Abstract
Osteoarthritis (OA) is one of the most common skeletal disease, which seriously affects the quality of life of patients, particularly in the middle-aged and elderly individuals. We aimed to explore whether rs9340799 [estrogen receptor alpha (ER-α) XbaI A/G] polymorphism was associated with OA using a meta-analysis. A literature search for eligible studies published before March 28, 2014 was conducted in the PubMed, Web of Science, Embase, Cochrane database, Current Controlled Trials, Clinicaltrials.gov, Chinese Clinical Trial Registry, CBMdisc, CNKI, Google Scholar and Baidu Library. The association between the rs9340799 polymorphism and OA risk was assessed by odds ratios (ORs) together with their 95 % confidence intervals (CIs). A total of 663 articles were found. After article review and quality assessment, 10 articles involving 2,924 OA cases and 5,868 controls were included in the final meta-analysis. The combined evidence suggested that rs9340799 polymorphism contributed significantly to an increased risk of OA (for G allele vs. A allele: OR = 1.21, 95 % CI 1.03–1.43, p = 0.02; for G/G vs. A/A: OR = 1.30, 95 % CI 1.07–1.57, p = 0.009). In the subgroup analyses, significant associations were found between the rs9340799 polymorphism and the OA risk in the European group, Asian group, and knee osteoarthritis group, respectively. These results suggested that the rs9340799 polymorphism might be associated with the risk of OA. However, the results should be interpreted with caution because of the publication bias.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Osteoarthritis (OA) is one of the most common skeletal disease, which is characterized by the progressive loss of articular cartilage in the synovial joints and changes in the adjacent bone (Poole et al. 1994). OA also known as degenerative arthritis or degenerative joint disease or osteoarthrosis, commonly affects the hands, feet, spine, and large weight-bearing joints, such as the hips and knees. The worldwide prevalence of OA was 11.3 % with approximately 85 million people affected in 2009, which is expected to increase 122 million in 2017. Thus, economic burden of OA on the national healthcare system is enormous, both in the developed countries and the developing countries (Chen et al. 2012; Cross et al. 2014). Despite the well established fact that advanced age, excess body weight, repeated trauma or surgery to the joint structures, abnormal joints at birth, gout, diabetes, and other hormone disorders are associated with the increased risk of OA (Brandt et al. 2009), a detailed etiology underlying OA is still obscure. A single factor is not generally sufficient to cause OA, but about half of the variation in susceptibility has been assigned to genetic factors (Spector and MacGregor 2004). Previous twin-pair and family-risk studies have shown that there is a greater prevalence of the disease among siblings and especially identical twins, indicating that the genetic factors play a crucial role in the development of OA (Spector et al. 1996; Chitnavis et al. 1997; Valdes and Spector 2008).
The prevalence of OA in both men and women is comparable up to the age of 50 years; however, the prevalence increases considerably among the postmenopausal women (Tanamas et al. 2011; Spector and Campion 1989), which suggests a link between OA and loss of ovarian function. Estrogens are hormones that are important for sexual and other body functions. Estrogens also play critical roles in bone formation and homeostasis (Syed and Khosla 2005). In the estrogen endocrine system, estrogen receptor alpha (ER-α) is an important mediator in the signal transduction pathway. Previous studies both in vitro and in vivo have suggested that ER-α plays an important role in the pathological process of OA (Roman-Blas et al. 2009). Meanwhile, a variety of epidemiological studies have focused on the association between rs9340799 (ER-α XbaI A/G) polymorphism and OA risk (Ushiyama et al. 1998; Bergink et al. 2003; Jin et al. 2004; Xue et al. 2004; Lian et al. 2007; Kang et al. 2007; Yang et al. 2009; Wise et al. 2009; Tian et al. 2009; Borgonio-Cuadra et al. 2012). However, despite intensive research efforts, results of different studies have been inconsistent. Therefore, whether the rs9340799 polymorphism is related to the risk of OA is still under debate. Most studies reported a negative association between the rs9340799 polymorphism and the risk of OA progression, while other studies generated positive results. To better clarify the association between the rs9340799 polymorphism and the risk of OA, we designed this meta-analysis by collecting and sorting the previous published studies.
Materials and methods
Data sources
This meta-analysis followed the preferred reporting items for systematic reviews and meta-analyses (PRISMA) criteria (Moher et al. 2009). We performed a comprehensive computer-based search of the PubMed, Web of Science, Embase, Cochrane database, Current Controlled Trials, Clinicaltrials.gov, Chinese Clinical Trial Registry, CBMdisc, CNKI, Google Scholar and Baidu Library (up to March 28, 2014) to identify studies analyzing the association of the rs9340799 polymorphism with OA. The following keywords were used for searching: (“estrogen receptor alpha” OR “ER alpha” OR “ER α” OR “ESR alpha” OR “ESR α” OR “rs9340799”) AND (“polymorphism” OR “mutation” OR “variant” OR “variation” OR “genotype”) AND (“osteoarthritis” OR “OA” OR “degenerative joint disease”). These literature searches were limited to the English and Chinese language articles. Additionally, the references of the eligible articles were searched to identify citations to other studies that were not identified initially.
Inclusion criteria
The studies included in the current meta-analysis had to meet the following criteria: (1) human studies; (2) studies on the relationship between the rs9340799 polymorphism and OA; (3) unrelated case–control or cohort studies; (4) sufficient published data on the genotypes or allele frequencies for estimating an odds ratio (OR) with 95 % confidence interval (CI); (5) not republished data.
Data extraction
The data were independently extracted by two authors of this article (Yin YW and Hu AM), and the result was reviewed by a third author (Sun QQ). The following data were collected from each study: last name of the first author, year of publication, country and ethnicity of the studied population, source of controls, the number of cases and controls, and information of genotypes and alleles. In addition, evidence of Hardy–Weinberg equilibrium (HWE) was also collected (p < 0.05 of HWE was considered significant).
Statistical analysis
The strength of association between the rs9340799 polymorphism and the OA risk was measured by ORs with 95 % CIs. The pooled ORs were estimated for four genetic models (allelic model: G allele vs. A allele, additive model: G/G vs. A/A, recessive model: G/G vs. A/G + A/A, and dominant model: G/G + A/G vs. A/A). Heterogeneity between studies was formally tested by using Cochran’s Q statistic and considered statistically significant when p < 0.10. Heterogeneity was also measured with I 2 statistic (I 2 > 50 % indicated evidence of heterogeneity) (Higgins et al. 2003). The fixed-effects model was used in the absence of the between-study heterogeneity; otherwise, the random-effects model was used (Mantel and Haenszel 1959; Berkey et al. 1995). Subgroup analyses were performed based on ethnicity and diseases. Furthermore, in order to evaluate the stability of the results, sensitivity analyses were performed by limiting the meta-analysis to studies conforming to HWE. An estimate of potential publication bias was assessed with Begg’s funnel plot and Egger’s regression test (p < 0.05 was considered representative of statistically significant publication bias) (Egger et al. 1997). If there was some evidence of publication bias, the Trim and Fill method was used to adjust the meta-analysis results by inputting data from the potential missing studies. Data of meta-analysis were analyzed using Review Manager 5.1.4 (Cochrane Collaboration, The Nordic Cochrane Centre, Copenhagen) and Stata 11.0 (StataCorp LP, College Station, TX, USA).
Results
Study characteristics
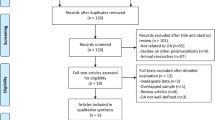
The study selection process is detailed in Fig. 1. A total of 10 eligible studies were identified in a systematic search of the PubMed, Web of Science, Embase, Cochrane database, Current Controlled Trials, Clinicaltrials.gov, Chinese Clinical Trial Registry, CBMdisc, CNKI, Google Scholar, Baidu Library and reference cited in the researches (Ushiyama et al. 1998; Bergink et al. 2003; Jin et al. 2004; Xue et al. 2004; Lian et al. 2007; Kang et al. 2007; Yang et al. 2009; Wise et al. 2009; Tian et al. 2009; Borgonio-Cuadra et al. 2012), which contained 2,924 OA cases and 5,868 controls. Tables 1, 2 show the studies identified and their main characteristics. Four studies were performed in Europeans (Bergink et al. 2003; Lian et al. 2007; Wise et al. 2009; Borgonio-Cuadra et al. 2012), and six studies were performed in Asians (Ushiyama et al. 1998; Jin et al. 2004; Xue et al. 2004; Kang et al. 2007; Yang et al. 2009; Tian et al. 2009). The countries of these studies included China, Japan, Korea, Mexico, Netherlands and USA. The genotype distributions among the controls of all studies followed the HWE except for the studies of Kang et al. (2007) and Yang et al. (2009) (p < 0.05).
Quantitative synthesis
According to the presence or absence of the heterogeneity (Table 2), the random-effects model was used to calculate the pooled ORs of the allelic model and the recessive model, and the fixed-effects model was used to calculate the pooled ORs of the additive model and the dominant model. The overall results showed that there was significant association between the rs9340799 polymorphism and OA, suggesting that the rs9340799 G allele may be a risk factor for OA (for G allele vs. A allele: OR = 1.21, 95 % CI 1.03–1.43, p = 0.02; for G/G vs. A/A: OR = 1.30, 95 % CI 1.07–1.57, p = 0.009; for G/G vs. A/G + A/A: OR = 1.21, 95 % CI 0.99–1.47, p = 0.07; for G/G + A/G vs. A/A: OR = 1.19, 95 % CI 0.99–1.43, p = 0.06). The main results of the meta-analysis are shown in Table 2 and Fig. 2, respectively.
In the subgroup analyses based on ethnicity and diseases, significant associations were also found between the rs9340799 polymorphism and OA in the Europeans (for G allele vs. A allele: OR = 1.11, 95 % CI 1.01–1.22, p = 0.03; for G/G vs. A/G + A/A: OR = 1.15, 95 % CI 1.02–1.31, p = 0.03), Asians (for G/G vs. A/A: OR = 2.18, 95 % CI 1.29–3.68, p = 0.004; for G/G + A/G vs. A/A: OR = 1.79, 95 % CI 1.10–2.92, p = 0.02), and knee osteoarthritis (KOA) group (for G allele vs. A allele: OR = 1.47, 95 % CI 1.08–1.99, p = 0.01; for G/G vs. A/A: OR = 2.07, 95 % CI 1.11–3.87, p = 0.02; for G/G + A/G vs. A/A: OR = 1.40, 95 % CI 1.06–1.86, p = 0.02). Furthermore, no significant association was found between this variation and other OA group. The main results of the subgroup analyses are shown in Table 2.
Sensitivity analysis
Sensitivity analysis was performed to evaluate the stability of the overall results. The studies of Kang et al. (2007) and Yang et al. (2009) were excluded from the sensitivity analysis due to the genotype distribution in the control group of the study deviating from HWE. We found that the corresponding pooled ORs were materially altered in the allelic model and the additive model, indicating that the study without HWE should be considered as a factor influencing the overall results. The results of the sensitivity analysis are shown in Table 2.
Publication bias
Begg’s funnel plot and Egger’s regression test were performed to assess the publication bias. As shown in Fig. S1, the shapes of the funnel plots revealed obvious asymmetry in the additive model (Fig. S1 B) and the dominant model (Fig. S1 D), suggesting that there were obvious publication biases in these two genetic models. Moreover, the results of Egger’s regression test also provided sufficient evidence for publication bias (p = 0.046 for the additive model, p = 0.028 for the dominant model). Given that publication bias existed in the above two comparisons, the Trim and Fill method was applied to adjust the results (Fig. S2). The corresponding pooled OR was materially altered in the additive model (OR = 1.17, 95 % CI 0.97–1.41), but not in the dominant model (OR = 1.10, 95 % CI 0.92–1.31) which suggested the result of the additive model was not statistically robust. In addition, there was no evidence for publication bias in other genetic models (p = 0.154 for allelic model, and p = 0.257 for recessive model) (Fig. S1 A and C).
Discussion
Osteoarthritis is a complex disease with various involved factors including environmental and genetic factors (Spector and MacGregor 2004; Coggon et al. 2001). The association between the rs9340799 polymorphism and OA has been intensively studied. However, previous attempts to explore the existing evidence were always in the context of a case–control or cohort study (Ushiyama et al. 1998; Bergink et al. 2003; Jin et al. 2004; Xue et al. 2004; Lian et al. 2007; Kang et al. 2007; Yang et al. 2009; Wise et al. 2009; Tian et al. 2009; Borgonio-Cuadra et al. 2012), not having enough statistical power to explore the real association of the rs9340799 polymorphism with OA. Recently, meta-analysis has been widely used in genetic association studies because it has the potential to detect small effects between gene polymorphism and human disease (Yin et al. 2013, 2014a, b). By increasing the sample size, the meta-analysis obtains enough statistical power to explore the real association of the gene polymorphism with human disease. Here, we focused on the association between the rs9340799 polymorphism and OA, and designed this meta-analysis to derive a more precise association between this gene variation and OA.
To the best of our knowledge, this is the first comprehensive meta-analysis to date investigating the association between the rs9340799 polymorphism and OA. The overall results showed that there was significant association between the rs9340799 polymorphism and OA, suggesting that the G allele may be a risk factor for OA. Considering that potential ethnic difference might be associated with the distribution of genotypes, we also performed subgroup analysis by ethnicity. Similarly, significant associations were found between this variation and OA in both European and Asian populations. Furthermore, in the subgroup analysis by disease type, significant association was also found between the rs9340799 polymorphism and OA in the KOA group. For the other OA group, however, no significant association was found. This inconsistent result on subgroup analysis may partly resulted from the genetic diversity among disease. Moreover, as OA is a multifactorial disease, in addition to the genetic factors, environmental factors also play an important role in OA etiology. Thus, this discrepancy may also be caused by varied geographic distribution, linked to climate, diet, economic status and lifestyle. Nevertheless, the results of subgroup analyses further strengthened our conclusion that the G allele of the rs9340799 polymorphism may be a risk factor for OA. In addition to the subgroup analyses, we also performed sensitivity analyses restricted to the studies conforming to HWE. We found that the corresponding pooled ORs were materially altered in the allelic model and the additive model, indicating that the study without HWE should be considered as a factor influencing the overall results.
Heterogeneity should not be ignored in the interpretation of the final results (Ioannidis et al. 2007), which was also observed in the present meta-analysis. Common reasons of heterogeneity may be attributed to the diversity in design, study quality, characteristics of the cases and controls involved, sample-sizes, genotyping quality, and some studies without HWE, etc. After subgroup analyses, we effectively removed the heterogeneity in the European group and the other OA group. Therefore, the heterogeneity might partly result from the ethnicity difference or disease type. To further explore the sources of heterogeneity, we created a Galbraith plot to identify the potential outlier studies. Three studies (Ushiyama et al. 1998; Xue et al. 2004; Tian et al. 2009) were identified as the main contributors to heterogeneity (Fig. S3). After excluding the outlier studies, the heterogeneity was effectively removed in the allelic model and the dominant model. Furthermore, the corresponding pooled ORs were not materially altered in all comparisons (data not show), suggesting the overall results of this meta-analysis were statistically robust.
Some limitations of this meta-analysis should be acknowledged. Firstly, between-study heterogeneity in the present meta-analysis should be noted, which may affect the results of the present meta-analysis. Secondly, publication bias existed in the additive model and the dominant model. Only full-text articles published in English and Chinese were included in this meta-analysis, missing some eligible studies which were unpublished or reported in other languages. This may bias the present results. Therefore, the results should be interpreted with caution. Thirdly, some limitations of the meta-analysis are inherent (including this one), such as their retrospective nature that is subject to the methodological deficiencies of the included studies. In spite of these, this meta-analysis of the association of the rs9340799 polymorphism with the OA risk is statistically more convincing than any single study.
In conclusion, our meta-analysis including 8,792 subjects suggested that the rs9340799 polymorphism might be associated with the risk of OA. However, the results should be interpreted with caution because of the publication bias. Further studies with large sample size, especially with multicentric case–control studies, will be needed to confirm our findings.
Abbreviations
- OA:
-
Osteoarthritis
- ER-α:
-
Estrogen receptor alpha
- PRISMA:
-
Preferred reporting items for systematic reviews and meta-analyses
- OR:
-
Odds ratio
- CI:
-
Confidence interval
- HWE:
-
Hardy–Weinberg equilibrium
- KOA:
-
Knee osteoarthritis
References
Bergink AP, van Meurs JB, Loughlin J, Arp PP, Fang Y, Hofman A, van Leeuwen JP, van Duijn CM, Uitterlinden AG, Pols HA (2003) Estrogen receptor alpha gene haplotype is associated with radiographic osteoarthritis of the knee in elderly men and women. Arthritis Rheum 48:1913–1922
Berkey CS, Hoaglin DC, Mosteller F, Colditz GA (1995) A random-effects regression model for meta-analysis. Stat Med 14:395–411
Borgonio-Cuadra VM, González-Huerta C, Duarte-Salazár C, de Los Ángeles Soria-Bastida M, Cortés-González S, Miranda-Duarte A (2012) Analysis of estrogen receptor alpha gene haplotype in Mexican mestizo patients with primary osteoarthritis of the knee. Rheumatol Int 32:1425–1430
Brandt KD, Dieppe P, Radin E (2009) Etiopathogenesis of osteoarthritis. Med Clin North Am 93:1–24
Chen A, Gupte C, Akhtar K, Smith P, Cobb J (2012) The global economic cost of osteoarthritis: how the UK Compares. Arthritis 2012:698709
Chitnavis J, Sinsheimer JS, Clipsham K, Loughlin J, Sykes B, Burge PD, Carr AJ (1997) Genetic influences in end-stage osteoarthritis. Sibling risks of hip and knee replacement for idiopathic osteoarthritis. J Bone Jt Surg Br 79:660–664
Coggon D, Reading I, Croft P, McLaren M, Barrett D, Cooper C (2001) Knee osteoarthritis and obesity. Int J Obes Relat Metab Disord 25:622–627
Cross M, Smith E, Hoy D, Nolte S, Ackerman I, Fransen M, Bridgett L, Williams S, Guillemin F, Hill CL, Laslett LL, Jones G, Cicuttini F, Osborne R, Vos T, Buchbinder R, Woolf A, March L (2014) The global burden of hip and knee osteoarthritis: estimates from the Global Burden of Disease 2010 study. Ann Rheum Dis 73:1323–1330
Egger M, Smith GD, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315:629–634
Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ 327:557–560
Ioannidis JP, Patsopoulos NA, Evangelou E (2007) Heterogeneity in meta-analyses of genome-wide association investigations. PLoS One 2:e841
Jin SY, Hong SJ, Yang HI, Park SD, Yoo MC, Lee HJ, Hong MS, Park HJ, Yoon SH, Kim BS, Yim SV, Park HK, Chung JH (2004) Estrogen receptor-alpha gene haplotype is associated with primary knee osteoarthritis in Korean population. Arthritis Res Ther 6:R415–R421
Kang SC, Lee DG, Choi JH, Kim ST, Kim YK, Ahn HJ (2007) Association between estrogen receptor polymorphism and pain susceptibility in female temporomandibular joint osteoarthritis patients. Int J Oral Maxillofac Surg 36:391–394
Lian K, Lui L, Zmuda JM, Nevitt MC, Hochberg MC, Lee JM, Li J, Lane NE (2007) Estrogen receptor alpha genotype is associated with a reduced prevalence of radiographic hip osteoarthritis in elderly caucasian women. Osteoarthr Cartil 15:972–978
Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22:719–748
Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med 151:264–269
Poole AR, Ionescu M, Swan A, Dieppe PA (1994) Changes in cartilage metabolism in arthritis are reflected by altered serum and synovial fluid levels of the cartilage proteoglycan aggrecan implications for pathogenesis. J Clin Invest 94:25–33
Roman-Blas JA, Castañeda S, Largo R, Herrero-Beaumont G (2009) Osteoarthritis associated with estrogen deficiency. Arthritis Res Ther 11:241
Spector TD, Campion GD (1989) Generalised osteoarthritis: a hormonally mediated disease. Ann Rheum Dis 48:523–527
Spector TD, MacGregor AJ (2004) Risk factors for osteoarthritis: genetics. Osteoarthr Cartil 12(Suppl A):S39–S44
Spector TD, Cicuttini F, Baker J, Loughlin J, Hart D (1996) Genetic influences on osteoarthritis in women: a twin study. BMJ 312:940–943
Syed F, Khosla S (2005) Mechanisms of sex steroid effects on bone. Biochem Biophys Res Commun 328:688–696
Tanamas SK, Wijethilake P, Wluka AE, Davies-Tuck ML, Urquhart DM, Wang Y, Cicuttini FM (2011) Sex hormones and structural changes in osteoarthritis: a systematic review. Maturitas 69:141–156
Tian Z, Guo XF, Zhou F, Huang CX, Su M, Long XL, Liu LP (2009) Study on relationship between polymorphisms of estrogen a receptor gene and primary knee osteoarthritis among women in Huna. Pract Prev Med 16:1724–1727
Ushiyama T, Ueyama H, Inoue K, Nishioka J, Ohkubo I, Hukuda S (1998) Estrogen receptor gene polymorphism and generalized osteoarthritis. J Rheumatol 25:134–137
Valdes AM, Spector TD (2008) The contribution of genes to osteoarthritis. Rheum Dis Clin North Am 34:581–603
Wise BL, Demissie S, Cupples LA, Felson DT, Yang M, Shearman AM, Aliabadi P, Hunter DJ (2009) The relationship of estrogen receptor-alpha and -beta genes with osteoarthritis of the hand. J Rheumatol 36:2772–2779
Xue Y, Li D, Yao L, Zhou YX, Xiao D, Guo SQ, Wang Q (2004) Relationship between estrogen receptor gene polymorphism and osteoarthritis in Han women. Chin J Rheumatol 8:583–586
Yang JX, Fu SJ, Xiao FS (2009) Case–control study between estrogen receptor gene polymorphisms and osteoarthritis in southern Sichuan high fluoride areas. West China Med J 24:826–829
Yin YW, Sun QQ, Zhang BB, Hu AM, Liu HL, Wang Q, Hou ZZ (2013) Association between apolipoprotein E gene polymorphism and the risk of coronary artery disease in Chinese population: evidence from a meta-analysis of 40 studies. PLoS One 8:e66924
Yin YW, Li JC, Gao D, Chen YX, Li BH, Wang JZ, Liu Y, Liao SQ, Zhang MJ, Gao CY, Zhang LL (2014a) Influence of ATP-binding cassette transporter 1 R219K and M883I polymorphisms on development of atherosclerosis: a meta-analysis of 58 studies. PLoS One 9:e86480
Yin YW, Sun QQ, Wang PJ, Qiao L, Hu AM, Liu HL, Wang Q, Hou ZZ (2014b) Genetic polymorphism of apolipoprotein A5 gene and susceptibility to type 2 diabetes mellitus: a meta-analysis of 15,137 subjects. PLoS One 9:e89167
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Hohmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
438_2014_940_MOESM1_ESM.tif
Fig. S1 Funnel plots for the rs9340799 polymorphism and OA risk. Allelic model: G allele vs. A allele; Additive model: G/G vs. A/A. Recessive model: G/G vs. A/G + A/A; Dominant model: G/G + A/G vs. A/A (TIFF 205 kb)
438_2014_940_MOESM2_ESM.tif
Fig. S2 Filled funnel plots for the rs9340799 polymorphism and OA risk. Additive model: G/G vs. A/A; Dominant model: G/G + A/G vs. A/A (TIFF 124 kb)
438_2014_940_MOESM3_ESM.tif
Fig. S3 Galbraith plot for the rs9340799 polymorphism and OA risk. Allelic model: G allele vs. A allele; Recessive model: G/G vs. A/G + A/A (TIFF 129 kb)
Rights and permissions
About this article
Cite this article
Yin, YW., Sun, QQ., Hu, AM. et al. Association of rs9340799 polymorphism in estrogen receptor alpha gene with the risk of osteoarthritis: evidence based on 8,792 subjects. Mol Genet Genomics 290, 513–520 (2015). https://doi.org/10.1007/s00438-014-0940-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-014-0940-3