2C forms two types of dimers: one dimer class is formed through interactions between transmembrane region 1 (TM1) and the second class shows interaction between TM4 and TM5. Ligand binding does not change the structure in dimers between TM1 domains, but changes the structure of only one of the protomers in the TM4–TM5 dimer, suggesting that upon activation, TM4–TM5 is a conformational heterodimer (Mancia et al. 2008).
Importantly, there is heterodimerization between the 5HT2C_FL and 5HT2C_tr receptors. It is possible that this heterodimerization is mediated by the TM1 domains, present in both proteins. Reflecting the strict intracellular localization of 5HT2C_tr, the heterodimer between 5HT2C_tr and 5HT2C_FL is localized in the endoplasmic reticulum. Transfection experiments showed that 5HT2C_tr stops 5HT2C_fl signaling, measured by a dose-dependent decrease in diacylglycerol synthesis (Martin et al. 2013). Thus, the function of the 5HT2C_tr truncated isoform is to stop the constitutive activity of the full-length receptor, either through sequestration inside the cell or through changing the 5HT2C homodimer formation necessary for coupling to the Gq/11 heterotrimer (Fig. 3b).
Biological function of 5HT2C isoforms. a Signaling of 5HT2C in POMC neurons of the arcuate nucleus. The regulated dsRNA structure shown in Fig. 1c is indicated schematically in the nucleus (red). Yellow and red triangles indicate the proximal and distal splice sites. ADAR and SNORD115 promote the formation of the full-length receptor through inclusion of exon Vb. The full-length receptor shuttles to the plasma membrane where it is active as a dimer. A heterotrimeric G-protein containing Gq11 (red star, blue circles) binds to the second intracellular loop. The binding affinity of the heterotrimeric G-protein is regulated by the editing of three amino acids in the second intracellular loop (Fig. 2b, e). Gq11 activates PLC, leading to the formation of DAG and IP3, which activates c-fos via the ERK pathway. C-fos turns on the POMC promoter, and the resulting POMC is processed into alpha MSH, which signals to neurons in the paraventricular nucleus generating a satiety signal. ER endoplasmic reticulum, q11 Gq11 alpha subunit, PLC phospholipase C, IP3 inositol trisphosphate, PKC protein kinase C, DAG diacylglycerol, ERK extracellular regulated kinase, POMC proopiomelanocortin, MSH melanocyte stimulating hormone, MCR4 melanocortin receptor 4. b Regulation of 5HT2C signaling through heterdimerization of the splicing isoforms. The truncated 5HT2C receptor is localized in intracellular membranes, likely endoplasmic reticulum (orange circle). It heterodimerizes with full-length receptors likely through interaction of the first transmembrane domains. Thus, a high ratio of RNA1 to RNA2 leads to an internalization of full-length receptors, which diminishes signaling. In contrast, a low RNA1/RNA2 ratio leads to full-length receptors on the surface and signaling. c 5HT2C pre-mRNA as a therapeutic target. (i) The default splice site selection is usage of the proximal splice site (yellow triangle) due to the secondary structure blocking the distal site (red triangle). U1: U1 snRNP, the size of the circle indicates the binding strength. (ii) Oligo#5 binds to the intronic part of the dsRNA structure, activating the distal splice site, similar to SNORD115 that binds to the exonic part. (iii) ADAR action also decreases the stability of the dsRNA structure, leading to distal splice site activation. Helix-threading peptides block ADAR2 activity in cell free assays, showing that this process can also be modified in principle (Schirle et al. 2010)
In addition to the interaction with the 5HT2C_tr, the full-length receptor heterodimerizes with the ghrelin receptor GHSR1a, leading to a decrease of ghrelin’s orexigenic effect (Schellekens et al. 2015). Since 5HT2C_tr also binds to the 5HT2C_FL, the truncated isoform might indirectly influence other receptor systems.
Thus, the properties of the 5HT2C signaling system are strongly influenced by pre-mRNA processing events, leading to proteins with different signaling properties. Alternative splicing is used to switch off the constitutive activity of full-length receptor isoforms.
Function of serotonin receptors under physiological and pathophysiological conditions
Control of food intake
The mechanistically best-understood function of the 5HT2C is the regulation of food intake (Berglund et al. 2014; Cone 2005) through 5HT2C action in proopiomelanocortin (POMC) neurons in the hypothalamus (Fig. 3a). Serotonin binding to 5HT2C leads to the dissociation of a heterotrimeric G-protein that binds to the second intracellular loop of 5HT2C. Upon dissociation, the alpha subunit Gq11 activates phospholipase C, generating IP3 and diacylglycerol (DAG), which activates Protein Kinase C (PKC). PKC activates the extracellular regulated kinase (ERK) pathway, leading to the phosphorylation of c-fos. C-fos activation leads to POMC synthesis. POMC is processed into alpha-melanocyte stimulating hormone (alpha MSH) that activates neurons in the paraventricular nucleus (PVN) via melanocortin 4 receptors. Activation of PVN neurons induces satiety, i.e., cessation of eating as an anorexic response. Knock-out mice lacking the 5HT2C receptor thus exhibit hyperphagia and are obese (Tecott et al. 1995), whereas activation of the 5HT2C receptor via an agonist such as the FDA approved drug, lorcaserin, inhibits food intake (Shukla et al. 2015). The activity of the 5HT2C is regulated through the ratio of the truncated to full-length receptor, since an increase in the truncated receptor sequesters the full-length receptor intracellularly, decreasing 5HT2C signaling (Martin et al. 2013; Zhang et al. 2016) (Fig. 3b). In addition, the editing status of the full-length receptors is important for the signaling of POMC neurons, as overexpression of fully-edited receptors results in hyperphagia in mice, suggesting an impaired activity of POMC neurons (Morabito et al. 2010). As described above, SNORD115 promotes the formation of the non-edited full-length receptors and ADAR induces the formation of the edited full-length receptors. Thus, food intake is regulated in part by RNA processing of the 5HT2C.
Prader–Willi syndrome (PWS)
Prader–Willi syndrome (PWS) is one of the most frequent genetic causes of morbid obesity and intellectual disability in humans. PWS subjects have short stature, severe hypotonia at birth, genital hypoplasia, and characteristic facial features (Driscoll et al. 1993–2017). PWS has a frequency of about 1:10,000 individuals and is caused by loss of paternal allele expression at an imprinted region on chromosome 15q11.2-q13.1. The imprinted region missing in PWS is expressed only from the paternal allele and contains five protein-coding genes (MKRN3, MAGEL2, NECDIN, SNURF-SNRPN, and NPAP1/C15orf2) and six orphan C/D box snoRNAs (SNORDs): 107, 64, 108, 109, 116 (29 copies falling into five classes), and 115 (47 almost identical copies) (Cavaille 2017). As described above, SNORD115 regulates splicing of the serotonin receptor 2C (5HT2C) by promoting exon Vb inclusion (Kishore and Stamm 2006; Zhang et al. 2016), though the targets and functions of the other SNORDs absent in PWS are unknown. Mice overexpressing SNORD115 show autistic features (Nakatani et al. 2009), illustrating that SNORD expression levels must be under tight control to prevent disease. A knock-out model for PWS lacking all SNORDs, including SNORD115, shows an increase of the 5HT2C_tr receptor isoform in POMC neurons (Garfield et al. 2016), as predicted from earlier transfection studies showing that SNORD115 promotes exon Vb inclusion (Kishore and Stamm 2006). The increase of 5HT2C_tr in POMC neurons stops the anorexic response of these animals, as they do not induce efficiently c-fos after 5HT2C stimulation (Garfield et al. 2016) (Fig. 3a). The loss of this signal leads likely to hyperphagia characteristic for PWS. Thus, in Prader–Willi syndrome, an increase of the truncated 5HT2C isoform likely reduces the serotonin response through heterodimerization with the 5HT2C_FL forms, blocking the satiety response (Fig. 3b). A characteristic feature of babies with PWS is their low muscle tone (Driscoll et al. 1993–2017), which could also indicate a deregulation of the 5HT2C system that activates motoneurons.
Role of the 5HT2C in the spinal cord
Serotonin is important for regulating muscle tone through its actions in the spinal cord, which receives serotoninergic projections from the rostral ventromedial medulla, as well as the brainstem raphe nuclei (Jacobs et al. 2002). Normally, brainstem-derived serotonin sets spinal motoneurons and interneurons into an excitable state, ready to respond to fast glutamate synaptic inputs and causes appropriate muscle contractions by activating 5HT2C receptors that facilitate ionic currents intrinsic to the motoneurons, including voltage-gated persistent Ca2+ and Na+ currents (termed persistent inward currents: PICs) (Jacobs et al. 2002; Murray et al. 2010; Perrier and Delgado-Lezama 2005; Rekling et al. 2000). Such PICs have low thresholds to both amplify and prolong the action of synaptic inputs, ultimately enabling sustained muscle contractions (Carlin et al. 2000; Harvey et al. 2006a; Li et al. 2004).
5HT2C-mediated spasticity after spinal cord injury
In addition to resulting paralysis and loss of sensation of upper or lower extremities, spinal cord injury (SCI) often manifests in secondary abnormal spinal reflexes that can significantly affect quality of life. Importantly, it can induce debilitating muscle spasms below the injury level characterized by involuntary contraction of muscles in response to either a stretch or painful stimulus (Rabchevsky and Kitzman 2011; Rabchevsky et al. 2012; Walter et al. 2002). Shortly after SCI, motoneurons caudal to the injury are in an unexcitable state, resulting in paralysis, areflexia, and hyporeflexia (spinal shock). A few weeks after injury, however, these changes reverse to hyperreflexia, concomitant with hyperexcitability of spinal cord motoneurons, leading to exaggerated cutaneous reflexes and spasticity in the chronic phase of the injury (Nielsen et al. 2007; Rabchevsky and Kitzman 2011). A loss of brainstem-derived serotonin after SCI acutely reduces motoneuron excitability below the level of injury that depresses all motor functions (Harvey et al. 2006b; Hounsgaard et al. 1988; Li et al. 2007; Perrier and Delgado-Lezama 2005). This reduction in activity results not only from a loss of supraspinal tracts controlling voluntary initiation of movement but also from a loss of descending brainstem tracts that provide spinal motoneurons with their major source of neuromodulators, such as serotonin (Carlsson et al. 1963; Rekling et al. 2000). Weeks after SCI, motoneurons spontaneously recover their excitability despite the continued absence of brainstem-derived serotonin (Li et al. 2007; Murray et al. 2010), which can lead to uncontrolled and debilitating muscle spasms in both humans (Gorassini et al. 2004) and rats (Bennett et al. 1999; Li et al. 2004). How motoneurons adapt in the absence of brainstem-derived serotonin is unclear, but since constitutively active receptors spontaneously couple to their Gq proteins and initiate intracellular signaling without being bound to serotonin or any other ligand, an increase of constitutively active receptors is likely (Berg et al. 2001; Herrick-Davis et al. 1999).
RNA1 and RNA2 have been detected both in naïve and injured spinal cord (Kim et al. 2001; Schmidt and Jordan 2000), suggesting that the activity of motoneurons could also be influenced by 5HT2C pre-mRNA processing which controls the constitutive activity. These changes are concomitant with PICs in motoneurons and a change in 5HT2A and 5HT2C receptors. Studies of 5HT2C alternations after SCI have focused on the full-length receptor. RT-PCR experiments showed that the hypersensitivity of motoneurons and subsequent spasticity is caused by an upregulation of 5HT2C_FL_INI (Fouad et al. 2010; Murray et al. 2010), which shows the highest constitutive activity. ADAR expression is downregulated in some models of SCI, possibly reflecting an inflammatory response (Di Narzo et al. 2015). Thus, 5HT2C_FL_INI upregulation could be caused by the decrease in ADAR activity (Di Narzo et al. 2015), resulting in the loss of 5HT2C_FL_ed. However, a study that relied on RNA sequencing, which has more statistical power, observed no difference in editing (Navarrett et al. 2012). A detailed time course study using antibodies against the full-length receptor showed a steady 1.7 fold upregulation of the 5HT2C_FL receptor 60 days after SCI. The extent of these changes correlated well with the clinical scores of tail spasticity in a rat SCI model (Ren et al. 2013). In a different SCI model, a change in RNA1/RNA2 ratios has been observed as well, where RNA1 increases after the injury (Nakae et al. 2013). It is thus very likely that 5HT2C isoforms and ADAR activity change after SCI, which contributes to spasticity, but the exact molecular events are determined by the type, timepoint, and localization of the injury.
Suicide
Given the role of 5HT2C in food control and anxiety, associations of genetic variants with neurological disorders have been investigated. A polymorphism rs6318 that changes amino acids Cys23Ser in the extracellular domain has been associated with suicidal behavior in a Serbian and Slovenian (Karanovic et al. 2015; Videtic et al. 2009), but not Chinese population (Zhang et al. 2008). The minor allele variant, 23Ser, shows a larger surface occupancy and altered resensitation (Walstab et al. 2011) and human subjects carrying this allele show a larger dopamine release in brain areas responding to stress (Mickey et al. 2012). Since the variation is located in the N-terminus, it will affect both the truncated and full-length receptor isoforms. The analysis of post-mortem tissue also showed that editing patterns are altered in brains of suicide victims (Weissmann et al. 2016), (Di Narzo et al. 2014), (Dracheva et al. 2008; Gurevich et al. 2002b) and the ratio of RNA2 to RNA1 is increased in the dorsal prefrontal cortex of suicide subjects (Dracheva et al. 2008). These studies demonstrate the role of the 5HT2C in behavior leading to suicide, but since they rely on post-mortem tissue, the molecular details are obscure. Furthermore, there are quantitative differences between the studies, as they rely on post-mortem tissue and use methods of different sensitivity, ranging from semi-quantitative RT-PCR to modern RNAseq.
Changes in editing patterns
The editing of the 5HT2C is highly dynamic and changes under physiological conditions, such as water maze learning (Du et al. 2007), obesity (Schellekens et al. 2012), and cytokine treatment (Yang et al. 2004), generating 5HT2C isoforms with different signaling properties. The changes in 5HT2C editing after cytokine treatment could explain depression associated with interferon treatment (Yang et al. 2004). Numerous studies identified changes in 5HT2C editing under pathophysiological conditions, such as spinal nerve transection in a model of neuropathic pain (Uchida et al. 2017) and spinal cord injury (Di Narzo et al. 2015). Furthermore, alcohol intake (Watanabe et al. 2014) and treatment with anitdepressants changes editing patterns and in case of antidepressants also expression levels (Barbon et al. 2011).
The molecular cause for these changes remains speculative. ADAR2 is phosphorylated at its N-terminus by an unknown kinase, which is required for its nuclear localization and enzymatic activity, mediated by the peptidyl prolyl-isomerase Pin1 (Marcucci et al. 2011). A dephosphorylation of ADAR2 leads to its cytosolic accumulation and subsequent degradation, which will stop the editing activity, suggesting that signaling pathways can modulate RNA editing through ADAR2 phosphorylation.
Are changes in alternative splicing a possible therapy for 5HT2C-related disorders?
The central role of the 5HT2C in neuropsychiatric diseases is reflected by the number of drugs that were developed to change the activity of the receptor. For example, the agonist lorcaserin is approved for short-term treatment of obesity (Shukla et al. 2015), and Vabicaserin is in clinical trials against schizophrenia (Shen et al. 2014). Because of the similarities of the serotonin receptors, even drugs like Lorcaserin and Vabicaserin that show selectivity for the 5HT2C will also interact with other serotonin receptors. Numerous drugs against depression/anxiety (Fluoxetine, Desipramine) target several receptors including 5HT2C. Since the activity of the 5HT2C receptor system is regulated in large part through RNA processing, targeting the 5HT2C pre-mRNA could be an alternative (Fig. 3c).
Recently, Nusinersen (Spinraza), an antisense oligonucleotide has been approved as a splice site changing drug to treat spinal muscular atrophy (Ottesen 2017), underlining the potential of modifying pre-mRNA splicing for therapeutic purposes.
A deregulation of the 5HT2C pre-mRNA splicing is seen in Prader–Willi syndrome, where RNA1 is increased (Garfield et al. 2016), likely because the trans-acting factor SNORD115 that promotes inclusion of exon Vb is absent.
To find oligonucleotides that could substitute SNORD115, an oligo-walk (Singh et al. 2004) with reporter genes was performed. In these experiments, oligo#5, binding to the 3′ stem in the regulated dsRNA region (Fig. 1d) was identified. Oligo#5 strongly promotes exon Vb inclusion in cell culture (Zhang et al. 2016), similar to SNORD115. Due the chemistry as a 2′-O-methyl-monophosphothioate, the oligo is taken up from cells without adjuvans. Oligo#5 was then tested in mice, first by intracerebral injection into the third ventricle. There is no blood–brain barrier between POMC neurons located in the arcuate nucleus and the CSF in the ventricles, as the surrounding epithelium is fenestrated (Rodriguez et al. 2010). Thus, oligo#5 rapidly accumulates in the neurons of the arcuate nucleus, promotes formation of the non-edited full-length receptor 5HT2C_FL_INI, and decreases RNA1. POMC mRNA is induced, and as expected, food intake is reduced for about 75%. The oligo has no effect in MCR4 knock out mice, but does reduce the food intake in ob/ob mice, showing that it targets the POMC/alpha-MSH signaling pathway. Unexpectedly, the oligo crossed the blood–brain barrier after injection through a carotid catheter and accumulated rapidly in the choroid plexus and within several hours in other brain region, including the arcuate nucleus, where it increased the RNA2/RNA1 ratio and reduced food intake for up to three days (Zhang et al. 2016).
The sequence targeted by the oligo is unique in the genome (Fig. 1c, d), which is in contrast to the highly similar protein sequences of the various 5HT2 receptors. However, to allow crossing the blood–brain barrier, injection of high dosages is necessary that might lead to unspecific responses.
These findings show the principle that 5HT2C pre-mRNA is a drug target and that SNORD115 missing in PWS can be functionally substituted using an oligonucleotide. The data also indicate that the ratio between the two 5HT2C proteins: a full-length receptor containing exon Vb and a truncated receptor lacking it is involved in the regulation of food intake. Since the full-length receptor heterodimerizes with other transmembrane receptors (Schellekens et al. 2013, 2015), it is possible that this ratio, controlled by the non-methylating SNORD115, contributes to the formation of the complex Prader–Willi syndrome by affecting different receptor systems. Since targeting the 5HT2C pre-mRNA has a different mechanism of action than targeting the 5HT2C protein with ligands, oligonucleotide-based therapies could be used in cases where ligand-based therapies no longer work.
Chemical screens identified pyrvinium pamoate as substance that binds the regulated dsRNA region and causes a structural change that allows U1 access to the distal splice site, leading to exon Vb inclusion. Although pyrvinium pamoate is too toxic to be used in the brain, the experiments prove the principle that the 5HT2C pre-mRNA can be targeted by small drugs. It is possible that endogenous peptides exist that target the pre-mRNA, but such endogenous ligands remained to be identified.
References
Barbon A, Orlandi C, La Via L, Caracciolo L, Tardito D, Musazzi L, Mallei A, Gennarelli M, Racagni G, Popoli M, Barlati S (2011) Antidepressant treatments change 5-HT2C receptor mRNA expression in rat prefrontal/frontal cortex and hippocampus. Neuropsychobiology 63:160–168. doi:10.1159/000321593
Becamel C, Gavarini S, Chanrion B, Alonso G, Galeotti N, Dumuis A, Bockaert J, Marin P (2004) The serotonin 5-HT2A and 5-HT2C receptors interact with specific sets of PDZ proteins. J Biol Chem 279:20257–20266. doi:10.1074/jbc.M312106200
Bennett DJ, Gorassini M, Fouad K, Sanelli L, Han Y, Cheng J (1999) Spasticity in rats with sacral spinal cord injury. J Neurotrauma 16:69–84
Berg KA, Maayani S, Goldfarb J, Scaramellini C, Leff P, Clarke WP (1998) Effector pathway-dependent relative efficacy at serotonin type 2A and 2C receptors: evidence for agonist-directed trafficking of receptor stimulus. Mol Pharmacol 54:94–104
Berg KA, Cropper JD, Niswender CM, Sanders-Bush E, Emeson RB, Clarke WP (2001) RNA-editing of the 5-HT(2C) receptor alters agonist-receptor-effector coupling specificity. Br J Pharmacol 134:386–392. doi:10.1038/sj.bjp.0704255
Berglund ED, Liu C, Sohn JW, Liu T, Kim MH, Lee CE, Vianna CR, Williams KW, Xu Y, Elmquist JK (2014) Serotonin 2C receptors in pro-opiomelanocortin neurons regulate energy and glucose homeostasis. J Clin Investig 124:1868. doi:10.1172/JCI75669
Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB (1997) Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387:303–308. doi:10.1038/387303a0
Carlin KP, Jiang Z, Brownstone RM (2000) Characterization of calcium currents in functionally mature mouse spinal motoneurons. Eur J Neurosci 12:1624–1634
Carlsson A, Magnusson T, Rosengren E (1963) 5-Hydroxytryptamine of the spinal cord normally and after transection. Experientia 19:359
Carmel L, Koonin EV, Dracheva S (2012) Dependencies among editing sites in serotonin 2C receptor mRNA. PLoS Comput Biol 8:e1002663. doi:10.1371/journal.pcbi.1002663
Castensson A, Emilsson L, Sundberg R, Jazin E (2003) Decrease of serotonin receptor 2C in schizophrenia brains identified by high-resolution mRNA expression analysis. Biol Psychiatry 54:1212–1221
Castensson A, Aberg K, McCarthy S, Saetre P, Andersson B, Jazin E (2005) Serotonin receptor 2C (HTR2C) and schizophrenia: examination of possible medication and genetic influences on expression levels. Am J Med Genet B Neuropsychiatr Genet 134B:84–89. doi:10.1002/ajmg.b.30151
Cavaille J (2017) Box C/D small nucleolar RNA genes and the Prader–Willi syndrome: a complex interplay. Wiley Interdiscip Rev RNA. doi:10.1002/wrna.1417
Chagraoui A, Thibaut F, Skiba M, Thuillez C, Bourin M (2016) 5-HT2C receptors in psychiatric disorders: a review. Prog Neuropsychopharmacol Biol Psychiatry 66:120–135. doi:10.1016/j.pnpbp.2015.12.006
Cone RD (2005) Anatomy and regulation of the central melanocortin system. Nat Neurosci 8:571–578. doi:10.1038/nn1455
Deffit SN, Hundley HA (2016) To edit or not to edit: regulation of ADAR editing specificity and efficiency. Wiley Interdiscip Rev RNA 7:113–127. doi:10.1002/wrna.1319
Di Narzo AF, Kozlenkov A, Roussos P, Hao K, Hurd Y, Lewis DA, Sibille E, Siever LJ, Koonin E, Dracheva S (2014) A unique gene expression signature associated with serotonin 2C receptor RNA editing in the prefrontal cortex and altered in suicide. Hum Mol Genet 23:4801–4813. doi:10.1093/hmg/ddu195
Di Narzo AF, Kozlenkov A, Ge Y, Zhang B, Sanelli L, May Z, Li Y, Fouad K, Cardozo C, Koonin EV, Bennett DJ, Dracheva S (2015) Decrease of mRNA editing after spinal cord injury is caused by down-regulation of ADAR2 that is triggered by inflammatory response. Sci Rep 5:12615. doi:10.1038/srep12615
Dracheva S, Chin B, Haroutunian V (2008) Altered serotonin 2C receptor RNA splicing in suicide: association with editing. NeuroReport 19:379–382. doi:10.1097/WNR.0b013e3282f556d2
Driscoll DJ, Miller JL, Schwartz S, Cassidy SB (1993–2017) Prader–Willi Syndrome. In: Pagon RA, Adam MP, Ardinger HH et al (eds) GeneReviews® [Internet]. University of Washington, Seattle, WA
Du Y, Stasko M, Costa AC, Davisson MT, Gardiner KJ (2007) Editing of the serotonin 2C receptor pre-mRNA: effects of the Morris Water Maze. Gene 391:186–197. doi:10.1016/j.gene.2006.12.023
Falaleeva M, Welden JR, Duncan MC, Stamm S (2017) C/D-box snoRNAs form methylating and non methylating ribonucleoprotein complexes: old dogs show new tricks. Bioessays. doi:10.1002/bies.201600264 (in press)
Flomen R, Knight J, Sham P, Kerwin R, Makoff A (2004) Evidence that RNA editing modulates splice site selection in the 5-HT2C receptor gene. Nucleic Acids Res 32:2113–2122
Fouad K, Rank MM, Vavrek R, Murray KC, Sanelli L, Bennett DJ (2010) Locomotion after spinal cord injury depends on constitutive activity in serotonin receptors. J Neurophysiol 104:2975–2984. doi:10.1152/jn.00499.2010
Fukuda M, Oyama Y, Nishitarumizu A, Omura M, Nose K, Deshimaru M (2015) Identification of an RNA element for specific coordination of A-to-I RNA editing on HTR2C pre-mRNA. Genes Cells 20:834–846. doi:10.1111/gtc.12272
Garfield AS, Davies JR, Burke LK, Furby HV, Wilkinson LS, Heisler LK, Isles AR (2016) Increased alternate splicing of Htr2c in a mouse model for Prader–Willi syndrome leads disruption of 5HT2C receptor mediated appetite. Mol Brain 9:95. doi:10.1186/s13041-016-0277-4
Gorassini MA, Knash ME, Harvey PJ, Bennett DJ, Yang JF (2004) Role of motoneurons in the generation of muscle spasms after spinal cord injury. Brain 127:2247–2258. doi:10.1093/brain/awh243
Greenway FL, Shanahan W, Fain R, Ma T, Rubino D (2016) Safety and tolerability review of lorcaserin in clinical trials. Clin Obes 6:285–295. doi:10.1111/cob.12159
Gurevich I, Englander MT, Adlersberg M, Siegal NB, Schmauss C (2002a) Modulation of serotonin 2C receptor editing by sustained changes in serotonergic neurotransmission. J Neurosci 22:10529–10532
Gurevich I, Tamir H, Arango V, Dwork AJ, Mann JJ, Schmauss C (2002b) Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron 34:349–356
Hartner JC, Schmittwolf C, Kispert A, Muller AM, Higuchi M, Seeburg PH (2004) Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J Biol Chem 279:4894–4902. doi:10.1074/jbc.M311347200
Harvey PJ, Li X, Li Y, Bennett DJ (2006a) 5-HT2 receptor activation facilitates a persistent sodium current and repetitive firing in spinal motoneurons of rats with and without chronic spinal cord injury. J Neurophysiol 96:1158–1170. doi:10.1152/jn.01088.2005
Harvey PJ, Li Y, Li X, Bennett DJ (2006b) Persistent sodium currents and repetitive firing in motoneurons of the sacrocaudal spinal cord of adult rats. J Neurophysiol 96:1141–1157. doi:10.1152/jn.00335.2005
Heisler LK, Zhou L, Bajwa P, Hsu J, Tecott LH (2007) Serotonin 5-HT(2C) receptors regulate anxiety-like behavior. Genes Brain Behav 6:491–496. doi:10.1111/j.1601-183X.2007.00316.x
Herrick-Davis K (2013) Functional significance of serotonin receptor dimerization. Exp Brain Res 230:375–386. doi:10.1007/s00221-013-3622-1
Herrick-Davis K, Grinde E, Niswender CM (1999) Serotonin 5-HT2C receptor RNA editing alters receptor basal activity: implications for serotonergic signal transduction. J Neurochem 73:1711–1717
Herrick-Davis K, Grinde E, Mazurkiewicz JE (2004) Biochemical and biophysical characterization of serotonin 5-HT2C receptor homodimers on the plasma membrane of living cells. Biochemistry 43:13963–13971. doi:10.1021/bi048398p
Herrick-Davis K, Weaver BA, Grinde E, Mazurkiewicz JE (2006) Serotonin 5-HT2C receptor homodimer biogenesis in the endoplasmic reticulum: real-time visualization with confocal fluorescence resonance energy transfer. J Biol Chem 281:27109–27116. doi:10.1074/jbc.M604390200
Hounsgaard J, Hultborn H, Jespersen B, Kiehn O (1988) Bistability of alpha-motoneurones in the decerebrate cat and in the acute spinal cat after intravenous 5-hydroxytryptophan. J Physiol 405:345–367
Humphrey PP, Hartig P, Hoyer D (1993) A proposed new nomenclature for 5-HT receptors. Trends Pharmacol Sci 14:233–236
Jacobs BL, Martin-Cora FJ, Fornal CA (2002) Activity of medullary serotonergic neurons in freely moving animals. Brain Res Brain Res Rev 40:45–52
Jorjani H, Kehr S, Jedlinski DJ, Gumienny R, Hertel J, Stadler PF, Zavolan M, Gruber AR (2016) An updated human snoRNAome. Nucleic Acids Res 44:5068–5082. doi:10.1093/nar/gkw386
Karanovic J, Svikovic S, Pantovic M, Durica S, Brajuskovic G, Damjanovic A, Jovanovic V, Ivkovic M, Romac S, Savic Pavicevic D (2015) Joint effect of ADARB1 gene, HTR2C gene and stressful life events on suicide attempt risk in patients with major psychiatric disorders. World J Biol Psychiatry 16:261–271. doi:10.3109/15622975.2014.1000374
Kim D, Murray M, Simansky KJ (2001) The serotonergic 5-HT(2C) agonist m-chlorophenylpiperazine increases weight-supported locomotion without development of tolerance in rats with spinal transections. Exp Neurol 169:496–500. doi:10.1006/exnr.2001.7660
Kishore S, Stamm S (2006) The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science 311:230–232
Kishore S, Khanna A, Zhang Z, Hui J, Balwierz PJ, Stefan M, Beach C, Nicholls RD, Zavolan M, Stamm S (2010) The snoRNA MBII-52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing. Hum Mol Genet 19:1153–1164. doi:10.1093/hmg/ddp585
Li Y, Gorassini MA, Bennett DJ (2004) Role of persistent sodium and calcium currents in motoneuron firing and spasticity in chronic spinal rats. J Neurophysiol 91:767–783. doi:10.1152/jn.00788.2003
Li X, Murray K, Harvey PJ, Ballou EW, Bennett DJ (2007) Serotonin facilitates a persistent calcium current in motoneurons of rats with and without chronic spinal cord injury. J Neurophysiol 97:1236–1246. doi:10.1152/jn.00995.2006
Macbeth MR, Schubert HL, Vandemark AP, Lingam AT, Hill CP, Bass BL (2005) Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science 309:1534–1539. doi:10.1126/science.1113150
Mancia F, Assur Z, Herman AG, Siegel R, Hendrickson WA (2008) Ligand sensitivity in dimeric associations of the serotonin 5HT2c receptor. EMBO Rep 9:363–369. doi:10.1038/embor.2008.27
Marcucci R, Brindle J, Paro S, Casadio A, Hempel S, Morrice N, Bisso A, Keegan LP, Del Sal G, O’Connell MA (2011) Pin1 and WWP2 regulate GluR2 Q/R site RNA editing by ADAR2 with opposing effects. EMBO J 30:4211–4222. doi:10.1038/emboj.2011.303
Martin CB, Ramond F, Farrington DT, Aguiar AS Jr, Chevarin C, Berthiau AS, Caussanel S, Lanfumey L, Herrick-Davis K, Hamon M, Madjar JJ, Mongeau R (2013) RNA splicing and editing modulation of 5-HT2C receptor function: relevance to anxiety and aggression in VGV mice. Mol Psychiatry 18:656–665. doi:10.1038/mp.2012.171
Martin CB, Hamon M, Lanfumey L, Mongeau R (2014) Controversies on the role of 5-HT(2C) receptors in the mechanisms of action of antidepressant drugs. Neurosci Biobehav Rev 42:208–223. doi:10.1016/j.neubiorev.2014.03.001
McCorvy JD, Roth BL (2015) Structure and function of serotonin G protein-coupled receptors. Pharmacol Ther 150:129–142. doi:10.1016/j.pharmthera.2015.01.009
Mickey BJ, Sanford BJ, Love TM, Shen PH, Hodgkinson CA, Stohler CS, Goldman D, Zubieta JK (2012) Striatal dopamine release and genetic variation of the serotonin 2C receptor in humans. J Neurosci 32:9344–9350. doi:10.1523/JNEUROSCI.1260-12.2012
Mongeau R, Martin CB, Chevarin C, Maldonado R, Hamon M, Robledo P, Lanfumey L (2010) 5-HT2C receptor activation prevents stress-induced enhancement of brain 5-HT turnover and extracellular levels in the mouse brain: modulation by chronic paroxetine treatment. J Neurochem 115:438–449. doi:10.1111/j.1471-4159.2010.06932.x
Monti JM (2011) Serotonin control of sleep-wake behavior. Sleep Med Rev 15:269–281. doi:10.1016/j.smrv.2010.11.003
Morabito MV, Abbas AI, Hood JL, Kesterson RA, Jacobs MM, Kump DS, Hachey DL, Roth BL, Emeson RB (2010) Mice with altered serotonin 2C receptor RNA editing display characteristics of Prader–Willi Syndrome. Neurobiol Dis 39:169–180
Murray KC, Nakae A, Stephens MJ, Rank M, D’Amico J, Harvey PJ, Li X, Harris RL, Ballou EW, Anelli R, Heckman CJ, Mashimo T, Vavrek R, Sanelli L, Gorassini MA, Bennett DJ, Fouad K (2010) Recovery of motoneuron and locomotor function after spinal cord injury depends on constitutive activity in 5-HT2C receptors. Nat Med 16:694–700. doi:10.1038/nm.2160
Nakae A, Nakai K, Tanaka T, Hosokawa K, Mashimo T (2013) Serotonin 2C receptor alternative splicing in a spinal cord injury model. Neurosci Lett 532:49–54. doi:10.1016/j.neulet.2012.10.034
Nakatani J, Tamada K, Hatanaka F, Ise S, Ohta H, Inoue K, Tomonaga S, Watanabe Y, Chung YJ, Banerjee R, Iwamoto K, Kato T, Okazawa M, Yamauchi K, Tanda K, Takao K, Miyakawa T, Bradley A, Takumi T (2009) Abnormal behavior in a chromosome-engineered mouse model for human 15q11-13 duplication seen in autism. Cell 137:1235–1246
Navarrett S, Collier L, Cardozo C, Dracheva S (2012) Alterations of serotonin 2C and 2A receptors in response to T10 spinal cord transection in rats. Neurosci Lett 506:74–78. doi:10.1016/j.neulet.2011.10.052
Nielsen JB, Crone C, Hultborn H (2007) The spinal pathophysiology of spasticity—from a basic science point of view. Acta Physiol 189:171–180. doi:10.1111/j.1748-1716.2006.01652.x
Niswender CM, Copeland SC, Herrick-Davis K, Emeson RB, Sanders-Bush E (1999) RNA editing of the human serotonin 5-hydroxytryptamine 2C receptor silences constitutive activity. J Biol Chem 274:9472–9478
Ottesen EW (2017) ISS-N1 makes the first FDA-approved drug for spinal muscular atrophy. Transl Neurosci 8:1–6. doi:10.1515/tnsci-2017-0001
Perrier JF, Delgado-Lezama R (2005) Synaptic release of serotonin induced by stimulation of the raphe nucleus promotes plateau potentials in spinal motoneurons of the adult turtle. J Neurosci 25:7993–7999. doi:10.1523/JNEUROSCI.1957-05.2005
Rabchevsky AG, Kitzman PH (2011) Latest approaches for the treatment of spasticity and autonomic dysreflexia in chronic spinal cord injury. Neurother J Am Soc Exp NeuroTher 8:274–282. doi:10.1007/s13311-011-0025-5
Rabchevsky AG, Patel SP, Lyttle TS, Eldahan KC, O’Dell CR, Zhang Y, Popovich PG, Kitzman PH, Donohue KD (2012) Effects of gabapentin on muscle spasticity and both induced as well as spontaneous autonomic dysreflexia after complete spinal cord injury. Front Physiol 3:329. doi:10.3389/fphys.2012.00329
Rekling JC, Funk GD, Bayliss DA, Dong XW, Feldman JL (2000) Synaptic control of motoneuronal excitability. Physiol Rev 80:767–852
Ren LQ, Wienecke J, Chen M, Moller M, Hultborn H, Zhang M (2013) The time course of serotonin 2C receptor expression after spinal transection of rats: an immunohistochemical study. Neuroscience 236:31–46. doi:10.1016/j.neuroscience.2012.12.063
Rodriguez EM, Blazquez JL, Guerra M (2010) The design of barriers in the hypothalamus allows the median eminence and the arcuate nucleus to enjoy private milieus: the former opens to the portal blood and the latter to the cerebrospinal fluid. Peptides 31:757–776. doi:10.1016/j.peptides.2010.01.003
Schellekens H, Clarke G, Jeffery IB, Dinan TG, Cryan JF (2012) Dynamic 5-HT2C receptor editing in a mouse model of obesity. PLoS One 7:e32266. doi:10.1371/journal.pone.0032266
Schellekens H, van Oeffelen WE, Dinan TG, Cryan JF (2013) Promiscuous dimerization of the growth hormone secretagogue receptor (GHS-R1a) attenuates ghrelin-mediated signaling. J Biol Chem 288:181–191. doi:10.1074/jbc.M112.382473
Schellekens H, De Francesco PN, Kandil D, Theeuwes WF, McCarthy T, van Oeffelen WE, Perello M, Giblin L, Dinan TG, Cryan JF (2015) Ghrelin’s orexigenic effect is modulated via a serotonin 2C receptor interaction. ACS Chem Neurosci. doi:10.1021/cn500318q
Schirle NT, Goodman RA, Krishnamurthy M, Beal PA (2010) Selective inhibition of ADAR2-catalyzed editing of the serotonin 2c receptor pre-mRNA by a helix-threading peptide. Org Biomol Chem 8:4898–4904. doi:10.1039/c0ob00309c
Schmauss C, Zimnisky R, Mehta M, Shapiro LP (2010) The roles of phospholipase C activation and alternative ADAR1 and ADAR2 pre-mRNA splicing in modulating serotonin 2C-receptor editing in vivo. RNA 16:1779–1785. doi:10.1261/rna.2188110
Schmidt BJ, Jordan LM (2000) The role of serotonin in reflex modulation and locomotor rhythm production in the mammalian spinal cord. Brain Res Bull 53:689–710
Serra MJ, Smolter PE, Westhof E (2004) Pronounced instability of tandem IU base pairs in RNA. Nucleic Acids Res 32:1824–1828. doi:10.1093/nar/gkh501
Shams TA, Muller DJ (2014) Antipsychotic induced weight gain: genetics, epigenetics, and biomarkers reviewed. Curr Psychiatry Rep 16:473. doi:10.1007/s11920-014-0473-9
Shen M, Eyras E, Wu J, Khanna A, Josiah S, Rederstorff M, Zhang MQ, Stamm S (2011) Direct cloning of double-stranded RNAs from RNase protection analysis reveals processing patterns of C/D box snoRNAs and provides evidence for widespread antisense transcript expression. Nucleic Acids Res 39:9720–9730. doi:10.1093/nar/gkr684
Shen M, Bellaousov S, Hiller M, de La Grange P, Creamer TP, Malina O, Sperling R, Mathews DH, Stoilov P, Stamm S (2013) Pyrvinium pamoate changes alternative splicing of the serotonin receptor 2C by influencing its RNA structure. Nucleic Acids Res 41:3819–3832. doi:10.1093/nar/gkt063
Shen JH, Zhao Y, Rosenzweig-Lipson S, Popp D, Williams JB, Giller E, Detke MJ, Kane JM (2014) A 6-week randomized, double-blind, placebo-controlled, comparator referenced trial of vabicaserin in acute schizophrenia. J Psychiatr Res 53:14–22. doi:10.1016/j.jpsychires.2014.02.012
Shukla AP, Kumar RB, Aronne LJ (2015) Lorcaserin Hcl for the treatment of obesity. Expert Opin Pharmacother 16:2531–2538. doi:10.1517/14656566.2015.1096345
Singh NN, Androphy EJ, Singh RN (2004) In vivo selection reveals combinatorial controls that define a critical exon in the spinal muscular atrophy genes. RNA 10:1291–1305
Soeno Y, Taya Y, Stasyk T, Huber LA, Aoba T, Huttenhofer A (2010) Identification of novel ribonucleo-protein complexes from the brain-specific snoRNA MBII-52. RNA 16:1293–1300
Tecott LH, Sun LM, Akana SF, Strack AM, Lowenstein DH, Dallman MF, Julius D (1995) Eating disorder and epilepsy in mice lacking 5-HT2c serotonin receptors. Nature 374:542–546
Uchida H, Matsumura S, Okada S, Suzuki T, Minami T, Ito S (2017) RNA editing enzyme ADAR2 is a mediator of neuropathic pain after peripheral nerve injury. FASEB J. doi:10.1096/fj.201600950R
Videtic A, Peternelj TT, Zupanc T, Balazic J, Komel R (2009) Promoter and functional polymorphisms of HTR2C and suicide victims. Genes Brain Behav 8:541–545. doi:10.1111/j.1601-183X.2009.00505.x
Walstab J, Steinhagen F, Bruss M, Gothert M, Bonisch H (2011) Differences between human wild-type and C23S variant 5-HT2C receptors in inverse agonist-induced resensitization. Pharmacol Rep 63:45–53
Walter JS, Sacks J, Othman R, Rankin AZ, Nemchausky B, Chintam R, Wheeler JS (2002) A database of self-reported secondary medical problems among VA spinal cord injury patients: its role in clinical care and management. J Rehabil Res Dev 39:53–61
Wang Q, O´Brien PJ, Chen C-X, Cho D-SC, Murray JM, Nishikura K (2000) Altered G protein-coupling functions of RNA editing isoform and splicing variant serotonin 2C receptors. J Neurochem 74:1290–1300
Watanabe Y, Yoshimoto K, Tatebe H, Kita M, Nishikura K, Kimura M, Tanaka M (2014) Enhancement of alcohol drinking in mice depends on alterations in RNA editing of serotonin 2C receptors. Int J Neuropsychopharmacol 17:739–751. doi:10.1017/S1461145713001545
Weissmann D, van der Laan S, Underwood MD, Salvetat N, Cavarec L, Vincent L, Molina F, Mann JJ, Arango V, Pujol JF (2016) Region-specific alterations of A-to-I RNA editing of serotonin 2c receptor in the cortex of suicides with major depression. Transl Psychiatry 6:e878. doi:10.1038/tp.2016.121
Yang W, Wang Q, Kanes SJ, Murray JM, Nishikura K (2004) Altered RNA editing of serotonin 5-HT2C receptor induced by interferon: implications for depression associated with cytokine therapy. Brain Res Mol Brain Res 124:70–78. doi:10.1016/j.molbrainres.2004.02.010
Zhang J, Shen Y, He G, Li X, Meng J, Guo S, Li H, Gu N, Feng G, He L (2008) Lack of association between three serotonin genes and suicidal behavior in Chinese psychiatric patients. Prog Neuropsychopharmacol Biol Psychiatry 32:467–471. doi:10.1016/j.pnpbp.2007.09.019
Zhang Z, Falaleeva M, Agranat-Tamir L, Pages A, Eyras E, Sperling J, Sperling R, Stamm S (2013) The 5′ untranslated region of the serotonin receptor 2C pre-mRNA generates miRNAs and is expressed in non-neuronal cells. Exp Brain Res 230:387–394. doi:10.1007/s00221-013-3458-8
Zhang Z, Shen M, Gresch P, Ghamari-Langroudi M, Rabchevsky AG, Emeson RB, Stamm S (2016) Oligonucleotide-induced alternative splicing of serotonin 2C receptor reduces food intake. EMBO molecular medicine 8:878–894. doi:10.15252/emmm.201506030
Acknowledgements
The work was supported by the NIH (R21NS098186) and the Foundation for Prader–Willi Research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Stamm, S., Gruber, S.B., Rabchevsky, A.G. et al. The activity of the serotonin receptor 2C is regulated by alternative splicing. Hum Genet 136, 1079–1091 (2017). https://doi.org/10.1007/s00439-017-1826-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-017-1826-3