Abstract
Pseudomonas aeruginosa (PA) is an important opportunistic pathogen that causes different infections on immunocompromised patients. Within PA accessory genome, differences in virulence, antibiotic resistance and biofilm formation have been described between strains, leading to the emergence of multidrug-resistant strains. The genome sequences of 17 strains isolated from patients with healthcare-associated infections in a Mexican hospital were genomically and phylogenetically analyzed and antibiotic resistance genes, virulence genes, and biofilm formation genes were detected. Fifteen of the 17 strains were resistant to at least two of the carbapenems meropenem, imipenem, and the monobactam aztreonam. The antibiotic resistance (mexA, mexB, and oprM) and the biofilm formation (pslA and pslD) genes were detected in all strains. Differences were found between strains in accessory genome size. The strains had different sequence types, and seven strains had sequence types associated with global high risk epidemic PA clones. All strains were represented in two groups among PA global strains. In the 17 strains, horizontally acquired resistance genes to aminoglycosides and beta-lactams were found, mainly, and between 230 and 240 genes that encode virulence factors. The strains under study were variable in terms of their accessory genome, antibiotic resistance, and virulence genes. With these characteristics, we provide information about the genomic diversity of clinically relevant PA strains.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Currently, most antibiotics have become ineffective for the control of bacterial infections, as bacteria have acquired multiresistance to these kinds of drugs due to the indiscriminate and widespread use of antibiotics (Shahi and Kumar 2015). These multidrug-resistant (MDR) pathogens have been called "superbugs" and have become one of the most critical public health problems causing more than 1 million deaths per year worldwide (CDC. Antibiotic Resistance Threats in the United States 2019; Antimicrobial Resistance Collaborators 2022). The term "ESKAPE" encompasses six of these pathogens with increasing multidrug resistance and virulence: Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa (PA), and Enterobacter spp. ESKAPE pathogens are responsible for most nosocomial infections and are able to “escape” the biocidal action of antimicrobial agents (Mulani et al. 2019).
The World Health Organization (WHO) reports that 50% of infections caused by Escherichia coli, Staphylococcus aureus, Klebsiella pneumoniae, and Pseudomonas aeruginosa show resistance to antibiotics such as cephalosporin, an improved beta-lactam antibiotic. This group of bacteria induces healthcare-associated infections (HAIs) or nosocomial infections, which are defined as infections acquired by a patient during treatment in a hospital that a patient does not have at the time of admission (PAHO 2012). HAIs affect 1 in 20 hospitalized patients, and in Mexico the yearly death toll attributed to HAI is 37,000 by this kind of infections (IMSS 2019).
In Mexico, the epidemiological surveillance of HAIs is under the charge of the Hospital Epidemiological Surveillance Network. Additionally, the Institute for Social Security and Services for State Workers (ISSSTE) carries out its control and surveillance through the Committee for the Detection and Control of Nosocomial Infections, finding PA among the first microorganisms causing HAIs (RHOVE 2021).
Pseudomonas aeruginosa is a Gram-negative bacterium established in water, soil, and various host organisms. In humans, it is considered the fourth most isolated nosocomial pathogen, accounting for 10% of all HAIs, and the second most common cause of pneumonia (IMSS 2019). PA is characterized by its ability to generate resistance to antibiotics, through gene mutation or by transferring horizontally acquired resistance genes (Ozer et al. 2014). Both clinical and environmental strains can thrive in a wide variety of ecological niches and cause damage in different hosts, as it is a versatile bacterium with easy adaptability due to the maintenance of virulence and metabolic genes in its genome. The importance of this bacterium lies in the fact that it causes significant morbidity and mortality among immunocompromised patients and those requiring mechanical ventilation, such as burn patients and those with cystic fibrosis (CF). Some multilocus sequence types (ST) referred to as high-risk clones are extensively issued throughout the world and frequently found in PA (Valot et al. 2015).
The increased availability of genomes has made it possible to perform comparative genomic analyses of PA strains in distinct parts of the world (Ozer et al. 2014; Jeukens et al. 2014; Subedi et al. 2018). These studies have defined the pangenome of strains with a "core genome", which is all the genes present, and an "accessory genome" which is the set of genes not shared among all members of the species (Ozer et al. 2014). The PA genome has a size of 5.5–7 Mbp; this variation is due to the presence of an accessory genome that is specific to each strain and can occupy up to 20% of the total genome. This accessory genome consists of horizontally transferable elements including prophages, plasmids, insertion sequences (IS), genomic islands (GI), and transposons. The accessory genome is significant for bearing acquired antibiotic resistance and virulence genes, with lateral transfer of these genes among strains contributing to the development of more virulent strains. Additionally, mutational changes may promote antibiotic resistance and virulence. Genome characterization studies of PA isolates such as those by Aggarwal et al. (2015) and Subedi et al. (2018) have shown that the pangenome contains large heterogeneity in accessory genome size among 22 strains which was correlated with the quantity of prophages, insertion sequences, and genomic islands. The strains from these studies had different sequence types and were distinct from the worldwide PA epidemic clones. The isolates previously characterized have genes that confer resistance to aminoglycosides, beta-lactams, quaternary ammonium compounds, sulfonamides, chloramphenicols and tetracyclines. We know that PA genome aids in comprehending the genetic modifications that are related to more resistant and pathogenic strains, however there is little information on comparative genomic analysis of PA strains isolated in Mexico. Hence, the objective of this study was to compare the genomic variability within PA strains isolated from hospitalized patients with different HAIs from the Centro Medico Nacional 20 de Noviembre in Mexico City, for which the complete genomes of 17 strains were sequenced and genomic analysis was performed by comparing evolutionary relationships, genomic divergence, antibiotic resistance, and virulence factors.
Materials and methods
Bacterial strains and antibiotic susceptibility testing
We studied 17 clinical strains of Pseudomonas aeruginosa provided by the Central Epidemiological Surveillance Laboratory, Centro Medico Nacional 20 de Noviembre ISSSTE of Mexico City, isolated only from this hospital from samples of different patients with HAIs between the years 2019 and 2021, as shown in Table 1. The disk diffusion method was performed based on the guidelines recommended by the Clinical and Laboratory Standards Institute (CLSI) to determine antibiotic susceptibility patterns, using PA ATCC 27853 as control strain. The antibiotic disks (BD BBL, USA) used in this study included meropenem (10 µg), imipenem (10 µg), and aztreonam (30 µg). The isolates with resistance to any of the two carbapenems (meropenem and imipenem) were considered as PA resistant to carbapenems, and the isolates with resistance to aztreonam were considered as PA resistant to monobactams (Magiorakos et al. 2012).
DNA extraction and PCR detection of resistance and biofilm formation genes
The strains were grown in 5 ml of TSB liquid medium at 37 °C for 24 h and genomic DNA was extracted following the method described by Chen and Kuo (1993). PCR reactions for detection of efflux pump (mexA, mexB, and oprM) and biofilm-forming (pslA and pslD) genes were done using specific primers listed in Table S1. Reactions were carried out in 0.2 mL microcentrifuge tubes in a reaction volume of 25 μL using Go Taq PCR CORE systems kit (Promega) containing 5× buffer, 25 mM MgCl2, 10 mM nucleotide triphosphates (dNTPs), 0.1 μM primers and1-unit Taq DNA polymerase (GoTaq Promega). PCR conditions consisted of one cycle at 95 °C for 5 min; 30 cycles of 30 s at 95 °C, 40 s at Tm specific for each primer, 1 min at 72 °C and a final elongation at 72 °C for 10 min. The PCR products were separated on a 1% ethidium bromide agarose gel with 1X TAE (70 v, 300 mA, 1 h) and visualized using UV light transilluminator (Ugwuanyi et al. 2021).
Whole genome sequencing
Once a 3-µg sample of genomic DNA with a purity of 1.8 (absorbance ratio of 260/280 nm) by the method described previously was obtained and its integrity verified by 1% agarose gel electrophoresis, the samples were sequenced by service at the Laboratorio Nacional de Genómica para la Biodiversidad del Centro de Investigación y de Estudios Avanzados del IPN (LANGEBIO, CINVESTAV-IPN, Irapuato Unit). Seventeen of 137 (12.4%) of the isolates collected as part of surveillance were sequenced. The selection criteria for the isolates to be sequenced were strains that had different susceptibility to the carbapenems and aztreonam tested and that they were strains isolated from different years. The sequencing of the genetic material was carried out by the sequencing-by-synthesis method using a Truseq DNA nano kit with the Illumina Miseq system in 2 × 150 bp format.
Genomes assembly and analysis
MiSeq sequencing resulted in a range of 653,270 to 2,199,951 reads per sample with an average coverage of 140×. With FastQC version 0.11.9 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc), the quality of the raw reads was evaluated, and Trimmomatic version 0.36 (Bolger et al. 2014) was used to remove adapters with default parameters (SLIDINGWINDOW:4:30 MINLEN:36). A de novo genome assembly was done using SPAdes version 3.13.1 (Bankevich et al. 2012) using the default settings. Prokka version 1.14.5 was then used for annotations of the assembled genomes (Seemann 2014). The PA PAO1 genome (RefSeq accession number NC_002516.2) was used as a reference in the present work. The preliminary genome contigs were rearranged and aligned to the reference genome with the MAUVE multiple genome alignment software (Darling et al. 2010; Rissman et al. 2009). Genomic islands were determined using IslandViewer (Bertelli et al. 2017), and PHASTER to identify and annotate prophages (Arndt et al. 2016). The pubMLST database was used to determine the multilocus sequence type, selecting Pseudomonas aeruginosa as an organism, single sequence and uploading the fasta file of each genome (Jolley et al. 2018). The completed genome sequence of the strains has been deposited in the GenBank database under accession number PRJNA1040412 (http://www.ncbi.nlm.nih.gov/bioproject/1040412).
Pangenome and phylogenetics
Pangenome analysis was done with Roary version 3.13.0 (Page et al. 2015). Common genes in at least 99% of the strains were considered core genes and cloud, shell and soft core genes corresponding to gene families with presence in < 15%, 15–95%, 95–99%, respectively. The accessory genome was obtained by subtracting the core genes from the total genes of the genome of each strain. Two hundred fifty-two genomes of PA strains (235 previously reported complete genomes downloaded from The Pseudomonas Genome Database and 17 genomes from this work) were aligned with Parsnp version 1.2 having the PAO1 strain sequence as a reference, and then it was used to make a maximum likelihood tree in Mega X, taking in account the core genome single nucleotide polymorphisms (SNPs).
Screening of antibiotic resistance and virulence genes
The genomes of PA isolates were screened to identify acquired antibiotic resistance genes by using Resfinder 3.0 (Centre for Genomic Epidemiology, DTU, Denmark) (Zankari et al. 2012). Virulence genes associated with adherence, quorum sensing, type IV secretion system, alginate production, toxins and protease production were searched in the genomes using the virulence factor database (VFDB) uploading the fasta file of each genome and selecting the genus Pseudomonas (Chen et al. 2005).
Results
Antibiotic susceptibility profile
Seven of the 17 strains had antibiotic resistance to imipenem and meropenem, 8 strains had intermediate resistance to aztreonam, while 2 strains were susceptible to the three antibiotics tested. The strains showed greater resistance to imipenem and meropenem (carbapenems), compared to aztreonam (monobactam), irrespective of the year of collection (Table 1).
General characteristics of genomes and detection of antibiotic resistance and biofilm formation genes
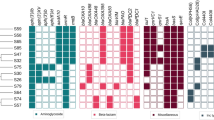
The contigs generated by de novo assembly of the 17 PA strain genomes ranged from 96 in strain 19,256 to 754 in strain 20,019. The genomes in this study had a size of 6.6–7.3 Mbp and a mean C+G content of 65.95%. Genome size ranged extensively between strains, with up to 1.0 Mbp more DNA than the PAO1 strain. The number of coding sequences (CDS) oscillates from 5969 (strain 19,029) to 6844 (strain 19,133). The general characteristics of the genomes are shown in Table 2. Genotypic evaluation showed that all antibiotic resistance genes (mexA, mexB and oprM) and biofilm formation genes (pslA and pslD) were detected in the 17 strains as shown in Fig. S1 and these five PCR-amplified genes presented above (mexA, mexB, oprM, pslA and pslD) were manually searched and found to be present in all the genomes of the strains.
A total of 12,273 orthologs were obtained from analyzed strains in this study. Additionally, of the 12,273 genes shared by all strains, this study described 4813 genes that formed the core genome which was common in at least 99% of the strains, no soft-core genes were found (common in 95–99% of strains), and there were 2938 shell genes (common in15-95% of strains) and 4521 cloud genes (common in 0–15% of strains) (Fig. 1).
Pangenome of the 17 PA strains studied. a Pie chart of the breakdown of genes. The pangenome is subdivided into core and accessory genes. The accessory genome is further subdivided into cloud and shell genomes, which are based on the number of genomes that contain the gene(s). The number of genes present in each category are shown. The number of strains necessary for a gene to meet the defined category is shown in parentheses. b A tree compared to a matrix that represents the gene content, in terms of presence/absence of core and accessory genes of the pangenome inferred from the comparison of the 17 PA complete genomes
The accessory genome ranged from 19 to 29% of the complete genome. In these accessory genomes, genes were found associated with antibiotic resistance and virulence (beta-lactamases, multidrug efflux pumps, toxins, transposases, integrases), as well as genes that aid strains to survive in hostile environments like strains cultured from sites containing toxic organic compounds and heavy metals which are unsuited for PA habitation (hypothetical proteins mainly).
As elements of accessory genome, the results indicate that the predicted number of genomic islands (GI) ranged from 27 to 90 and that of of prophages from 1 to 5 in the 17 genomes (Table S2).
According to the multilocus sequence typing (MLST) analysis, ten different sequence types (ST) were found in the 17 genomes, and only one constituted a new type. The results indicate that five isolates belonged to ST 233, three isolates to ST 309 and two strains to ST 3045 (sequence types determined by MLST database are listed in Table 2), while the remaining strains corresponded to seven different STs.
Phylogenetics
The strains were not clonally related and were well represented in two groups, nine strains in group 1 and eight in group 2 (Fig. 2). Interestingly, three groups of strains that were grouped by clade had the same ST in each clade. Strains 19,121, 19,241, 19,256, 19,299 and 21,049 were grouped within group 1 and had ST 233 reported as high-risk clones, also within the group. Group 1 strains 20,061 and 21,068 had ST 3045 and, within group 2, strains 20,019, 19,071 and 19,291 with ST 309 were grouped in the same clade. This could indicate potential transmission, but it would be necessary to analyze both phylogenetic distance and epidemiological link.
Genetic profiles of antibiotic resistance and virulence genes
A total of 45 different acquired antibiotic resistance genes were found in the present work using ResFinder database (Fig. 3). Most resistance genes associated with aminoglycosides and beta-lactams were observed in the strains from this study. The genes blaPAO (resistance to beta-lactams), fosA (resistance to fosfomycin), aph(3′)-IIb (resistance to aminoglycosides) and catB7 (resistance to chloramphenicol) were detected in the 17 strains. The aph(3')-IIb gene was the most common aminoglycoside, being found in all strains except 19,256 strain. One strain (19,291) had five resistance genes that were not detected in other strains; aph(6)-Id, aph(3')-VIa, aph(3'')-Ib aminoglycoside, blaIMP-15 and catA1 that confer resistance to aminoglycosides, beta-lactams and phenicols.
Virulence genes related with adherence, quorum sensing, type IV secretion system, alginate production, toxins and protease production were searched and found between 230 and 240 genes that encode virulence factors in our 17 PA genomes using the Virulence Factor Database (VFDB). Differences in virulence genes were found for genes involved in adherence, iron uptake and secretion systems (Table 3).
Discussion
A total of 17 strains of PA isolated from different samples of hospitalized patients with HAIs were obtained. Although most of the strains were resistant to more than one antibiotic, it would be appropriate to use a greater number of strains from 2020 and 2021 to make a fair comparison since most of the strains were isolated from the year 2019. Also, carbapenems are described as the most active drugs against PA (Bajpai et al. 2019), so it is important to highlight that most of the strains in this study were resistant to this class of antibiotics, which points to the need to find new alternatives to control this pathogen. Jeukens et al. (2017) reported that seven strains from CF patients were mostly susceptible to imipenem and meropenem, but resistant to ampicillin and amoxicillin; however, these strains were isolated in 1993, so it can be inferred that the bacterium have become more resistant over the years. The emergence of Pseudomonas species resistant to carbapenems and monobactams has been reported in several studies. Bajpai et al. (2019) found 126 isolates resistant to multiple kinds of antibiotics, with greater resistance to meropenem, 92/126 (73%); aztreonam, 87/126 (69%); and imipenem, 78/126 (61.9%). In another work, Subedi et al. (2018) reported 22 susceptible and intermediate resistance strains to imipenem and aztreonam (carbapenem and monobactam, respectively), but resistant to gentamicin (aminoglycoside) and ciprofloxacin (quinolone); so, susceptibility to antibiotics may vary according to the classes of antibiotics.
Given the high levels of resistance present in the strains under study, the detection of some genes involved in antibiotic resistance and biofilm formation was sought. For example, the mexAB-oprM efflux pump system has been reported to be very common and aids in PA intrinsic resistance to β-lactams (including carbapenems); thus, the genes (mexA, mexB and oprM) related to this antibiotic resistance system were searched for in the 17 strains. Also were searched the biofilm formation genes (pslA and pslD) that have been reported to enhance biofilms formation that promote resistance to antibiotics (Ugwuanyi et al. 2021). Our results coincide with the previous work of Ugwuanyi et al. (2021) where they describe that the mexA, pslA and pslD genes were detected in 39 (100%) isolates, but not the mexB and oprM genes, which were identified in 34–36 isolates (88–92%), respectively. However, Abbas et al. (2018) reported the detection of mexAB-R in 100% of PA isolates from urinary tract infections in Egypt.
In the genomic characterization, the number of contigs and genome size were similar to those in other published PA genomes (Jeukens et al. 2014, 2017; Subedi et al. 2018; Hao et al. 2021). The genomes of the 17 strains isolated in this work contain more ortholog genes than those in the study of Subedi et al. (2018), which isolated 22 PA strains from various origins and tfound 9786 orthologs in the pangenome. The larger pangenome found in the present study may be due to the diverse type of samples of patients that were used to isolate the strains, since the size of the pangenome can increase depending on the diversity of the strains in the study and depicts the accumulated genetic information between a group of bacterial genomes.
Previous reports have informed core genomes of 5316 (6406 pangenes) (Ozer et al. 2014), 5233 (9344 pangenes) (Valot et al. 2015), and 4910 (9786 pangenes) (Subedi et al. 2018) in different strains of PA. Even though these studies used less (less than 17) genomes, the results can be compared. Finding the smallest core genomes in this study may be due to the use of more genomes for the alignment, as well as the use of incomplete draft genomes and definition of the core genome (≥ 99% similarity in each strain) (Subedi et al. 2018). Within the core genome were genes related to respiratory and metabolic genes that give the bacterium the ability to survive on a variety of carbon sources to obtain energy (Valot et al. 2015).
Besides the core genome, PA has an accessory genome that varies across strains, where most of these genes are acquired horizontally. In this work, accessory genes were calculated by removing the core genes (4813) from the number of CDS. The accessory genome of the strains in this study, which is larger than previously reported may be attributed to the use of draft genomes that may overestimate the number of accessory genes giving a seemingly larger genome (Subedi et al. 2018). Therefore, the increase in the number of accessory genes found in the present work may be because the strains have acquired many genes to be able to grow and survive in the hospital environment where, thanks to the contribution of hospital equipment and personnel, there is greater dissemination of resistance genes (Subedi et al. 2018). According to previously reported accessory genomes, a large number of undetermined genes were also found in this study, either because their function is unknown, or they are pseudogenes. It is important to highlight that in these accessory genomes, there were many genes that participate in replication, recombination, and repair, mainly integrases, recombinases and transposases, whose function is DNA mobility (Valot et al. 2015). As important elements of accessory genome, genomic islands (GI) and prophages were also identified and found in the genomes (Kung et al. 2010).
The ST was allocated to every strain; therefore if a strain did not coincide with the MLST database, it was taken as a new ST and is stored in the database after verification. These results indicate that the strains in the study belong to a wide variety of STs and coincide with previously clinical epidemic isolates reported worldwide (Aguilar-Rodea et al. 2017; Miyoshi-Akiyama et al. 2017; Witney et al. 2014; Woodford et al. 2011). The ST 233 was the most common ST found in this work, which has been reported in MDR hospital strains in different countries (Kocsis et al. 2021). Most frequently reported international high-risk MDR PA clones are ST 111, 175, 233, 235, 277, 357, 654, and 773 (Woodford et al. 2011; Kos et al. 2015; Kocsis et al. 2019). ST 233 has been described as a high-risk clone in Mexico, the USA, South Africa, and Japan. An ST 111 strain reported in Bulgarian hospitals was resistant to carbapenems and aztreonam (Kocsis et al. 2021), which coincides with the 19,133 strain that had the same ST and presented resistance to the three antibiotics.
The phylogenetic analysis coincides with previous studies where it has been described that PA strains from different sources fall into two main groups, where group 1 is the largest and includes the most investigate worldwide strains DK2, PAO1, LESB58 and PAK, while group 2 is slightly smaller and contains the studied virulent strain PA14 (Stewart et al. 2014).
In the identification of acquired antibiotic resistance genes, as in the 22 PA strains reported in the work by Subedi et al. (2018), the genes blaPAO (resistance to b-lactams), fosA (resistance to fosfomycin), aph(3′)-IIb (resistance to aminoglycosides) and catB7 (resistance to chloramphenicol) were also detected in the 17 strains (100%) analyzed.
Differences in virulence genes were more evident for genes with the type III secretion system (exoS, exoU, exoY and exoT). The ExoT and ExoS toxins were the first to be identified in PA, participating in related functions such as altering the host cell actin cytoskeleton, inducing host cell rounding, inhibiting phagocytosis, and even causing cell death (Derakhshan et al. 2021). ExoY cleaves the intracellular cAMP in eukaryotic cells and causes cell rounding (Derakhshan et al. 2021). ExoU is considered the most virulent effector, as it disrupts eukaryotic membranes in different cell types. Based on these characteristics, PA strains can be classified into two groups: strains expressing ExoT and exoU cause rapid host cell death, while strains expressing exoS and exoT cause slow cell death. With the presence of exoS in ten strains in this study (59%), the strains can evade the action of antibiotics due to their ability to invade mammalian cells, while the presence of exoU in seven strains (41%) gives them a survival advantage by increasing resistance (Derakhshan et al. 2021). The exoS and exoU genes are mutually exclusive which was verified in the strains under study, since one of the two genes was found in each strain (Berthelot et al. 2003). The exoU gene is part of genomic islands, so the studied strains that possess exoU showed larger accessory genomes. The exoY and exoT genes have been described as the most prevalent secretory toxins in strains, being found in all (100%) and 15 (88%) of the strains, respectively (Fig. 4) (Subedi et al. 2018).
Regarding the type VI secretion system in PA, it has been reported that the phospholipase D gene (pldA) promotes chronic infections (Subedi et al. 2018); however, pldA was absent in 8 (47%) of the 17 isolates in this study (Fig. 4). Adhesion-associated genes such as type IV pili (pilA) are involved in adherence to and invasion of mucosal surfaces; consequently, pilA mutants exhibit less adherence and invasion of airway epithelial cells (Okuda et al. 2013). In this study, pilA gene was found in 10 (59%) of the 17 strains (Fig. 4), including some associated with respiratory tract infections.
Conclusion
In this study, a comparative genomic analysis was performed between PA isolates from patients with HAIs. The strains under study were different in terms of their accessory genome, antibiotic resistance, and virulence genes, and phylogenetically, the 17 strains were distributed throughout the two PA groups described. However, all strains have all the characteristics to become resistant to antibiotics used in hospitals, as well as possessing the necessary pathogenicity to cause infections in humans. Knowledge of the genes shared by most PA isolates makes it possible to search for, propose, and approve new therapies to combat the great diversity of human infections caused by this bacterium. Therefore, this information can be used to elucidate mechanisms or search for alternatives that help combat this virulent and multiresistant bacterium in the hospital environment.
Conflict of interest
The authors declare no competing interests.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Abbas H, El-Ganiny A, Kamel H (2018) Phenotype and genotypic delection of antibiotic resistance of Pseudomonas aeruginosa isolated from urinary tract infections. Afri Health Sci 18(1):11–21. https://doi.org/10.4314/ahs.v18i1.3
Aggarwal R, Dawar C, Das S, Sharma S (2015) Draft genome sequences of two drug resistant isolates of Pseudomonas aeruginosa obtained from keratitis patients in India. Genome Announc 3. https://doi.org/10.1128/genomeA.01404-14
Aguilar-Rodea P, Zúñiga G, Rodríguez-Espino B, Olivares-Cervantes A, Gamiño-Arroyo A, Moreno-Espinosa S et al (2017) Identification of extensive drug resistant Pseudomonas aeruginosa strains: New clone ST1725 and high-risk clone ST233. PLoS ONE 12:e0172882
Antimicrobial Resistance Collaborators (2022) Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399:629–655. https://doi.org/10.1016/S0140-6736(21)02724-0
Arndt D, Grant J, Marcu A, Sajed T, Pon A, Liang Y et al (2016) PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44:W16-21. https://doi.org/10.1093/nar/gkw387
Bajpai V, Govindaswamy A, Khurana S, Batra P, Aravinda A, Katoch O et al (2019) Phenotypic and genotypic profile of antimicrobial resistance in Pseudomonas species in hospitalized patients. Indian J Med Res pp 216–221. https://doi.org/10.4103/ijmr.IJMR_1_18
Bankevich A, Nurk S, Antipov D, Gurevich A, Dvorkin M, Kulikov A et al (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. https://doi.org/10.1089/cmb.2012.002
Bertelli C, Laird M, Williams K, Simon Fraser University Research Computing Group, Lau B, Hoad G et al (2017) IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res 45:W30–W35. https://doi.org/10.1093/nar/gkx343
Berthelot P, Attree I, Plesiat P, Chabert J, de Bentzmann S, Pozzetto B et al (2003) Genotypic and phenotypic analysis of type III secretion system in a cohort of Pseudomonas aeruginosa bacteremia isolates: evidence for a possible association between O serotypes and exogenes. J Infect Dis 188:512–518. https://doi.org/10.1086/377000
Bolger M, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
CDC. Antibiotic Resistance Threats in the United States (2019) Atlanta, GA, U.S. Department of Health and Human Services
Chen L, Yang J, Yu J (2005) VFDB: a reference database for bacterial virulence factors. 33:D325–D328. https://doi.org/10.1093/nar/gki008
Chen W, Kuo T (1993) A simple and rapid method for the preparation of gram-negative bacterial genomic DNA. Nucleic Acids Res 21:22–60. https://doi.org/10.1093/nar/21.9.2260
Darling A, Mau B, Perna N (2010) progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 5:e11147. https://doi.org/10.1371/journal.pone.0011147
Derakhshan S, Rezaee A, Mohammadi S (2021) Relationship between type III secretion toxins, biofilm formation, and antibiotic resistance in clinical Pseudomonas aeruginosa isolates. Russ J Infect Immun 11(6):1075–1082
Hao M, Ma W, Dong X, Li X, Cheng F, Wang Y (2021) Comparative genome analysis of multidrugresistant Pseudomonas aeruginosa JNQHPA57, a clinically isolated mucoid strain with comprehensive carbapenem resistance mechanisms. BMC Microbiol 21:133. https://doi.org/10.1186/s12866-021-02203-4
IMSS (Instituto Mexicano del Seguro Social) (2019) Manual para la implementación de los paquetes de acciones para prevenir y vigilar las infecciones asociadas a la atención de la salud (IAAS). Primera edición, México, 68 pp
Jeukens J, Boyle B, Kukavica-Ibrulj I, Ouellet M, Aaron S, Charette S et al (2014) Comparative genomics of isolates of a Pseudomonas aeruginosa epidemic strain associated with chronic lung infections of cystic fibrosis patients. PLoS ONE 9(2):e87611. https://doi.org/10.1371/journal.pone.0087611
Jeukens J, Kukavica I, Emond J, Freschi L, Levesque R (2017) Comparative genomics of a drug-resistant Pseudomonas aeruginosa panel and the challenges of antimicrobial resistance prediction from genomes. FEMS Microbiol Lett 364:18. https://doi.org/10.1093/femsle/fnx161
Jolley KA, Bray JE, Maiden MCJ (2018) Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 24(3):124. https://doi.org/10.12688/wellcomeopenres.14826.1
Kocsis B, Toth A, Gulyas D, Ligeti B, Katona K, Rokusz L et al (2019) Acquired qnrVC1 and blaNDM-1 resistance markers in an international high-risk Pseudomonas aeruginosa ST773 clone. J Med Microbiol 68:336–338
Kocsis B, Gulyás D, Szabó D (2021) Diversity and distribution of resistance markers in Pseudomonas aeruginosa international high-risk clones. Microorganisms 9:359. https://doi.org/10.3390/microorganisms9020359
Kos V, Déraspe M, McLaughlin R, Whiteaker J, Roy P, Alm R et al (2015) The resistome of Pseudomonas aeruginosa in relationship to phenotypic susceptibility. Antimicrob Agents Chemother 59:427–436
Kung V, Ozer E, Hauser A (2010) The accessory genome of Pseudomonas aeruginosa. Microbiol Mol Biol Rev 74:621–641. https://doi.org/10.1128/MMBR.00027-10
Magiorakos A, Srinivasan A, Carey R, Carmeli Y, Falagas M, Giske C et al (2012) Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18(3):268–281. https://doi.org/10.1111/j.1469-0691.2011.03570.x
Miyoshi-Akiyama T, Tada T, Ohmagari N et al (2017) Emergence and Spread of Epidemic Multidrug-Resistant Pseudomonas aeruginosa. Genome Biol Evol 9:3238–3245
Mulani M, Kamble E, Kumkar S, Tawre M (2019) Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: a review. Front Microbiol 10(539):1–24. https://doi.org/10.3389/fmicb.2019.00539
Okuda J, Hayashi N, Arakawa M, Minagawa S, Gotoh N (2013) Type IV pilus protein PilA of Pseudomonas aeruginosa modulates calcium signaling through binding the calcium modulating cyclophilin ligand. J Infect Chemother 19:653–664. https://doi.org/10.1007/s10156-012-0536-456-y
Ozer E, Allen J, Hauser A (2014) Characterization of the core and accessory genomes of Pseudomonas aeruginosa using bioinformatic tools Spine and AGEnt. BMC Genomics 15:737. https://doi.org/10.1186/1471-2164-15-737
Page A, Cummins C, Hunt M, Wong V, Reuter S, Holden M et al (2015) Roary: rapid large-scale prokaryote pangenome analysis. Bioinformatics 31:3691–3693. https://doi.org/10.1093/bioinformatics/btv421
PAHO (Pan American Health Organization) (2012) Epidemiological Surveillance of Healthcare Associated Infections. Washington DC, 60 pp.
RHOVE (Red Hospitalaria de Vigilancia Epidemiológica) (2021) Health Care Associated Infections Bulletin. México, 23 pp
Rissman A, Mau B, Biehl B, Darling A, Glasner J, Perna N (2009) Reordering contigs of draft genomes using the Mauve aligner. Bioinformatics 25:2071–2073. https://doi.org/10.1093/bioinformatics/btp356
Seemann T (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. https://doi.org/10.1093/bioinformatics/btu153
Shahi S, Kumar A (2015) Isolation and genetic analysis of multidrug resistant bacteria from diabetic foot ulcers. Front Microbiol 6. https://doi.org/10.3389/fmicb.2015.01464
Stewart L, Ford A, Sangal V, Jeukens J, Boyle B, Kukavica-Ibrulj I et al (2014) Draft genomes of 12 host-adapted and environmental isolates of Pseudomonas aeruginosa and their positions in the core genome phylogeny. Pathog Dis 71(1):20–25. https://doi.org/10.1111/2049-632X.12107
Subedi D, Vijay A, Kohli G, Rice S, Willcox M (2018) Comparative genomics of clinical strains of Pseudomonas aeruginosa strains isolated from different geographic sites. Sci Rep 8:15668. https://doi.org/10.1038/s41598-018-34020-7
Ugwuanyi F, Ajayi A, Ojo D, Adeleye A, Smith S (2021) Evaluation of efflux pump activity and biofilm formation in multidrug resistant clinical isolates of Pseudomonas aeruginosa isolated from a Federal Medical Center in Nigeria. Ann Clin Microbiol Antimicrob 20:11. https://doi.org/10.1186/s12941-021-00417-y
Valot B, Guyeux C, Rolland J, Mazouzi K, Bertrand X, Hocquet D (2015) What it takes to be a Pseudomonas aeruginosa? The core genome of the opportunistic pathogen updated. PLoS ONE 10(5):e0126468. https://doi.org/10.1371/journal.pone.0126468
Witney A, Gould K, Pope C, Bolt F, Stoker N, Cubbon M et al (2014) Genome sequencing and characterization of an extensively drug-resistant sequence type 111 serotype O12 hospital outbreak strain of Pseudomonas aeruginosa. Clin Microbiol Infect 20:O609–O618
Woodford N, Turton J, Livermore D (2011) Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev 35:736–755
Zankari E, Hasman H, Cosentino S et al (2012) Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. https://doi.org/10.1093/jac/dks261
Funding
This research was funded by E05-ciencia y tecnología ISSSTE.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Juana Salazar-Salinas, Omar Fernando Mendoza-Vázquez, and Gabriel Damazo-Hernández provided clinical isolates. Josefina León-Félix and María José Martínez-Gallardo designed the experiments. Experiment execution was performed by María José Martínez-Gallardo. Data analysis was performed by Josefina León-Félix, María José Martínez-Gallardo, and Claudia-Villicaña. The first draft of the manuscript was written by María José Martínez-Gallardo. Martha Yocupicio-Monroy and Sofía Lizeth Alcaraz-Estrada critically revised the work. All authors read and approved the final manuscript.
Corresponding author
Additional information
Communicated by Martine Collart.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Martínez-Gallardo, M.J., Villicaña, C., Yocupicio-Monroy, M. et al. Comparative genomic analysis of Pseudomonas aeruginosa strains susceptible and resistant to carbapenems and aztreonam isolated from patients with healthcare-associated infections in a Mexican hospital. Mol Genet Genomics 299, 29 (2024). https://doi.org/10.1007/s00438-024-02122-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00438-024-02122-9