Abstract
Next generation sequencing tests are used routinely as first-choice tests in the clinic. However, systematic performance comparing the results of exome sequencing as a single test replacing Sanger sequencing of targeted gene(s) is still lacking. Performance comparison data are critically important for clinical case management. In this study, we compared Sanger-sequencing results of 258 genes to those obtained from next generation sequencing (NGS) using two exome-sequencing enrichment kits: Agilent-SureSelectQXT and Illumina-Nextera. Sequencing was performed on leukocytes and buccal-derived DNA from a single individual, and all 258 genes were sequenced a total of 11 times (using different sequencing methods and DNA sources). Sanger sequencing was completed for all exons, including flanking ± 8 bp regions. For the 258 genes, NGS mean coverage was > 20 × for > 98 and > 91% of the regions targeted by SureSelect and Nextera, respectively. Overall, 449 variants were identified in at least one experiment, and 407/449 (90.6%) were detected by all. Of the 42 discordant variants, 23 were determined as true calls, summing-up to a truth set of 430 variants. Sensitivity of true-variant detection was 99% for Sanger sequencing and 97–100% for the NGS experiments. Mean false-positive rates were 3.7E-6 for Sanger sequencing, 2.5E-6 for SureSelect-NGS and 5.2E-6 for Nextera-NGS. Our findings suggest a high overall concordance between Sanger sequencing and NGS performances. Both methods demonstrated false-positive and false-negative calls. High clinical suspicion for a specific diagnosis should, therefore, override negative results of either Sanger sequencing or NGS.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Next generation sequencing (NGS) has taken the fast lane from the bench to the bedside. What was once a “last resort” technique for exceptional cases in which a molecular diagnosis could not be achieved by standards means has become a routine clinical test allowing physicians to translate genomic information into clinically actionable decisions (Goodwin et al. 2016). NGS-based testing has thus largely replaced Sanger sequencing, long considered to be the gold standard of sequencing tests. This shift has occurred despite lack of bidirectional performance comparisons of Sanger sequencing and NGS. Data on such comparisons can directly impact the decision-making process of clinicians aiming to determine the genetic basis of various conditions.
NGS has been applied for a variety of purposes, including exome/genome sequencing for diagnosis of genetic diseases (Gilissen et al. 2014; Lee et al. 2014), cancer genomics (Shen et al. 2015), discovery of transcription factor binding sites (Mundade et al. 2014), transcriptome profiling (Li et al. 2014), DNA methylation sequencing (Brunner et al. 2009) and noncoding RNA expression profiling (Ching et al. 2015). In the clinical context, the wide scope of testing enabled by NGS has facilitated the deciphering of the genetic bases of various human diseases for which the molecular mechanism was unknown (Ng et al. 2010), has expanded the number of genes known to be associated with defined phenotypes (De Rubeis and Buxbaum 2015) and has led to detection of novel mutations in genes with well described functions (Lerat et al. 2016; Maksemous et al. 2016).
Molecular investigation of genetically heterogeneous phenotypes often mandates the sequential or simultaneous sequencing of multiple genes which can be achieved through Sanger sequencing or NGS. Sanger sequencing entails the design of specific primers to guarantee successful amplification of regions of interest and may overcome difficulties such as sequencing within GC-rich regions and separating sequencing of genes from their pseudogenes (Wenzel et al. 2009; Sumner et al. 2014). However, Sanger sequencing of multiple genes for diagnostic purposes is costly and time consuming, whereas it is eminently feasible using NGS (Rabbani et al. 2016). The advantages of NGS platforms over Sanger sequencing with respect to genotyping capacity and cost have been demonstrated repeatedly (Goodwin et al. 2016). Indeed, NGS applications are readily available as gene panels or through exome sequencing (Iglesias et al. 2014) for analysis of various phenotypes, e.g. non-syndromic deafness, neuro-muscular diseases and Noonan-spectrum syndromes (Ankala et al. 2015; Neveling et al. 2013; Wang et al. 2016; Yang et al. 2013). However, NGS also has shortcomings. NGS solutions have been previously associated with higher error rates as compared to Sanger sequencing (Liu et al. 2012), which led to the common practice of confirming NGS results by Sanger sequencing. Other common problems include unequal coverage throughout the targeted region, sequencing of pseudogenes, and difficulty in sequencing GC-rich regions (Chen et al. 2013). It is therefore surprising that unbiased, two-way systematic comparisons of performance between various NGS platforms and Sanger sequencing is still rare, despite the important implications of such comparisons for genetic diagnostics.
Previous studies examined the ability of NGS to detect variants identified by Sanger sequencing. For a panel comprised of a limited number of genes for hereditary colon cancer, arrhythmias, cardiomyopathies, and other cardiovascular‐related genes, it was shown that all 919 variants previously observed by Sanger sequencing were also identified by NGS (Baudhuin et al. 2015). Another study, however, demonstrated that up to 18% of 137 pathogenic variants identified by Sanger sequencing, in the context of neuromuscular diseases, were not identified by exome sequencing due to low coverage (Ankala et al. 2015). In another study, variants identified by clinical Sanger sequencing was compared to results obtained from exome sequencing for the same patients (Hamilton et al. 2016). Exome sequencing identified 97.3% of the coding variants and 81.8% of the non-coding variants detected by Sanger sequencing. One of the discordant variants was proven to be Sanger-sequencing false-positive call, while the rest was exome-sequencing false-negative calls, despite sufficient coverage (30–60×). Nine genes were excluded from the comparison due to consistently low coverage in exome sequencing. These results demonstrated molecular circumstances in which Sanger sequencing performed better than NGS (Hamilton et al. 2016). Neither study examined performance of NGS independently of the Sanger-sequencing results, e.g. if there were additional variants identified by NGS that were missed by Sanger sequencing. In fact, all these studies were one-way comparisons that assessed sensitivity of NGS for variants identified by Sanger sequencing. None provided data regarding NGS false positives or Sanger-sequencing false negatives.
We present a comparative, unbiased evaluation of sequencing results of 258 genes comprising ~ 1.3% of the human exome. DNA from a single individual was extracted from peripheral blood leukocytes and buccal swabs. The platforms assessed were Sanger sequencing of the coding exons ± 8 bp of the 258 genes, the Agilent SureSelectQXT exome capture kit (SureSelect) and the Illumina Nextera Rapid Capture Expanded Exome (Nextera). We aimed to shed light on the performance and accuracy of each of the sequencing methods, from the two DNA sources, and to assess the implications of replacing Sanger sequencing with exome sequencing, using standard capture kits. Although only one individual was sequenced, to ensure a single reference sequence, we performed 11 different sequencing experiments for each of 258 genes, using different sequencing strategies and different DNA sources. This enabled in-depth comparisons between Sanger sequencing and NGS results.
Materials and methods
Samples and sequencing
Samples from blood and buccal cells were taken the same day from one healthy individual following informed consent. All DNA handling and sequencing procedures were completed at Gene by Gene’s CAP accredited laboratory in Houston, Texas. DNA was extracted from blood (peripheral leukocytes) and buccal swabs following standard protocols, and quantitated with a SpectraMax190 (Molecular Devices, Sunnyvale, CA). Sanger sequencing was completed for 258 genes on DNA extracted from buccal cells by amplifying the coding exons and their flanking regions (approximately 20 bp from each side) using conventional PCR techniques. PCR products were purified using magnetic-particle technology (Seradyn, Inc.). After purification, all fragments were sequenced with forward and reverse primers. Sequencing was performed on a 3730xl DNA Analyzer (Applied Biosystems), and the resulting sequences were analyzed with the Sequencher software (Gene Codes Corporation). Variants were analyzed relative to the reference sequences deposited in the National Center for Biotechnology Information.
Exome sequencing was performed using the Agilent enrichment capture kit (SureSelectQXT), and the Nextera enrichment rapid capture kit (FC-140-1006) on DNA extracted from both peripheral leukocytes and buccal cells. exome-sequencing data were generated using the Illumina HiSeq2500 with manufactures’ protocols for all runs. A coverage of 70 × was considered the minimal threshold for clinical grade sequencing.
Sequencing quality reassurance
For Sanger sequencing, each nucleotide was covered by at least two different sequences, preferably by one forward and one reverse sequences. In cases where this was not possible due to poly-regions, two sequences from the same direction were generated independently. The interpretation of the data was done by two operators independently. We used a Phred score of 30 as our confidence threshold.
Exome-sequencing data analysis and quality reassurance
Each exome-sequencing sample was analyzed using the Genoox platform (http://www.genoox.com). The NGS pipeline was based on BWA aligner (Li and Durbin 2009), and the two variant callers: GATK HaplotypeCaller (Mckenna et al. 2010) and FreeBayes (Garrison and Marth 2012). Coverage reports were obtained from Genoox platform. The minimal Mapping Quality (MQ) was MQ > 0 for each read. The main parameters used for filtering were identical for both variant callers and comprised a quality score > 100, a depth (DP) > 4 and the quality/depth ratio (QD) of 7.
Variants comparison
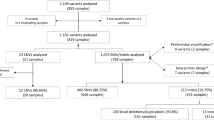
We included 258 genes in our analysis (Table S1). The list of genes was assembled from the genes previously Sanger-sequenced at Gene by Gene’s CAP accredited laboratory. The only genes excluded were those expected to perform less efficiently in NGS, due to issues involving pseudo genes or GC-rich regions (i.e. GBA (OMIM 606463), MAPT (OMIM 157140), PMS2 (OMIM 600259)). The analyzed regions include coding regions (exons) ± 8 bp flanking introns attempting to cover the splice sites. Following quality reassurance, we established the list of all the variants identified by any of the different platforms and experiments, and checked systematically how many experiments detected each variant. Variants that were detected by all experiments were classified as true variants. Variants that were not concordant between all experiments were considered potential false positives or false negatives. The Sanger sequencing suspected false positive or negative variants were re-sequenced with an alternative primer pair. The NGS suspected false positive variants were checked for their quality parameters (DP < 20; QD < 7; QUALITY < 100).
Results
We compared the results of Sanger sequencing and exome sequencing for 258 genes. We analyzed two exome-sequencing runs. In the first run, exome sequencing was performed on DNA from both sources (leukocyte and buccal), once using the SureSelect kit and once using the Nextera kit. Each exome sequencing was performed in duplicate, so this run included eight exome-sequencing tests, enabling within-run comparisons. The second run included two exome-sequencing tests performed on leukocyte-extracted DNA, one using the SureSelect kit and one using the Nextera kit. This enabled between-run comparisons.
Targeted genomic region
The targeted genomic region, as noted above, included 258 genes (Table S1), with a total of 4629 exons. The genomic region encompassed by these exons is 729,724 bps and 803,788 bps including ± 8 bp of the intron/exon boundaries. Table 1 summarizes the targeted genomic region, the expected overlap according to the SureSelect and Nextera BED files, and the actual adequately enriched regions (> 20×) obtained from the experiments. More than 98%/91% of the region was covered with > 20 × coverage for the SureSelect/Nextera experiments.
Overall NGS performance
We performed ten NGS experiments. Table 2 highlights the pivotal metrics including the mean coverage obtained for the exome sequencing as well as the targeted region, and the percentage of bps obtained at a coverage of 0x, ≤ 10x, ≤ 20 × and > 20×.
Comparison of duplicate experiments within run 1, and comparison of run 1 and run 2 demonstrated no statistically significant differences for the major displayed metrics. For the entire exome, mean coverage was 166 × with the SureSelect kit and 90 × with the Nextera kit. In the targeted region (258 genes, including the exon ± 8 bp intron/exon boundaries), mean coverage of the SureSelect kit was about double that of the Nextera kit (Table 2). Notably, the total number of aligned reads was similar in both capture kits (> 99%), but the SureSelect kit mean coverage was about double that of the Nextera kit mean coverage since a large proportion of Nextera reads did not align to the target region. For both the SureSelect and the Nextera capture kits, less than 1% of the nucleotides had 0 × coverage in any of the experiments. The mean proportion of nucleotides with 0 × coverage across all experiments of the same kit were 0.1% and 0.62% for the SureSelect and Nextera, respectively.
Identified variants
We analyzed the exome-sequencing data using standard NGS analytic pipelines with predefined quality thresholds (Methods). A total of 449 variants were identified at least once, in a total of 258 genes (range of 0–13 variants per gene) (Table 3, Table S2, Table S3). The majority of those variants, 407/449 (90.6%), were detected by all platforms and experiments. The remaining 42/449 (9.4%) discordant variants were examined further to determine whether they represent true or false variants (Table 3, Table S3).
Discordant variants were classified into two groups: variants discordant between Sanger sequencing and NGS (Sanger–NGS discordance) and variants discordant within NGS experiments (within-NGS discordance). Sanger–NGS discordance was defined as discordance between Sanger sequencing and most (≥ 5) NGS experiments. Within-NGS discordance was defined as discordance between NGS experiments, where the majority of NGS results (> 5) were concordant with Sanger-sequencing results. Thirteen variants were Sanger–NGS discordant and 29 variants were within-NGS discordant (Table 3, Table S3). Among the 13 Sanger–NGS discordant variants, four variants (in the DLL3 (OMIM 602,768), CACNA1A (OMIM 601,011) and PIK3R2 (OMIM 603,157) genes) represent NGS false negatives due to coverage failure (< 10x), since they were identified by Sanger sequencing and by all NGS experiments with sufficient coverage. In the other nine Sanger–NGS discordant variants, there was sufficient coverage in all NGS experiments. For these variants, we repeated Sanger sequencing using an alternative primer pair. Discrepancies were resolved for seven of the nine variants, showing that the original Sanger sequencing included four false-negative and three false-positive calls. All three false positives were in the same PCR amplicon of the same gene (ABCC6 (OMIM 603,234)), which lies within a segmental duplication region. The two remaining variants (CFTR (OMIM 602,421) and NOTCH3 (OMIM 600,276) genes) were not detected by repeated Sanger sequencing despite several primer redesign attempts, and were thus considered as NGS false positive calls. Twenty-nine variants were within-NGS discordant. Of these, 14 variants were not detected by Sanger sequencing and detected by only one of all NGS experiments: eight in leukocyte-derived DNA and four in buccal-swab DNA with Nextera capture; one in leukocyte DNA and one in buccal-swab DNA with SureSelect capture. These variants barely passed the quality filters, and had coverage < 20 and/or Quality < 100 and/or QD < 7. Therefore, they are considered as NGS false positives due to low quality. Of the remaining 15 variants, 13 were detected by the Sanger sequencing, the SureSelect experiments and at least one Nextera experiment. The other two variants were detected by the Sanger sequencing, the Nextera experiments and at least one SureSelect experiment. All these 15 variants are thus considered as NGS false negatives. In 12 of them, lack of detection is explained by low coverage (< 20×).
Cumulatively, we determined that of the 42 discordant variants, 23 were true variant calls, including four Sanger-sequencing false negatives and 19 NGS false negatives. The remaining 19 variants were false positives, including three Sanger-sequencing calls and 16 NGS experiments calls (Table 3, Table S3). False-positive NGS calls included 9/16 with poor coverage (< 20x), and 7/16 variants with > 20 × coverage, but borderline quality (QD < 7). Notably, 12 of the 16 NGS false positives (75%) have rs IDs.
Thus, together with the 407 variants detected by all experiments, a total of 430/449 (95.8%) variants were defined as “true variants”, and regarded as the truth set for further analysis.
Comparative sequencing performance
As shown in Fig. 1, overall, sensitivity was > 97% in all platforms and experiments. Sanger sequencing had a detection rate of 99%. In NGS experiments, higher coverage resulted in better detection rates. The mean sensitivity of SureSelect and Nextera experiments was 99.5% and 98.5%, respectively.
Figure 2 presents the false-positive and false-negative calls for each of the platforms and experiments. In general, the number of false-negative calls in the NGS experiments decreased with higher coverage. None of the differences was statistically significant using Chi-square test (p > 0.05). The PPV (positive predictive value) and NPV (negative predictive value) for all experiments were both > 0.99.
Number of false calls. The number of false-positive and false-negative variants is shown for each experiment. False-negative calls (solid bars). False-positive calls (striped bars). Number of each type of call is indicated within each bar. Bu: buccal; bl: blood; D: duplicate. None of the differences are statistically significant
Zygosity discordance between platforms and experiments
We observed zygosity discordance for a total of ten variants (Table 4). In 5/10 cases, the discordance was between Sanger sequencing and all NGS experiments (Sanger–NGS discordance). In four of those five cases, Sanger sequencing suggested a homozygous state while all NGS experiments suggested heterozygosity. In the fifth case, the opposite was noted. The NGS coverage for these variants ranged from 27 × to 307× (average 183×). For the other 5/10 variants there was within-NGS zygosity discordance: Sanger sequencing and SureSelect experiments were concordant but there was discordance with one of the Nextera experiments. Coverage was < 20 × in four of the cases, and 41 × in the fifth case (Table 4).
We repeated Sanger sequencing using an alternative primer pair for all 10 variants. The zygosity status changed for the first 5 variants, which indicates Sanger sequencing error due to primer design in the first sequencing. The zygosity status of the other five variants did not change, which indicates a single NGS experiment error.
Discussion
Accuracy of NGS platforms and the ability to obtain full exome/genome-sequencing information are critical elements of clinical genetic testing and of precision medicine initiatives (Ashley 2016). We compared the performance of Sanger sequencing and NGS for 258 genes that are commonly sequenced in a commercial laboratory. Sequencing these genes is generally requested as part of routine clinical testing for well-defined OMIM (https://omim.org/) phenotypes or during the investigation of less defined, orphan phenotypes with unknown molecular bases. OMIM phenotypes are assigned to 257 of 258 genes included in our study and 30 are among the 59 genes listed in the American College of Medical Genetics and Genomics recommendations for reporting of incidental findings in clinical exome and genome-sequencing (Kalia et al. 2017). Accordingly, the genes we examined can be regarded as representative of common clinical scenarios mandating single or multiple gene sequencing. These scenarios often present the dilemma of choosing between Sanger sequencing of specific gene(s), NGS of a gene panel including the requested gene(s), or full exome sequencing that might serve both immediate and future needs of the patient. Compared to exome sequencing, sequencing specific gene(s) by either Sanger or NGS, involves higher sequencing costs per gene, and incurring additional costs of further genetic testing if a molecular diagnosis is not confirmed in the first round of testing. However, sequencing of single genes or gene panels are more likely to provide complete coverage of the targeted genes.
Although we only sequenced one individual in this study, the study we performed is unique in providing bidirectional comparison of Sanger sequencing and NGS. Using different sequencing strategies, different NGS solutions and different DNA sources, each gene was sequenced a total of 11 times. This way we could determine with confidence whether each variant observed represents a true or false call, examine false negative and false positive rates of the methods, and evaluate characteristics of both true and false calls.
We first examined the effect of the DNA source and capture kit on NGS results, as well as consistency of NGS results between duplicates and between different runs. NGS experiments were performed using the exact guidelines of the respective manufacturers. Within each of the kits, it is evident that no statistically significant differences were noted between duplicates in the same run or different sequencing runs (Table 2). Differences in performance between capture kits have been previously reported (Asan et al. 2011; Chilamakuri et al. 2014; Clark et al. 2011). Our results demonstrate that the mean coverage obtained by the SureSelect kit was higher, reaching 182×/183 × for DNA extracted from leukocytes or buccal cells, respectively. There were no significant differences in the performance of NGS on DNA extracted from these two different sources.
We then determined the rates and types of false calls in both Sanger sequencing and NGS. The detection rate of true variants was 99.1% (426/430) for Sanger sequencing, and ranged between 97.9 and 100% in the NGS experiments (Fig. 1). Although the sensitivity, false-positive and false-negative rates calculated for all experiments were not statistically different, probably due to the small number of discordant variants, the differences observed offer important insights into the strengths and weaknesses of each sequencing method.
For Sanger sequencing, the false-positive rate was 3.7E-6, and the false-negative rate was 0.009. Sanger false positives all occurred in a single amplicon of one gene (ABCC6 (OMIM 603234)), whereas false negatives occurred in three different genes. All false Sanger calls were resolved using alternative primer pairs. The false-negative calls could be idiosyncratic to polymorphisms in the tested individual, while the false positive calls are most likely explained by insufficiently specific primers in an amplicon located in a segmental duplicated region.
For NGS, the mean false positive rate was 2.5E-6 in the SureSelect experiments, and 5.2E-6 in the Nextera experiments. The mean false negative rates were 0.005 for SureSelect experiments, and 0.01 for Nextera experiments. Most false positive NGS calls were singular events, i.e. they occurred in only one of 10 NGS experiments. These variants had barely passed the quality filter, and failed one or more of the basic quality parameters (DP > 20; QUALITY > 100; QD > 7). Changing the quality filter thresholds might obviously decrease the number of false positives, but this would come at the cost of an increase in false negatives. Interestingly 15/19 (78.9%) of false-positive calls, including all three Sanger-sequencing false-positives, have rs IDs. This may indicate that current databases contain false positive calls presented as true variants. Conversely, all 430 true variants were detectable by NGS, as long as coverage was adequate and quality parameters were fulfilled, consistent with previous reports that there is no need for Sanger-sequencing confirmation of such high-coverage/high-quality variants (Goldfeder et al. 2016).
NGS false negatives were all associated with low (< 20×) coverage. In some cases, low coverage was a singular event (e.g. COL5A (OMIM 120215) or COL5A2 (OMIM 120190)), but in other cases low coverage was systematic to a specific kit (e.g. PIK3R2 (OMIM 603157) in SureSelect or FRMD7 (OMIM 300628) in Nextera), or to both capture methods (e.g. DLL3 (OMIM 602768)). Systematic false negatives indicate problematic genomic regions in which variant detection is hampered. This is a concern, since it was previously shown that large areas of medically actionable genes fall within low confidence regions (Baudhuin et al. 2015), which might explain consistently low coverage for some variants. In this study, using exome sequencing to target a specific set of genes encompassing ~ 1.3% of the exome, resulted in adequate coverage (> 20×) of > 98% and > 91%, of exons ± 8 bp with the SureSelect and the Nextera kits, respectively (Table 1). This suggests that ~ 16,075 and ~ 72,340 bp (of a total of 803,788 bp in 258 genes), would not be adequately covered. Clearly, disease causing variants could be located in such regions with insufficient coverage. Taken together, our data suggest that exome sequencing could miss ~ 2–3% of the coding variants and up to 7% of the non-coding variants. These results are in agreement with Hamilton et al. (Hamilton et al. 2016) who demonstrated a 92.3% concordance between exome sequencing and Sanger sequencing in the ± 20 bp intronic/exon boundaries.
We also found zygosity status errors for both Sanger sequencing and NGS. As presented in Table 4, Sanger sequencing determined a false zygosity status of five variants (four false homozygotes and one false heterozygote) that was correctly assessed in repeated Sanger sequencing using an alternative primer pair. In five additional variants, there was a zygosity status error in a single NGS experiment, which in 4/5 of cases was associated with low coverage (< 20×).
In the clinical setting, false negatives are more concerning than false positives. Whereas false positives can be re-evaluated, detecting false negatives requires complete re-testing, which is unlikely to be routinely performed. The number of false negatives indicated are for ~ 1.3% of the exome, so over an entire exome the absolute number of false negatives will be correspondingly higher. We also note that these results overestimate the performance of NGS over the entire exome, since we purposely did not include genes in which the NGS is expected a priori to be less effective, e.g. genes that are GC rich or known to have pseudogenes.
In summary, our results confirm the notion that neither Sanger sequencing nor NGS can be regarded as a “gold-standard” method, as both had false-positive and false-negative calls. We have demonstrated that the performances of both NGS and Sanger sequencing are satisfactory, and that no statistically significant differences were noted between the two sequencing methods with respect to the ability to sequence the targeted regions, and the observed, high (> 98%) variant detection rates. Within NGS, it is evident that while clear differences were noted in the obtained sequencing coverage for the different capture kits, the detection rates of the true calls were not significantly different, at least when reaching the preset threshold of 70 × coverage. Bearing in mind the potential caveats that might be associated with pseudogenes or GC-rich regions, our data cautiously suggests that off-the-shelf exome-sequencing solutions might serve as a viable and practical alternative to gene(s) or gene-panel sequencing, albeit with a false negative rate of at least 1–3%. Nevertheless, physicians must be aware of the limitations of both Sanger sequencing and NGS. As demonstrated herein, Sanger-sequencing primer binding-site polymorphisms and chance or systematic NGS-coverage failure are integral hurdles of these methods. We show that Sanger sequencing and different NGS solutions are synergistic, as any combination of the Sanger sequencing, SureSelect and Nextera experiments yielded an overall greater detection rate. Accordingly, high index of suspicious for a given clinical diagnosis must overcome negative molecular results of either Sanger sequencing or NGS and repeated investigation with an alternative method should be considered.
Availability of data and material
All data are available upon request.
References
Ankala A, Da Silva C, Gualandi F et al (2015) A comprehensive genomic approach for neuromuscular diseases gives a high diagnostic yield. Ann Neurol 77:206–214. https://doi.org/10.1002/ana.24303
Asan XuY, Jiang H et al (2011) Comprehensive comparison of three commercial human whole-exome capture platforms. Genome Biol. https://doi.org/10.1186/gb-2011-12-9-r95
Ashley EA (2016) Towards precision medicine. Nat Rev Genet 17:507–522. https://doi.org/10.1038/nrg.2016.86
Baudhuin LM, Lagerstedt SA, Klee EW et al (2015) Confirming variants in next-generation sequencing panel testing by sanger sequencing. J Mol Diagnostics 17:456–461. https://doi.org/10.1016/j.jmoldx.2015.03.004
Brunner AL, Johnson DS, Si WK et al (2009) Distinct DNA methylation patterns characterize differentiated human embryonic stem cells and developing human fetal liver. Genome Res 19:1044–1056. https://doi.org/10.1101/gr.088773.108
Chen YC, Liu T, Yu CH et al (2013) Effects of GC bias in next-generation-sequencing data on de novo genome assembly. PLoS ONE. https://doi.org/10.1371/journal.pone.0062856
Chilamakuri CSR, Lorenz S, Madoui MA et al (2014) Performance comparison of four exome capture systems for deep sequencing. BMC Genomics. https://doi.org/10.1186/1471-2164-15-449
Ching T, Masaki J, Weirather J, Garmire LX (2015) Non-coding yet non-trivial: a review on the computational genomics of lincRNAs. BioData Min 8:1–12. https://doi.org/10.1186/s13040-015-0075-z
Clark MJ, Chen R, Lam HYK et al (2011) Performance comparison of exome DNA sequencing technologies. Nat Biotechnol 29:908–916. https://doi.org/10.1038/nbt.1975
De Rubeis S, Buxbaum JD (2015) Recent advances in the genetics of autism spectrum disorder. Curr Neurol Neurosci Rep. https://doi.org/10.1007/s11910-015-0553-1
Garrison E, Marth G (2012) Haplotype-based variant detection from short-read sequencing. Nat Commun 5:1–9
Gilissen C, Hehir-Kwa JY, Thung DT et al (2014) Genome sequencing identifies major causes of severe intellectual disability. Nature 511:344–347. https://doi.org/10.1038/nature13394
Goldfeder RL, Priest JR, Zook JM et al (2016) Medical implications of technical accuracy in genome sequencing. Genome Med 8:1–12. https://doi.org/10.1186/s13073-016-0269-0
Goodwin S, McPherson JD, McCombie WR (2016) Coming of age: Ten years of next-generation sequencing technologies. Nat Rev Genet 17:333–351. https://doi.org/10.1038/nrg.2016.49
Hamilton A, Tétreault M, Dyment DA et al (2016) Concordance between whole-exome sequencing and clinical sanger sequencing: Implications for patient care. Mol Genet Genomic Med 4:504–512. https://doi.org/10.1002/mgg3.223
Iglesias A, Anyane-Yeboa K, Wynn J et al (2014) The usefulness of whole-exome sequencing in routine clinical practice. Genet Med 16:922–931. https://doi.org/10.1038/gim.2014.58
Kalia SS, Adelman K, Bale SJ et al (2017) Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): A policy statement of the American College of Medical Genetics and Genomics. Genet Med 19:249–255. https://doi.org/10.1038/gim.2016.190
Lee H, Deignan JL, Dorrani N et al (2014) Clinical exome sequencing for genetic identification of rare mendelian disorders. JAMA 312:1880–1887
Lerat J, Jonard L, Loundon N et al (2016) An Application of NGS for Molecular Investigations in Perrault Syndrome: Study of 14 Families and Review of the Literature. Hum Mutat 37:1354–1362. https://doi.org/10.1002/humu.23120
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Li S, Tighe SW, Nicolet CM et al (2014) Multi-platform assessment of transcriptome profiling using RNA-seq in the ABRF next-generation sequencing study. Nat Biotechnol 32:915–925. https://doi.org/10.1038/nbt.2972
Liu L, Li Y, Li S et al (2012) Comparison of next-generation sequencing systems. J Biomed Biotechnol. https://doi.org/10.1155/2012/251364
Maksemous N, Smith RA, Haupt LM, Griffiths LR (2016) Targeted next generation sequencing identifies novel NOTCH3 gene mutations in CADASIL diagnostics patients. Hum Genomics 10:1–12. https://doi.org/10.1186/s40246-016-0093-z
Mckenna A, Hanna M, Banks E et al (2010) The Genome analysis toolkit : a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303
Mundade R, Ozer HG, Wei H et al (2014) Role of ChIP-seq in the discovery of transcription factor binding sites, differential gene regulation mechanism, epigenetic marks and beyond. Cell Cycle 13:2847–2852. https://doi.org/10.4161/15384101.2014.949201
Neveling K, Feenstra I, Gilissen C et al (2013) A Post-Hoc comparison of the utility of sanger sequencing and exome sequencing for the diagnosis of heterogeneous diseases. Hum Mutat 34:1721–1726. https://doi.org/10.1002/humu.22450
Ng SB, Bigham AW, Buckingham KJ et al (2010) Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet 42:790–793. https://doi.org/10.1038/ng.646
Rabbani B, Nakaoka H, Akhondzadeh S et al (2016) Next generation sequencing: Implications in personalized medicine and pharmacogenomics. Mol Biosyst 12:1818–1830. https://doi.org/10.1039/c6mb00115g
Shen T, Pajaro-Van de Stadt SH, Yeat NC, Lin JCH (2015) Clinical applications of next generation sequencing in cancer: from panels, to exomes, to genomes. Front Genet 6:1–9. https://doi.org/10.3389/fgene.2015.00215
Sumner K, Swensen JJ, Procter M et al (2014) Noncontinuously binding loop-out primers for avoiding problematic DNA sequences in PCR and Sanger sequencing. J Mol Diagnostics 16:477–480. https://doi.org/10.1016/j.jmoldx.2014.04.005
Wang Y, Peng W, Guo HY et al (2016) Next-generation sequencing-based molecular diagnosis of neonatal hypotonia in Chinese Population. Sci Rep 6:1–11. https://doi.org/10.1038/srep29088
Wenzel JJ, Rossmann H, Fottner C et al (2009) Identification and prevention of genotyping errors caused by G-quadruplex- and i-motif-like sequences. Clin Chem 55:1361–1371. https://doi.org/10.1373/clinchem.2008.118661
Yang Y, Muzny DM, Reid JG et al (2013) Clinical whole-exome sequencing for the diagnosis of Mendelian disorders. N Engl J Med 369:1502–1511. https://doi.org/10.1056/nejmoa1306555
Acknowledgements
We thank the donor of the control sample.
Author information
Authors and Affiliations
Contributions
HF, DMB, and ELL conceived and designed the study. CB, AB, BM, and LFS provided the DNA samples to the study, managed and completed the genotyping campaign. ME, YP, and DA completed the NGS analysis. HF and DMB wrote the paper. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The lab work presented in this study was conducted in Gene by Gene Ltd, in which Doron M. Behar, Concetta Bormans, and Arjan Bormans declare stock ownership, and Daniel Au, Brent Manning, and Luisa Fernanda Sanchez are employees. The analytic pipeline used in this study was conducted in Genoox Ltd, in which Moshe Einhorn and Yuval Porat declare stock ownership. The other authors claim no competing financial interests associated with this paper.
Ethics approval
Written informed consent was obtained from the participant included in the study.
Additional information
Communicated by Stefan Hohmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fridman, H., Bormans, C., Einhorn, M. et al. Performance comparison: exome sequencing as a single test replacing Sanger sequencing. Mol Genet Genomics 296, 653–663 (2021). https://doi.org/10.1007/s00438-021-01772-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-021-01772-3