Abstract
Theileriosis is a tick-borne disease that causes enormous losses in the dairy industry. There are several species of Theileria that can infect bovines. Generally, more than one species are prevalent in any geographical area; thus, chances of co-infections are high. Differentiation of these species may not be possible by microscopic examination or serological tests. Therefore, in this study, a multiplex PCR assay was standardized and evaluated for rapid and simultaneous differential detection of two species of Theileria viz., Theileria annulata and Theileria orientalis. Species-specific primers were designed to target the merozoite piroplasm surface antigen gene (TAMS1) of T. annulata and the major piroplasm surface protein gene of T. orientalis, yielding specific amplicon of 229 bp and 466 bp, respectively. The sensitivity of multiplex PCR was 102 and 103 copies for T. annulata and T. orientalis, respectively. The simplex and multiplex PCRs were specific and showed no cross-reactivity with other hemoprotozoa for either primer. For comparative evaluation, blood samples from 216 cattle were tested by simplex and multiplex PCR for both species. Using multiplex PCR, 131 animals were found infected for theileriosis, of which 112 were infected with T. annulata, five were infected with T. orientalis, and 14 had mixed infections. This is the first report of T. orientalis from Haryana, India. Representative sequences of T. annulata (ON248941) and T. orientalis (ON248942) were submitted in GenBank. The standardized multiplex PCR assay used in this study was specific, sensitive, for the screening of field samples.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ticks are a major problem in tropical and sub-tropical countries, including India, and so are tick-borne diseases. Theileriosis is the most predominant among these diseases, causing substantial economic losses to the livestock sector (Rashid et al. 2019). Several Theileria species can infect bovines, including Theileria annulata, T. orientalis, T. parva, and T. mutans. Tropical theileriosis (T. annulata) and east coast fever (T. parva) are important clinical manifestations of cattle, while T. orientalis (oriental theileriosis) causes benign and mild disease. However, recently, fatal outbreaks caused by T. orientalis have been documented in various parts of the world where it was previously considered non-pathogenic (McFadden et al. 2011; Thompson et al. 2020), including India (Aparna et al. 2011; Patial et al. 2021). If the animal is already infected with T. mutans or T. annulata, then the infection of benign Theileria species such as T. orientalis is much more pathogenic. (Junlong et al. 2015).
Two hundred fifty million cattle are affected globally by tropical theileriosis caused by T. annulata (Erdemir et al. 2007). In India, losses of approximately 8426.7 crore INR in the dairy sector have been attributed to Tropical theileriosis (Narladkar 2018). High-yielding crossbred cattle, exotic breeds, and young calves are highly susceptible to the clinical form of the disease and have a high mortality rate. In contrast, mild or subclinical form of disease prevails in indigenous breeds. In addition, sub-clinically infected or carrier animals often go undiagnosed and pose a potential risk of infection in susceptible animals. Subclinical theileriosis is associated with invisible production losses due to decreased milk production (Rashid et al. 2019; Singh et al. 2022), poor body scores, and increased disease susceptibility (Kolte et al. 2017).
Another important prevalent species of Theileria in India and other parts of Asia and Australia is T. orientalis which causes oriental theileriosis. It is a non-transforming species of Theileria and is known to cause mild or sometimes no disease. However, clinical diseases and outbreaks with high morbidity and severe clinical signs have been reported worldwide (McFadden et al. 2011; Kamau et al. 2011; Thompson et al. 2020). Clinical signs such as abortion, hemolytic anemia, jaundice, fever, and arthritis are observed in clinical infection of T. orientalis (Kamau et al. 2011; Patial et al. 2021). Economic loss in oriental theileriosis occurs because of decreased milk production and abortion (Perera et al. 2014; Gebrekidan et al. 2020). In addition, chronic subclinical infections with T. orientalis may also lead to anemia (Choi et al. 2016).
The diagnosis of theileriosis under field conditions primarily relies on clinical signs, history, tick infestation, and reported disease prevalence. Many laboratory diagnostic methods are available, ranging from simple microscopic examinations to complex serological and molecular techniques. Microscopic examination of lymph node biopsies and blood smears is the most common laboratory diagnostic technique (Chauhan et al. 2015). Although economical, it is not possible to detect subclinical carriers. In addition, expertise is required to identify organisms correctly. Moreover, it is difficult to distinguish between different Theileria species or to determine co-infection based on microscopic characteristics (Liu et al. 2010). Serological investigations, such as enzyme-linked immunosorbent assay and indirect fluorescent antibody tests, are more specific and sensitive than microscopic examination. However, these tests cannot differentiate between T. annulata and T. orientalis because of cross-reactivity between these species (Mans et al. 2015).
Molecular techniques such as PCR have the highest specificity and sensitivity among all diagnostic tests. In addition, PCR assays can easily differentiate closely related species. Furthermore, PCR is beneficial for detecting carrier animals, as parasitemia in such animals is reported to be above the detection limit of PCR (Mans et al. 2015). However, screening a large population for individual pathogens using PCR is laborious and resource-intensive. This limitation is overcome by multiplex PCR assays, in which multiple pathogens can be detected simultaneously in one reaction, thus saving time and money. Therefore, they are best suited for epidemiological investigation.
Therefore, in this study, we aimed to develop a multiplex PCR assay for the rapid and simultaneous detection of two important Theileria species, viz., Theileria annulata and Theileria orientalis.
Material and methods
Positive control
A partial gene sequence of the major surface piroplasm protein (MPSP) cloned in the pBluescript II ks + vector synthesized by Bio Basic Inc., Canada, was used as a positive control for T. orientalis. The plasmid was reconstituted according to the manufacturer’s instructions and was inserted into competent DH5αcells. Similarly, for the positive control of T. annulata, the partial gene sequence merozoite-piroplasm surface antigen (TAMS1) cloned in the pBluescript II ks + vector (Bio Basic Inc., Canada) was purchased and processed further as mentioned above. DNA extracted from these DH5αcells was used as a positive control.
Primer design
After in silico screening and analysis of sequences from the GenBank® sequence database (http://www.ncbi.nlm.nih.gov/), two novel primer sets intended to identify the TAMS1 gene of T. annulata and MPSP gene of T. orientalis were designed using Primer3Plus software. Both primer sets had comparable annealing temperatures and were analyzed for self-annealing, hair loop formation, and self- and cross-dimer formation using the OligoAnalyzer software (https://www.idtdna.com/pages/tools/oligoanalyzer). The specificity of the primers was analyzed by BLASTn. The sequences of the primer sets and their corresponding product sizes are listed in Table 1.
Standardization of simplex and multiplex PCR
Simplex PCR assay was standardized for T. annulata and T. orientalis individually using the positive controls as a template. The PCR reaction mix in both cases had a total volume of 12.5µL consisting of 6.25µL of 2 × TopTaq PCR master mix (Qiagen, Germany), 0.4µL (10 µM) of each forward and reverse primers, and 1µL of template DNA and nuclease-free water to make up the final volume. The PCR was performed in Agilent SureCycler 8800 (USA). The gradient PCR was performed to optimize temperature (48–58 °C), primer concentration, and the number of cycles. For multiplex PCR assay, the reaction mix consisted of 6.25µL of 2 × TopTaq PCR master mix (Qiagen, Germany), 0.4µL (10µ M) of each forward and reverse primers for MPSP and TAMS1 gene, 1µL of template DNA, and 3.65 µL of nuclease-free water. Multiplex PCR was standardized to optimize the annealing temperature, thermo-cycling conditions, and primer concentration. PCR products were resolved in 1.8% agarose gel and visualized using a UV transilluminator (GeNei™ UVITEC imaging systems, Cambridge).
Determination of the minimum detection limits for simplex and multiplex PCR
The PCR amplicons of T. annulata and T. orientalis were purified from agarose gel using a gel purification kit (Qiagen, USA) and cloned into the pJET1.2 cloning vector. Plasmid DNA was extracted from the clones using a Zymo Plasmid Miniprep kit. The concentration of extracted DNA was estimated using a NanoDrop™ 2000 spectrophotometer. The estimation of the gene copy numbers was based on the assumption that the average weight of a base pair is 650 Da, thus calculating the number of plasmid copies using the following formula:
To determine the sensitivity of simplex PCR for T. annulata and T. orientalis, the stock solution of purified recombinant plasmids containing the target sequence TAMS1 gene (pJET1.2/TAMS1) and MPSP gene (pJET1.2/MPSP) with the copy number 1010 were taken and tenfold dilution of these stock solution was made fold till copy number equals to 10. For determination of sensitivity of multiplex PCR assay, stock solutions of T. annulata and T. orientalis were mixed and diluted tenfold till copy number equals to 10.
Specificity assay for simplex and multiplex PCR
DNA was extracted from blood samples that were positive for Babesia spp., Anaplasma spp., and Trypanosoma spp. by microscopic examination. These positive DNA samples were used to determine the specificity of simplex PCR and multiplex PCR for T. orientalis and T. annulata. In addition, the cross-reactivity between T. orientalis and T. annulata was checked using their positive controls in both simplex and multiplex PCR.
Collection of samples
Blood samples from cattle suspected of hemoprotozoan disease having any of the clinical signs, i.e., fever, enlarged superficial lymph nodes, tick infestation, and anemia, were used to validate and evaluate the efficacy of the designed multiplex PCR. Blood samples were collected from 216 suspected cattle from the Veterinary Clinical Complex, LUVAS, Hisar, and organized dairy farms near Hisar, Haryana, India.
Microscopic examination of blood smears
The blood smears were prepared by fresh whole blood and stained with Giemsa stain as per standard procedure. Each slide was examined for presence of Theileria piroplasm in erythrocytes and schizonts in lymphocytes.
Evaluation of multiplex PCR using field samples
The DNA was extracted from the blood using DNeasy blood and tissue kit (Qiagen, Germany) by following the method recommended by the manufacturer. The extracted DNA was then stored at − 20 °C until further used. First, simplex PCR was performed for each sample to detect presence of T. annulata and T. orientalis. Samples were then screened by multiplex PCR, and the results of all tests were compared.
Sequencing of samples
Sequencing was performed using the PCR amplicons from representative samples positive for T. annulata and T. orientalis, to further confirm the PCR results. PCR amplicons were purified for selected samples from agarose gel using a gel purification kit (Qiagen, USA) and sequenced using an array-based automatic capillary ABI 3130xL machine (Applied Biosystems™, USA).
Results
Standardization of simplex and multiplex PCR
PCR conditions were individually optimized for the T. annulata TAMS1 primer pair and the T. orientalis MPSP primer pair. The optimum annealing temperature was 58 °C based on the gradient PCR results for both primer pairs. The final optimized thermo-cycling conditions for both simplex PCR were as follows: initial denaturation at 95 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 58 °C for 30 s, and 72 °C for 40 s, and a final extension at 72 °C for 10 min. The TAMS1 and MPSP primer pairs amplified amplicons of predicted sizes of 229 bp and 466 bp, respectively.
The optimum conditions for multiplex PCR were initial denaturation at 95 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 58 °C for 30 s, and 72 °C for 40 s and then a final extension at 72 °C for 10 min. In a multiplex PCR assay, the specific amplicons (229 bp and 466 bp for T. annulata and T. orientalis, respectively) belonging to individual species could be readily differentiated in a 1.8% agarose gel. No non-specific amplification was detected in the multiplex PCR assay.
Minimum detection limits of simplex and multiplex PCR
The simplex PCR for T. annulata was able to detect template up to 102 copies whereas the detection limit of simplex PCR for T. orientalis was 103 copies. The detection limit of multiplex PCR was 102 and 103 copies for T. annulata and T. orientalis, respectively.
Specificity assay for simplex and multiplex PCR
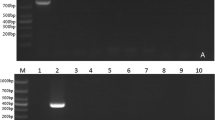
Each primer set was checked for cross-reactivity with other hemoprotozoa such as Babesia spp., Anaplasma spp., and Trypanosoma spp. Primers were species-specific, as no amplification was observed for DNA from other parasites (Fig. 1). In addition, T. annulata was not amplified by T. orientalis primers and vice versa. These results indicated that both primer sets were highly specific.
Specificity assay of Multiplex PCR for detecting T. annulata (229 bp) and T. orientalis (466 bp). Lane L: 100 bp ladder; Lane 1: positive control of T. annulata; 2: positive control of T. orientalis; 3: Positive sample for T. annulata and T. orientalis; 4: Positive sample for Babesia spp.; 5: Positive sample for Anaplasma spp.; 6: Positive sample for Trypanosoma spp., N: negative control
Microscopic examination of blood smears
Blood smears of 216 animals were examined, out of which 77 (35.64%) animals were found positive for theileriosis. The piroplasm of various shapes such as round dot like, signet-ring shaped, oval, elongated, and rod-shaped were observed in erythrocytes. All the animals that were positive by blood smear examination were also positive in PCR examination. Microscopic smears for blood samples positive for PCR for T. annulata and T. orientalis were compared and are shown in Fig. 2 (T. annulata), Fig. 3 (T. orientalis), and Fig. 4 (mixed infection). From comparison, it was found difficult to distinguish the two infection and almost impossible to identify the mixed infections.
Evaluation of field sample using multiplex PCR
DNA samples from 216 cattle were examined using both simplex and multiplex PCR (Fig. 5). On examination by simplex PCR for T. annulata, 128 (59.3%) animals out of 216 were found positive for T. annulata. In simplex PCR for T. orientalis, 19 (8.8%) animals out of 216 were found positive for T. orientalis. Multiplex PCR showed that out of 216 animals, 131 (60.64%) were found to be infected, of which 112 were infected with T. annulata, five were infected with T. orientalis, and 14 had mixed infection. The results of multiplex PCR were similar to those of simplex PCR except for two animals that were positive for T. annulata in simplex PCR while negative in multiplex PCR (Table 2). This is the first report of T. orientalis in Haryana, India.
Sequencing of samples
Representative samples were sequenced and submitted to NCBI (accession no. ON248942 for T. orientalis, and ON248941 for T. annulata). These nucleotide sequences were analyzed using the NCBI BlastN software. The submitted sequence of T. orientalis showed 98.95% identity with isolate of cattle from China (MT517305.1) and 97–98.95% similarity with other T. orientalis sequences. The submitted sequence of TAMS1 gene of T. annulata showed 97.9% with T. annulata isolates of cattle from Ankara, Turkey (AJ276654.1), and 95–97% similarity with other T. annulata sequences.
Discussion
Owing to the high tick vector population in tropical and subtropical regions, haemoprotozoan diseases are a major concern for dairy animals in these regions (Li et al. 2016; Fukushima et al. 2021; Krishnamoorthy et al. 2021). However, due to climatic changes and global warming, the vectors such as ticks have expanded to new regions, thus increasing the risk of haemoprotozoan diseases in these areas (Medlock et al. 2013; Rojas-Downing et al. 2017). Theileriosis is one such haemoprotozoan disease responsible for huge economic losses in the dairy industry (Narladkar 2018). There are several species of Theileria infecting bovines, but T. annulata is the most widely prevalent and causes severe clinical infections. Therefore, epidemiological studies and preventive strategies primarily focused on the T. annulata. But in past few decades, outbreaks and clinical reports of T. orientalis had been reported worldwide particularly from Australia and Asia including India (Kamau et al. 2011; Aparna et al. 2011; Kakati et al. 2015). Thus, these areas have prevalence of both the species, which necessitates the inclusion of T. orientalis also in the epidemiological studies, vaccination for formulation of better prevention, and control strategies for theileriosis.
The conventional approach of diagnosis by microscopic examination is neither sensitive (Nayel et al. 2012) nor can it differentiate between these two species (Bilgiç et al. 2013). Similarly, serological tests have the problem of cross-reactivity between different species of Theileria (Renneker et al. 2009) and also have chances of false-negative results in chronic infections as immune responses in such cases wane away over time (Ranjan et al. 2015). Molecular tests such as PCR are highly sensitive and specific and multiplexing can reduce the cost and time of diagnosis for multiple pathogens, especially in epidemiological studies where many samples are to be analyzed (El Damaty et al. 2021; Tuli et al. 2015). Many scientific reports of multiplex PCR for detecting different hemoprotozoa such as Theileria, Babesia, Anaplasma, and Trypanosoma are available (Bilgiç et al. 2013; Ashuma et al. 2014; Ganguly et al. 2020; Charaya et al. 2021). However, literature describing multiplex PCR assays for diagnosing different species of Theileria in cattle is very scarce (Orkun et al. 2012; Junlong et al. 2015). Therefore, in this study, multiplex PCR was standardized for rapid differential diagnosis of T. annulata and T. orientalis. The multiplex PCR standardized in the present study can differentially detect both the species of Theileria in less than 5 h of receiving a blood sample.
Highly specific and sensitive primers are required for efficient PCR. Primers based on Tams1 and MPSP genes were used for T. annulata and T. orientalis, respectively. Both primers were specific as they showed no amplification for other hemoprotozoa and one another in the specificity assay. TAMS1 gene is widely used for the detection of T. annulata. Kirvar et al. (Kirvar et al. 2000) also reported that primers designed from the TAMS1 gene showed no cross-reactivity with other hemoprotozoa. The present and previous studies have shown that there is no cross-reactivity of the TAMS1 gene T. annulata with other Theileria species, making it a suitable candidate for multiplex PCR (Orkun et al. 2012; Kundave et al. 2018; Ganguly et al. 2020). Similarly, the MPSP gene is also widely described for molecular identification of T. orientalis (Ota et al. 2009; Kamau et al. 2011). The specificity of the multiplex PCR was further elucidated by the fact that amplicons of only the expected size were observed in the analysis of field samples.
The standardized multiplex PCR assay in the present study had a very low minimum detection limit for both species; therefore, the assay had high sensitivity. The high sensitivity of this assay ensures that it can efficiently detect subclinical infections (Khatoon et al. 2015). The detection of subclinical carriers is of utmost importance in theileriosis control as they serve as source of infection for healthy animals (Abdela and Bekele 2016). Additionally, subclinical infections are linked to anemia, poor immunity, and production loss (Kolte et al. 2017).
The prevalence by microscopic examination (35.64%) was very low when compared to that of multiplex PCR (60.64%). The lower sensitivity of blood smear examination was reported by other workers also (Nourollahi-Fard et al. 2015; Kundave et al. 2015; Charaya et al. 2016). The sensitivity of the test was lower than PCR because of the low parasitemia in chronic and subclinical carrier animals (Nayel et al. 2012). There is lot of polymorphism in piroplasmic form of Theileria which creates confusion and requires a great expertise to correctly identify the organism (Acharya et al. 2017). The distinction of the two species was not possible by microscopic examination; thus, the prevalence of specific species of theielria and mixed infections cannot be determined. In the present study also, microscopic smears for blood samples positive for PCR for T. annulata and T. orientalis were compared; it was difficult to distinguish the two infection and almost impossible to identify the mixed infections.
On screening 216 animals using simplex PCR, 128 (59.25%) were positive for T. annulata, while 19 (8.79%) were positive for T. orientalis. The results of multiplex PCR were almost the same as those of simplex PCR except for two cases, which were positive for T. annulata with simplex PCR while negative with multiplex PCR. This minute difference may be because parasitemia might be very low in those cases, as the detection ability of multiplex PCR is compromised by the extremely low level of parasitemia (Ganguly et al. 2020). The multiplex PCR used in the present study was as efficient as the simplex PCR, highlighting its potential usefulness in epidemiological investigations.
The results indicated that the prevalence of T. annulata was higher than that of T. orientalis in the study region. A high prevalence of T. annulata has been previously reported in the studied geographical area (Ganguly et al. 2020; Charaya et al. 2021). However, there have been no previous studies regarding the prevalence of T. orientalis in the studied geographical area. To the best of our knowledge, this is the first report of T. orientalis in Haryana, India. T. orientalis is generally considered mild or virulent, but recent outbreaks with high mortality and morbidity from different parts of the country suggest the importance of epidemiological investigation of the organism. The present study also indicated that mixed infections are also prevalent. Multiple species infections of parasites can alter their pathogenicity and disease pattern and can also lead to cross-immunity (Awad et al. 2020). Competition between the two species of parasites in the same host leads to genetic changes in both parasites (Bashey 2015). Thus, periodic molecular epidemiology of both Theileria spp. is necessary in areas such as India where both species co-exist. The multiplex PCR assay standardized in the present study could potentially be useful in such investigations.
Conclusion
T. annulata and T. orientalis are both important species of Theileria that affect cattle and both these species co-exist in the same geographical area. Thus, to formulate better prevention and control strategies, the epidemiology of both the species should be investigated. The multiplex PCR assay developed in the present study can rapidly and efficiently detect both the Theileria species, i.e., T. annulata and T. orientalis. The assay is highly specific and sensitive. Thus, it may be useful in epidemiological studies.
Data availability
Data are available from the authors upon reasonable request; sequences are available via GenBank.
References
Abdela N, Bekele T (2016) Bovine Theileriosis and its control: a review. Adv Biol Res (Rennes) 10:200–212. https://doi.org/10.5829/idosi.abr.2016.10.4.103107
Acharya AP, Panda SK, Prusty BK et al (2017) Diagnosis and confirmation of Theileria annulata infection in cattle in Odisha, India. J Entomol Zool Stud 5:1543–1546
Aparna M, Ravindran R, Vimalkumar MB et al (2011) Molecular characterization of Theileria orientalis causing fatal infection in crossbred adult bovines of South India. Parasitol Int 60:524–529. https://doi.org/10.1016/j.parint.2011.08.002
Ashuma SA, Kaur P et al (2014) Application of multiplex PCR for the simultaneous detection of natural infection of theileriosis, babesiosis and trypanosomosis in cattle. J Vet Parasitol 28:112–116
Awad H, Gadalla AAH, Postigo M et al (2020) Dynamics and within-host interaction of Theileria lestoquardi and T. ovis among naive sheep in Oman. Sci Rep 10:1–9. https://doi.org/10.1038/s41598-020-76844-2
Bashey F (2015) Within-host competitive interactions as a mechanism for the maintenance of parasite diversity. Philos Trans R Soc Lond B Biol Sci 370:20140301. https://doi.org/10.1098/rstb.2014.0301
Bilgiç HB, Karagenç T, Simuunza M et al (2013) Development of a multiplex PCR assay for simultaneous detection of Theileria annulata, Babesia bovis and Anaplasma marginale in cattle. Exp Parasitol 133:222–229. https://doi.org/10.1016/j.exppara.2012.11.005
Charaya G, Rakha NK, Maan S et al (2016) Comparative evaluation of polymerase chain reaction assay with microscopy for detection of asymptomatic carrier state of theileriosis in a herd of crossbred cattle. Vet World 9:1039–1042. https://doi.org/10.14202/vetworld.2016.1039-1042
Charaya G, Rakha NK, Kumar A et al (2021) End point multiplex PCR for diagnosis of haemoprotozoan diseases in cattle. Acta Parasitol 66:91–97. https://doi.org/10.1007/s11686-020-00259-2
Chauhan HC, Patel BK, Bhagat AG et al (2015) Comparison of molecular and microscopic technique for detection of Theileria annulata from the field cases of cattle. Vet World 8:1370–1374. https://doi.org/10.14202/VETWORLD.2015.1370-1374
Choi KS, Yu DH, Chae JS et al (2016) Seasonal changes in hemograms and Theileria orientalis infection rates among Holstein cattle pastured in the mountains in the Republic of Korea. Prev Vet Med 127:77–83. https://doi.org/10.1016/j.prevetmed.2016.03.018
El Damaty HM, Yousef SG, Mahmmod YS et al (2021) Sensitivity and specificity of piroplasm indirect fluorescent antibody test and PCR for Theileria annulata infection in clinically asymptomatic large ruminants using Bayesian latent class analysis. Veterinary Parasitology: Regional Studies and Reports 24100563. https://doi.org/10.1016/j.vprsr.2021.100563
Erdemir A, Aktas M, Dumanli N, Turgut-Balik D (2007) Isolation, cloning and sequence analysis of the lactate dehydrogenase gene from Theileria annulata may lead to design of new antitheilerial drugs. Vet Med (Praha) 57:559–567. https://doi.org/10.17221/6368-VETMED
Fukushima Y, Horii Y, Honkawa K, Sasaki Y (2021) A large-scale survey of Theileria orientalis infection in grazing dairy heifers in Kyushu, Japan. J Vet Med Sci 83:36–41. https://doi.org/10.1292/JVMS.20-0567
Ganguly A, Maharana BR, Ganguly I (2020) Pentaplex PCR assay for rapid differential detection of Babesia bigemina, Theileria annulata, Anaplasma marginale and Trypanosoma evansi in cattle. Biologicals 63:81–88. https://doi.org/10.1016/j.biologicals.2019.10.011
Gebrekidan H, Perera PK, Ghafar A et al (2020) An appraisal of oriental theileriosis and the Theileria orientalis complex, with an emphasis on diagnosis and genetic characterisation. Parasitol Res 119:11–22. https://doi.org/10.1007/s00436-019-06557-7
Junlong L, Li Y, Liu A et al (2015) Development of a multiplex PCR assay for detection and discrimination of Theileria annulata and Theileria sergenti in cattle. Parasitol Res 114:2715–2721. https://doi.org/10.1007/s00436-015-4478-z
Kakati P, Sarmah PC, Ray D et al (2015) Emergence of oriental theileriosis in cattle and its transmission through Rhipicephalus (Boophilus) microplus in Assam, India. Vet World 8:1099–1104. https://doi.org/10.14202/vetworld.2015.1099-1104
Kamau J, de Vos AJ, Playford M et al (2011) Emergence of new types of Theileria orientalis in Australian cattle and possible cause of Theileriosis outbreaks. Parasit Vectors 4:22. https://doi.org/10.1186/1756-3305-4-22
Khatoon S, Kolte SW, Kurkure NV et al (2015) Detection of tropical bovine theileriosis by polymerase chain reaction in cattle. J Parasit Dis 39:53–56. https://doi.org/10.1007/s12639-013-0270-0
Kirvar E, Ilhan T, Katzer F et al (2000) Detection of Theileria annulata in cattle and vector ticks by PCR using the Tams1 gene sequences. Parasitology 120:245–254. https://doi.org/10.1017/S0031182099005466
Kolte SW, Larcombe SD, Jadhao SG et al (2017) PCR diagnosis of tick-borne pathogens in Maharashtra state, India indicates fitness cost associated with carrier infections is greater for crossbreed than native cattle breeds. PLoS One 12:e0174595. https://doi.org/10.1371/journal.pone.0174595
Krishnamoorthy P, Akshata LG, Jacob SS et al (2021) Theileriosis prevalence status in cattle and buffaloes in India established by systematic review and meta-analysis. Indian J Anim Sci 91:269–279
Kundave VR, Patel AK, Patel PV et al (2015) Detection of theileriosis in cattle and buffaloes by polymerase chain reaction. J Parasit Dis 39:508–513. https://doi.org/10.1007/s12639-013-0386-2
Kundave VR, Ram H, Banerjee PS et al (2018) Development of multiplex PCR assay for concurrent detection of tick borne haemoparasitic infections in bovines. Acta Parasitol 63:759–765. https://doi.org/10.1515/ap-2018-0090
Li Y, Liu Z, Liu J et al (2016) Seroprevalence of bovine theileriosis in northern China. Parasit Vectors 9:591–597. https://doi.org/10.1186/s13071-016-1882-x
Liu A, Guan G, Liu Z et al (2010) Detecting and differentiating Theileria sergenti and Theileria sinensis in cattle and yaks by PCR based on major piroplasm surface protein (MPSP). Exp Parasitol 126:476–481. https://doi.org/10.1016/j.exppara.2010.05.024
Mans BJ, Pienaar R, Latif AA (2015) A review of Theileria diagnostics and epidemiology. Int J Parasitol Parasites Wildl 4:104–118. https://doi.org/10.1016/j.ijppaw.2014.12.006
McFadden AMJ, Rawdon TG, Meyer J et al (2011) An outbreak of haemolytic anaemia associated with infection of Theileria orientalis in naïve cattle. N Z Vet J 59:79–85. https://doi.org/10.1080/00480169.2011.552857
Medlock JM, Hansford KM, Bormane A et al (2013) Driving forces for changes in geographical distribution of Ixodes ricinus ticks in Europe. Parasit Vectors 6:1–11
Narladkar BW (2018) Projected economic losses due to vector and vector-borne parasitic diseases in livestock of india and its significance in implementing the concept of integrated practices for vector management. Vet World 11:151–160. https://doi.org/10.14202/vetworld.2018.151-160
Nayel M, El-Dakhly KM, Aboulaila M et al (2012) The use of different diagnostic tools for babesia and theileria parasites in cattle in Menofia. Egypt Parasitol Res 111:1019–1024. https://doi.org/10.1007/s00436-012-2926-6
Nourollahi-Fard SR, Khalili M, Ghalekhani N (2015) Detection of Theileria annulata in blood samples of native cattle by PCR and smear method in Southeast of Iran. J Parasit Dis 39:249–252. https://doi.org/10.1007/s12639-013-0333-2
Orkun O, Deniz A, Guven E (2012) Survey of Theileria annulata and Theileria buffeli/orientalis complex in cattle in the Kırşehir region using multiplex-PCR. Turkish J Parasitol 36:9–11. https://doi.org/10.5152/tpd.20112.03
Ota N, Minzuno D, Kuboki N et al (2009) Epidemiological survey of theileria orientalis infection in grazing cattle in the Eastern Part of Hokkaido, Japan. J Vet Med Sci 71:937–944. https://doi.org/10.1292/jvms.71.937
Patial V, Gupta T, Angaria S et al (2021) Theileria orientalis outbreak in an organized cattle breeding farm. Vet Parasitol Reg Stud Reports 24:100572. https://doi.org/10.1016/j.vprsr.2021.100572
Perera PK, Gasser RB, Firestone SM et al (2014) Oriental theileriosis in dairy cows causes a significant milk production loss. Parasit Vectors 7:73. https://doi.org/10.1186/1756-3305-7-73
Ranjan K, Prasad M, Prasad G (2015) Application of molecular and serological diagnostics in veterinary parasitology. J Adv Parasitol 2:80–99. https://doi.org/10.14737/journal.jap/2015/2.4.80.99
Rashid M, Rashid MI, Akbar H et al (2019) A systematic review on modelling approaches for economic losses studies caused by parasites and their associated diseases in cattle. Parasitology 146:129–141. https://doi.org/10.1017/S0031182018001282
Renneker S, Abdo J, Ahmed JS, Seitzer U (2009) Field validation of a competitive ELISA for detection of Theileria annulata infection. Parasitol Res 106:47–53. https://doi.org/10.1007/S00436-009-1625-4
Rojas-Downing MM, Nejadhashemi AP, Harrigan T, Woznicki SA (2017) Climate change and livestock: impacts, adaptation, and mitigation. Clim Risk Manag 16:145–163
Singh K, Kumar S, Sharma AK et al (2022) Economic impact of predominant ticks and tick-borne diseases on Indian dairy production systems. Exp Parasitol 243:108408. https://doi.org/10.1016/j.exppara.2022.108408
Thompson AT, White S, Shaw D et al (2020) Theileria orientalis Ikeda in host-seeking Haemaphysalis longicornis in Virginia, U S A. Ticks Tick Borne Dis 11:101450. https://doi.org/10.1016/j.ttbdis.2020.101450
Tuli A, Das Singla L, Sharma A et al (2015) Molecular epidemiology, risk factors and hematochemical alterations induced by Theileria annulata in bovines of Punjab (India). Acta Parasitol 60:378–390. https://doi.org/10.1515/ap-2015-0053
Acknowledgements
The authors are thankful to Department of Veterinary Medicine, LLR University of Veterinary and Animal Sciences, Hisar, for providing financial support, and to the Department of Animal Biotechnology, LLR University of Veterinary and Animal Sciences, Hisar, for providing the infrastructural facility.
Author information
Authors and Affiliations
Contributions
Sumnil Marwaha performed the experiments, analyzed the data, prepared the figures and tables, and authored and reviewed the drafts of the paper. Basanti Brar assisted in conceptual discussions and revision of the manuscript. Vinod Kumar Jain and Minakshi Prasad conceived and designed the experiments, and reviewed and edited the manuscript. Rachna Poonia assisted in sampling and revision of manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics of approval
In this study, blood was collected from the animals by the veterinarians as per the procedure in compliance with the ethical standards and guidelines of the Institutional Animal Ethics Committee (IAEC), LUVAS, Hisar, India, and permission was obtained via. Order no. VCC/IEAC/2625–2647/21–12-19.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Section Editor: Leonhard Schnittger
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Marwaha, S., Brar, B., Jain, V.K. et al. Multiplex PCR for rapid differential diagnosis of co-prevalent species of Theileria (Theileria annulata and Theileria orientalis) in cattle. Parasitol Res 122, 1189–1197 (2023). https://doi.org/10.1007/s00436-023-07819-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-023-07819-1