Abstract
Cystic echinococcosis (CE) is a disease that can be transmitted from animals to humans, caused by the metacestode of Echinococcus granulosus. The disease has significant health and economic impacts worldwide, particularly in endemic areas. The study aimed to evaluate the prevalence of hydatid cysts in ruminants (cattle and sheep) (n = 2060) from the Setif Province of Algeria using microscopy. The results showed that hydatid cysts were detected in 9.6% (198/2060) of ruminants, with a higher prevalence in cattle (16.8%; 56/333) compared to sheep (8.2%; 142/1727). Molecular techniques were used to analyze a subset of animals consisting of 30 sheep and 4 cattle. Specifically, a fragment of the mitochondrial cytochrome c oxidase subunit 1 (mt-CO1) gene was sequenced and compared to sequences from seven humans from the same region. The results indicated that all isolates were identified as E. granulosus sensu stricto. Haplotype analysis identified 19 E. granulosus s.s. haplotypes arranged like a star, with the dominant haplotype (Hap04) at the center. Hap04 has been assigned a total of 17 positives, including positives from sheep, cattle, and two humans. This study is noteworthy for being the first to use a molecular approach to human and ruminant echinococcosis in Setif, a significant breeding region in Algeria.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cystic echinococcosis (CE), also known as hydatid disease, is a chronic zoonotic parasitic disease that has been known for centuries. It is widespread in countries around the Mediterranean basin, mainly in Algeria and North Africa. The definitive host of the parasite is dogs and some wild canids (Cai et al. 2017; Oudni-M’rad et al. 2016; Romig et al. 2017). The climate in these regions is also favorable for the resistance of embryophores outdoors (Joanny et al. 2021a, b). Farm animals serve as intermediate hosts. Although epidemiologically a dead-end host, humans can still be infected (Oudni-M’rad et al. 2016; Stoore et al. 2018). CE has a significant economic and social impact and ranks second most common foodborne diseases globally. The disease affects more than one million people worldwide and costs in excess of two billion US dollars annually. The World Health Organization has classified it among the 17 neglected tropical diseases (WHO 2015).
While Algeria exhibits a high endemicity of CE, there is a paucity of data, particularly in molecular studies. Previous research indicates a high prevalence of CE in Algeria and surrounding countries. A total of 174 Echinococcus isolates were analyzed, including those from wild boars (n = 13), camels (n = 7), cattle (n = 16), goats (n = 14), donkeys (n = 10), sheep (n = 33), humans (n = 22), dogs (n = 20), and jackals (n = 2). The analysis was based on the sequences of the cox1 (828 bp) and elongation factor 1-alpha (ef1a; 656 bp) gene regions. The study found that all host species examined were infected with E. granulosus s.s. Notably, Echinococcus canadensis (G6) was detected in a camel, and fertile cysts of E. granulosus s.s. and E. equinus were identified in equids (donkeys) for the first time. Additionally, E. granulosus s.s. was detected in wild boars and goats (Boufana et al. 2014). Lung cysts were obtained from sheep, camels, cattle, and humans across various provinces in Algeria. Subsequent molecular analyses showed that both sheep and camel strains of E. granulosus were present (Bardonnet et al. 2003). According to a study conducted in Algiers, the majority of the human samples examined (90.7%) were identified as E. granulosus s.s. (Zait et al. 2016). In the Djelfa region of the Algerian steppe, a study was conducted on 125 hydatid cyst isolates from cattle and sheep, which were identified as E. granulosus s.s. Additionally, the study found 73 haplotypes (Laatamna et al. 2019).
The purpose of this study is to assess the fertility rate and molecular characteristics of hydatid cysts collected from humans, sheep, and cattle in the Setif Province of Algeria. The aim is to determine the current haplotypic status and phylogenetic relationships of E. granulosus s.l. across various hosts in Algeria.
Material and methods
Sample collection and fertility assessment
The study was carried out in the highlands of northeastern district of Setif, Algeria, located at 36° 11′ 29″ N, 5° 24′ 34″ E with an area of 6549.64 km2 and an altitude of 1100 m above sea level. The region is predominantly semiarid. Surveys were conducted at municipal slaughterhouses in the northeast region of Algeria from November 2021 to April 2022. Hydatid cysts were collected three times a week. During the examination process, the internal organs of all slaughtered animals were carefully observed, palpated, and incised, including the lung, liver, spleen, kidney, and heart, in order to detect any potential presence of hydatid cysts. A total of 2060 ruminants were examined during the sampling period, comprising 333 cattle and 1727 sheep. Of these, 94 female and 239 male cattle, as well as 471 female and 1256 male sheep, underwent postmortem inspection for hydatid cysts. Additionally, 23 human’s hydatid cyst samples were obtained post-surgical resection from patients admitted to the University Hospital Center of Setif (UHC). The samples were transported in coolers directly after surgery to the parasitology laboratory. Only one of the fluid-filled, non-calcified, prominent cyst was excised each organ then transported to the laboratory in chilled containers, for fertility analysis.
The fertility assessment was performed in the parasitology laboratory. For this purpose, the entire fluid from an individual cyst sample was aseptically aspirated and transferred to Eppendorf tubes. Additionally, the germinal membranes of the cysts were removed, immersed, and repeatedly washed in the cyst’s fluid to dislodge the protoscoleces. Following, the fluid was then centrifuged at 1000 g for 5 min, and the pellet was scrutinized for the presence of protoscoleces and hooks using a light microscope (× 40 magnification) (Nikon, Japan) (Daryani et al. 2009).
For molecular analysis, small pieces of the germinal layer of each isolated cyst and/or protoscoleces were washed with 1× PBS (pH = 7.4) and stored in sterile Eppendorf tubes containing 70% ethanol at −20 °C for preservation.
A total of, 41 individual protoscoleces and/or germinal membrane tissue were selected, including 30 from sheep, four from cattle, and seven from humans. Genomic DNA was extracted from individual hydatid cyst samples using the Hibrigen Tissue Kit (Hibrigen, Turkey) with minor modifications to the manufacturer's protocol. Before being transferred to new 1.5 mL Eppendorf tubes, germinal membranes were homogenized. After washing the protoscoleces with 1X PBS (pH = 7.4) at least five times, excess ethanol was removed, and they were used without homogenization. To each tube, 400 μL of lysis buffer and 20 μL of Proteinase-K were added. The tubes were then vortexed and incubated at 65°C overnight in a water bath. The gDNA was subsequently extracted the following day using kit procedures and stored at -20°C until further analysis.
PCR amplification
PCR amplifications were carried out to amplify the genomic DNA targeting a specific mitochondrial cytochrome oxidase subunit 1 (mt-CO1) gene. The length of the resulting amplicon was 875 base pairs. The amplification processused the forward-CO1 primer (5′-TTGAATTTGCCACGTTTGAATGC-3′) and reverse-CO1 primer (5′-GAACCTAACGACATAACATAATGA-3′), as previously described by Nakao et al. (2000). The PCR mixture consisted of 5 µL of 10× PCR buffer, 5 µL of MgCl (25 mM), 400 µM of each dNTP, 20 pmol of each primer, 0.2 µL (1.25 IU) of TaqDNA polymerase (Hibrigen, Ankara, Turkey), and 28.8 µL of PCR-grade water. To achieve the final volume, 5 µL of genomic DNA from each individual sample was added to the PCR mixture. As a positive control, we used gDNA that was available in our laboratory and verified as E. granulosus s.s. by sequence analysis. Distilled water was used as a negative control. The PCR amplification was carried out using a thermal cycler (SensoQuest, GmbH, Germany) with the following conditions: an initial denaturation at 94 °C for 10 min, 30 cycles of denaturation at 94 °C for 30 s, annealing at 52 °C for 45 s, elongation at 72 °C for 1 min, and a final extension step at 72 °C for 10 min (Kesik et al. 2021). The resulting PCR products were visualized on a 1.4% agarose gel stained with ethidium bromide. The PCR bands were extracted from the gels and purified using the QIAquick Gel Extraction Kit (Qiagen). Subsequently, the PCR products were commercially sequenced by BM Labosis (Ankara, Turkey) using a unidirectional forward-CO1 primer. All sequences obtained during the study were submitted to the NCBI database.
Sequence analysis
Each isolate’s chromatogram for sequence analysis was checked for misread nucleotide errors individually using the FinchTV 1.4.0 software (Geospiza, Seattle, WA, USA) (http://www.geospiza.com). The identity of each isolate was confirmed using the “BLAST” tool (http://www.ncbi.nlm.nih.gov/BLAST/) by comparing their nucleotide sequence with previously published sequences and trimming them to 811 bp. The trimmed sequences were uploaded into MEGA X software, followed by alignment with the reference sequence (NC_044548).
Phylogenetic analysis
Nucleotide diversities were analyzed by comparing them with the reference sequence using phylogenetic analysis. Outgroups for the phylogenetic analysis were reference sequences from NCBI PubMed. Sequence alignment was performed using ClustalW in the MEGA X program. The Tamura-Nei gamma distribution (TN93+G) model was selected as the most fitting phylogenetic tree model for the sequences. The maximum likelihood statistical method with a bootstrap test of 1000 replicates (Kumar et al. 2018) was used to generate the evolutionary tree. A phylogenetic tree was constructed using reference sequences of E. granulosus s.s. (NC_044548, MK774655, MG682522), E. equinus (NC_020374), E. ortleppi (NC_011122), and E. canadensis (G6/G7, G8, and G10) (MT380299, AB235848, OQ161122), along with outgroup sequences from E. multilocularis (NC_000928) and Taenia saginata (NC_009938). The phylogenetic tree was constructed using the TN93+G model and the maximum likelihood method in MEGA X. The reliability of the tree was confirmed by 1000 bootstrap tests (Fig. 1). The reference sequence (Accession No. NC044548) revealed 21 point mutations. Furthermore, a conserved fragment ranging from 220 to 385 bp was identified in all sequenced isolates.
Relationship of mt-CO1 gene sequences of the isolates of E. granulosus s.s. (NC_044548, MK774655, MG682522), E. equinus (NC_020374), E. ortleppi (NC_011122), E. canadensis (G6/G7, G8, and G10) (MT380299, AB235848, OQ161122), and outgroup sequences, E. multilocularis (NC_000928) and Taenia saginata (NC_009938)
Haplotype network analysis
To investigate haplotype variation among isolates, DnaSP v.6 software was used following the methodology reported by Rozas et al. (2017). Polymorphic site analysis of the mt-CO1 gene fragment was conducted for various isolates using DnaSP. Population diversity indices, including haplotype numbers (h), nucleotide diversity (π), and haplotype diversity (Hd), were then calculated for different values. Neutrality indices, such as Tajima’s D statistics, as well as Fu’s statistics and Fu and Li’s D and F values, were calculated (Tashani et al. 2002). The minimum spanning networks (MSN) method was used to create the network with the PopART 1.7 software (Leigh and Bryant 2015).
Results
The overall prevalence of infection was 9.6% (198/2060), with a higher prevalence in cattle 16.8% (56/333) than in sheep 8.2% (142/1727). Prevalence was higher in females than males for both cattle 87.5% (82/94) and sheep 90.8% (428/471). The lungs exhibited a high infection rate in both sheep and cattle (42.4%, n = 84), followed by the liver (26.3%, n = 52), and multiple organ infections, including both the liver and lungs (31.3%, n = 62). Additionally, during surgery, human hepatic hydatid cyst samples (n = 23) were collected, (one cyst material from each patient). The fertility rate of the hydatid cysts was 76.7% (109/142) in sheep, 25% (14/56) in cattle, and 95.6% (22/23) in humans.
Due to limitations in isolating gDNA from germinal membranes, complete success could not be attained. Therefore, only 41 samples (sheep, n = 30; humans, n = 7; and cattle, n = 4) underwent molecular analyses. A fragment of the mt-CO1 gene has been amplified by PCR, and a band of 875 bp has been visualized in all the samples analyzed. The PCR products were subsequently sequenced one-way. The nucleotide sequences were compared with previously published reference sequences and subsequently trimmed. The final size of the trimmed sequences was 811 bp for 41 sequences. According to BLAST searches, all of the sequences were identified as E. granulosus s.s. (G1/G3). The sequences from this study were deposited in the NCBI database with the following accession numbers: OQ269889-OQ269918, OQ269919-OQ269922, and OQ269923-OQ269929 (Table 1).
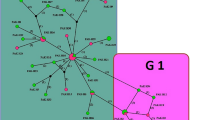
After analyzing haplotypes and identifying a primary focal haplotype, E. granulosus s.s. was oriented in a star-like configuration. It differed from other haplotypes by 1–17 mutation steps. Nineteen haplotypes were detected in 41 samples, with 17 (41.46%) isolates belonging to the primary haplotype (Hap04) (Fig. 2 and Table 2). In the mt-CO1 sequences, 21 polymorphic sites were observed, with 10/21 (47.61%) being parsimony informative. Table 3 provides evidence of higher haplotype diversity and low nucleotide diversity in the gene region under examination, confirming the findings of this study. Tajima’s D indicative of population expansion and/or purification of selection was negative, while observed Fu’s Fs values revealed the presence of rare haplotypes anticipated by hitchhiking or recent population expansion. Crucially, 78.94% (15/19) of haplotype groups were made up of single haplotypes, reinforcing the study’s conclusions.
Discussion
The high plains of Sétif are well-regarded for their meat production. Breeders utilize one or more ruminant species, depending on local practices and the availability of food resources. Specifically, only 5.4% of agricultural units raise all three species (cattle, sheep, and goats); 39.3% focus on cattle and sheep exclusively, while cattle are raised alone on 55.3% of farms (Bir et al. 2014). Consequently, this region represents a high-risk area for the spread of various zoonotic diseases. CE remains a significant economic and public health concern in Algeria. Studies on the prevalence and molecular characteristics of E. granulosus s.l. isolates in Algeria have been limited, as evidenced by the works of Zait et al. (2016), Laatamna et al. (2019), Bardonnet et al. (2003), Maillard et al. (2007), Moussa et al. (2021), and Samari et al. (2022). Reports on the prevalence indicate variations in both intermediate hosts, represented by various ruminant species, and the organ predilection of cyst localization. The overall prevalence rate was 9.6% (198/2060), with cattle having a rate of 16.8% (56/333) and sheep having a rate of 8.2% (142/1727) among the total ruminant population of 2060 (cattle, n = 333; sheep, n = 1727). The prevalence rate of CE in animals found in this study is almost the same that reported in Souk Ahras province (10%), higher than the prevalence rate reported in Algiers (4.9%) (El fegoun et al. 2020). However, it is lower than that reported by Laatamna et al. (2019) in Djelfa with a prevalence of 13.9% in cattle and 5.7% in sheep, taking into account the total number of animals (n = 1278). It is overall, the diverse prevalence rates of CE across the globe are challenging to assess after the fact due to various factors such as focal transmission, age of livestock at slaughter, sample size, and diagnostic procedures as noted by Laatamna et al. (2019). As an example, the study revealed that the lungs had significantly high infection rates (42.4%, n = 84) present in both sheep and cattle, trailed only by liver infections (26.3%, n = 52). As for the double infection liver and lung in the same animal, it was estimated to (31.3%, n = 62). These findings appear to differ from previous research that suggested cysts were mainly found in the liver, followed by the lungs (Mehmood et al. 2020). However, Laatamna et al. (2019) seem to support the idea that E. granulosus s.s. can adapt to sheep as intermediate hosts, as cysts were found in both the liver and lungs with similar frequency and high fertility rates in both organs.
The present study reports a fertility rate of 25% (14/56) in cattle, which is consistent with Romig et al.’s (2015) findings. Additionally, our study found a higher frequency of human fertile cyst rates at 95.6% compared to other studies. This finding highlights the potential risk of secondary CE infections during surgical intervention. The results are consistent with previous studies conducted by Khalf et al. (2014) in Iraq, Piccoli et al. (2013) in Romania, and Issa et al. (2022) in Iraq, which reported fertility rates of 47%, 92%, and 82.81%, respectively.
In molecular studies, determining the haplotypes of hydatid cysts is considered crucial. This is especially important in Algeria, where there is limited knowledge of the genetic diversity of CE in the highlands of Setif. Therefore, this study represents the first molecular characterization of E. granulosus in both livestock and humans in the highlands of Setif, which could provide valuable insights for future research. Identifying the genotypes of E. granulosus found within intermediate hosts in the study region is an important step towards disrupting the parasite’s lifecycle and establishing more effective control strategies for CE. The genetic variation of Echinococcus spp can indicate differences in infectivity among various host groups or species. Therefore, it is essential to genetically characterize the population structure (Thompson and McManus 2002) in order to gain a better understanding of the epidemiology of the disease and to develop more targeted interventions. The purpose of this study was to analyze the E. granulosus isolates by sequencing mt-CO1. The findings confirmed the presence of E. granulosus s.s. in all samples, which is consistent with previously reported findings in Algeria. The analyzed genotype exhibited a frequency of 90.7 to 100% among CE cases. According to the literature, it has been found that the G1 genotype of E. granulosus s.s. is primarily responsible for the global prevalence of human CE. In some areas with high CE incidence, this genotype is the only cause of human CE, as noted by Alvarez Rojas et al. in 2014.
To gain a better understanding of PCR errors, much attention has been given to base substitution errors made by DNA polymerase. The fidelity of DNA polymerase replication has been extensively studied using various methods and assay conditions, such as blue/white screening, forward mutation, denaturing gradient gel electrophoresis, high-throughput Sanger sequencing, and next-generation sequencing. It is important to consider that differences in error rates may arise from a variety of factors, such as variations in assay methodology, reaction conditions, template sequences, and error reporting units (Potapov and Ong 2017). This may cause errors and false haplotypic findings, especially in long sequence readings. However, since the region analyzed in this study was not very long, we think that there was no case of misreading and haplotype detection.
The study identified 19 distinct haplotypes in 41 samples, with 17 (41.46%) belonging to the primary haplotype (Hap04). The same haplotype (Hap04) was shared by only two isolates from humans in Boumerdes and Laghouat (both cities located far from our study area) (Moussa et al. 2021). The second most frequent haplotype in the current work is Hap06; this haplotype was found in one isolate previously described by Moussa et al. (2021). From the results of our study and similar works, it can be concluded that a considerable intraspecific variation was observed in the mt-CO1 sequences (Casulli et al. 2012).
Conclusion
This study presents the molecular characterization data on hydatid cyst isolates of ruminants and humans from various regions of Setif Province (northeast Algeria) by successfully sequencing the mt-CO1 gene. It is important to identify the most common species to determine the mode of transmission of the parasite, its pathogenicity, diagnostic procedures, vaccine development, and treatment options. Identification of common species is considered crucial for implementing effective control measures. It is recommended to genetically analyze a wider range of isolates and, if possible, more animal species to identify the E. granulosus s.s. species circulating in Algeria and to implement appropriate control measures.
Data availability
No data was used for the research described in the article.
References
Alvarez Rojas CA, Romig T, Lightowlers MW (2014) Echinococcus granulosus sensu lato genotypes infecting humans – review of current knowledge. Int J Parasitol 44(1):9–18. https://doi.org/10.1016/j.ijpara.2013.08.008
Bardonnet K, Benchikh-Elfegoun MC, Bart JM et al (2003) Cystic echinococcosis in Algeria: cattle act as reservoirs of a sheep strain and may contribute to human contamination. Vet Parasitol 116(1):35–44. https://doi.org/10.1016/S0304-4017(03)00255-3
Bir A, Yakhlef H, Madani T (2014) Diversité des exploitations agricoles laitières en zone semi-aride de Sétif (Algérie). Livestock Res Rural Develop 26(2):26. http://www.lrrd.org/lrrd26/2/bir26026.htm
Boufana B, Lahmar S, Rebaï W et al (2014) Genetic variability and haplotypes of Echinococcus isolates from Tunisia. Trans R Soc Trop Med Hygiene 108(11):706–714. https://doi.org/10.1093/trstmh/tru138
Cai H, Guan Y, Ma X et al (2017) Epidemiology of Echinococcosis among schoolchildren in Golog Tibetan Autonomous Prefecture, Qinghai China. Am J Trop Med Hyg 96(3):674–9. https://doi.org/10.4269/ajtmh.16-0479
Casulli A, Interisano M, Sreter T et al (2012) Genetic variability of Echinococcus granulosus sensu stricto in Europe inferred by mitochondrial DNA sequences. Inf Gene Evol 12(2):377–383. https://doi.org/10.1016/j.meegid.2011.12.014
Daryani A, Sharif M, Amouei A, Nasrolahei M (2009) Fertility and viability rates of hydatid cysts in slaughtered animals in the Mazandaran Province, Northern Iran. Trop Anim Health Prod 41:1701–1705. https://doi.org/10.1007/s11250-009-9368-x
El Fegoun MCB, Kohil K, Benguesmia M et al (2020) Cystic Echinococcosis in Algeria: the role of cattle as reservoirs in the dynamics of transmission of Echinococcus granulosus to humans via dogs. Bull Soc Pathol Exot 113(3):130–135. https://doi.org/10.3166/bspe-2020-0130
Issa AR, Arif SH, Mohammed AA et al (2022) Insights into human cystic echinococcosis in the Kurdistan Region, Iraq: characteristics and molecular identification of cysts. Pathogens 11(4):408. https://doi.org/10.3390/pathogens11040408
Joanny G, Cappai MG, Nonnis F et al (2021a) Human cystic echinococcosis in Lebanon: a retrospective study and molecular epidemiology. Acta Parasitol 67(1):186–195. https://doi.org/10.1007/s11686-021-00453-w
Joanny G, Mehmood N, Dessi G et al (2021b) Cystic echinococcosis in sheep and goats of Lebanon. Parasitology 148:871–878. https://doi.org/10.1017/S0031182021000494
Kesik HK, Celik F, Simsek S et al (2021) Molecular characterization and haplotype analyses of lung hydatid cyst isolates of cattle and first report of Echinococcus canadensis (G6/G7) in cattle isolates in Turkey. Acta Parasitol 66(4):1538–1547. https://doi.org/10.1007/s11686-021-00432-1
Khalf MS, AlTaie LH, AlFaham MA (2014) The incidence of hydatid cyst in human in Baghdad governorate. IOSR J Pharm Biol Sci (IOSR-JPBS), 9(3):11–14
Kumar S, Stecher G, Li M et al (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Laatamna AE, Ebi D, Brahimi K et al (2019) Frequency and genetic diversity of Echinococcus granulosus sensu stricto in sheep and cattle from the steppe region of Djelfa. Algeria Parasitol Res 118(1):89–96. https://doi.org/10.1007/s00436-018-6118-x
Leigh JW, Bryant D (2015) POPART: full-feature software for haplotype network construction. Methods Ecol Evol 6:1110–1116
Maillard S, Benchikh-Elfegoun MC, Knapp J et al (2007) Taxonomic position and geographical distribution of the common sheep G1 and camel G6 strains of Echinococcus granulosus in three African countries. Parasitol Res 100:495–503. https://doi.org/10.1007/s00436-006-0286-9
Mehmood N, Arshad M, Ahmed H et al (2020) Comprehensive account on prevalence and characteristics of hydatid cysts in livestock from Pakistan. Korean J Parasitol 58(2):121–127. https://doi.org/10.3347/kjp.2020.58.2.121
Moussa D, Senouci K, Midoun N et al (2021) Genetic diversity of Echinococcus granulosus sensu stricto infecting humans in western Algeria. Parasitol Res 120(9):3195–3202. https://doi.org/10.1007/s00436-021-07223-7
Nakao M, Sako Y, Yokoyama N et al (2000) Mitochondrial genetic code in cestodes. Mol Biochem Parasitol 111:415–424
Oudni-M’rad M, Cabaret J, M’Rad S et al (2016) Genetic relationship between the Echinococcus granulosus sensu stricto cysts located in lung and liver of hosts. Infect Genet Evol 44:356–360. https://doi.org/10.1016/j.meegid.2016.07.024
Piccoli L, Bazzocchi C, Brunetti E et al (2013) Molecular characterization of Echinococcus granulosus in south-eastern Romania: evidence of G1–G3 and G6–G10 complexes in humans. Clin Microbiol Inf 19(6):578–582. https://doi.org/10.1111/j.1469-0691.2012.03993.x
Potapov V, Ong JL (2017) Examining sources of error in PCR by single-molecule sequencing. PLoS ONE 12(1):e0169774. https://doi.org/10.1371/journal.pone.0181128
Romig T, Ebi D, Wassermann M et al (2015) Taxonomy and molecular epidemiology of Echinococcus granulosus sensu lato. Vet Parasitol 213(3–4):76–84. https://doi.org/10.1016/j.vetpar.2015.07.035
Romig T, Deplazes P, Jenkins D et al (2017) Ecology and life cycle patterns of Echinococcus species. Adv Parasitol 95:213–314. https://doi.org/10.1016/bs.apar.2016.11.002
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC et al (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol 34:3299–3302. https://doi.org/10.1093/molbev/msx248
Samari H, Laurimäe T, Reghaissia N et al (2022) Molecular characterization of Echinococcus granulosus sensu lato genotypes in dromedary camels from extreme Sahara of Algeria based on analysis of nad2 and nad5 genetic markers. Acta Trop 234:106616. https://doi.org/10.1016/j.actatropica.2022.106616
Stoore C, Andrade C, Hidalgo C et al (2018) Echinococcus granulosus hydatid cyst location is modified by Fasciola hepatica infection in cattle. Parasit Vectors 11(1):542. https://doi.org/10.1186/s13071-018-3128-6
Tashani OA, Zhang LH, Boufana B et al (2002) Epidemiology and strain characteristics of Echinococcus granulosus in the Benghazi area of eastern Libya. Ann Trop Med Parasitol 96:369–381. https://doi.org/10.1179/000349802125000952
Thompson RA, McManus DP (2002) Towards a taxonomic revision of the genus Echinococcus. Trends Parasitol 18(10):452–457. https://doi.org/10.1016/S1471-4922(02)02358-9
World Health Organization (2015) Investing to overcome the global impact of neglected tropical diseases: third WHO report on neglected diseases 2015. Geneva, Switzerland: World Health Organization
Zait H, Kouidri M, Grenouillet FE et al (2016) Molecular characterization of Echinococcus granulosus sensu stricto and Echinococcus canadensis in humans and livestock from Algeria. Parasitol Res 115(6):2423–2431. https://doi.org/10.1007/s00436-016-4994-5
Acknowledgements
The authors thank the responsible veterinarian at the Setif slaughterhouse and the University Hospital Center of Setif (UHC) for their assistance and participation during the sampling of the material.
Funding
This work was financially supported by Firat University Scientific Research Project Management Unit (Project no: VF.22.25) in Elazig, Turkey.
Author information
Authors and Affiliations
Contributions
Amina Kheninef: investigation, methodology, data analyses, writing-original draft. Figen Celik: investigation, methodology, data analyses. Lynda Aissaoui: methodology, writing-review and editing. Sami Simsek: funding acquisition, conceptualization, supervision, writing-review and editing.
Corresponding author
Ethics declarations
Ethical approval
N/A.
Consent to participate
Informed consent was obtained from the slaughterhouse staff prior to sampling the hydatid cyst samples.
Consent for publication
All authors read and consent to the publication of the manuscript.
Conflict of interest
The authors declare no competing interests.
Additional information
Handling Editor: Una Ryan
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kheninef, A., Celik, F., Aissaoui, L. et al. Molecular characterization and haplotypes of hydatid cyst isolates collected from humans and ruminants in Setif Province (northeast of Algeria) based on mitochondrial cytochrome C oxidase subunit 1 (mt-CO1) gene sequences. Parasitol Res 123, 159 (2024). https://doi.org/10.1007/s00436-024-08176-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00436-024-08176-3