Abstract
Multiple Kudoa spp. (Myxozoa: Myxosporea: Multivalvulida) have been recorded in Japanese parrotfish (Calotomus japonicus) from the Philippine Sea (Northwest Pacific Ocean), off southwestern Japan; Kudoa yasunagai in the brain, and K. igami, K. lateolabracis, and K. thalassomi in the muscles. This study examined eight Philippine Sea Japanese parrotfish samples collected in January and February 2019 and found K. prunusi in the brain (3–57 plasmodia/fish; average 17.9) and K. lateolabracis plasmodia in the trunk muscle of all fish individuals examined. The K. prunusi in this study was characterized by myxospores predominatetly with six shell valves (SVs) and a corresponding number of polar capsules (PCs), contrasting with the original description of the species from farmed Pacific bluefin tuna (Thunnus orientalis) brain that characterized the species as having predominately five SVs/PCs. Molecular-genetic characterization of 18S and 28S ribosomal RNA genes and mitochondrial DNA genes (cytochrome c oxidase subunit 1 and small and large ribosomal RNA subunits) clearly differentiated the K. prunusi isolate from K. yasunagai, commonly characterized by six or seven, but rarely five, SVs/PCs myxospores. The Japanese parrotfish is a new host record for K. prunusi and speculated to be an important reservoir host in its natural waters. Kudoa lateolabracis myxospores isolated from pseudocysts in the myofiber were morphologically and phylogenetically close to a clade of the Kudoa spp. that exhibit cruciform myxospores similar to K. thyrsites. This study is the first to sequence a mitochondrial DNA of small and large subunit ribosomal RNA of K. lateolabracis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Multiple Kudoa spp. (Cnidaria: Myxozoa: Myxosporea: Multivalvulida) have been recorded in the Japanese parrotfish Calotomus japonicus (Valenciennes, 1840) from the Philippine Sea (Northwest Pacific Ocean), off southwestern Japan. Kudoa yasunagai (Hsieh et Chen, 1984) was recorded in the brain, and K. igami Shirakashi, Yamane, Ishitani, Yanagida et Yokoyama, 2014, K. lateolabracis Yokoyama, Whipps, Kent, Mizuno et Kawakami, 2004, and K. thalassomi Adlard, Bryant, Whipps et Kent, 2005 were recorded in the trunk muscle (Shirakashi et al. 2014; Sakai et al. 2019).
Kudoa igami was originally characterized by stellate myxospores with five or six shell valves (SVs) and a corresponding number of polar capsules (PCs), forming pseudocysts in the myofiber of the Japanese parrotfish (Shirakashi et al. 2014). Later, Shin et al. (2016) found the species in the trunk muscle of the olive flounder Paralichthys olivaceus (Temminck et Schlegel, 1846) farmed in the Philippine Sea, where the K. igami infections in Japanese parrotfish were recorded. More recently, Sakai et al. (2019) recorded the species in the trunk muscle of the Carolines parrotfish Calotomus carolinus (Valenciennes, 1840), African coris Coris gaimard (Quoy et Gaimard, 1824), and the pastel ring wrasse Hologymnosus doliatus (Lacepède, 1801) from the border of the Philippine and East China Seas, off Miyako Island, Okinawa, Japan. Sakai et al. (2019) observed, however, seven to nine SVs/PCs in myxospores of their K. igami isolates, in contrast to five or six SVs/PCs previously reported by Shirakashi et al. (2014) and Shin et al. (2016). Morphological variations in the SV and PC numbers in myxospores from the same or different plasmodia are also established among other Kudoa spp. characterized by myxospores with more than four SVs/PCs: e.g., K. yasunagai (Egusa 1986; Burger and Adlard 2010b; Miller and Adlard 2012; Sakai et al. 2019); K. septempunctata Matsukane, Sato, Tanaka, Kamata et Sugita-Konishi, 2010 (Matsukane et al. 2010; Kasai et al. 2016b; Yokoyama et al. 2017); K. thalassomi (Burger and Adlard 2011; Shirakashi et al. 2014; Sakai et al. 2019); K. neothunni (Arai et Matsumoto, 1953) (Kasai et al. 2017b); K. chaetodoni Burger, Cribb et Adlard, 2007 (Burger et al. 2007; Miller and Adlard 2012); K. lemniscati (Miller and Adlard 2012); and K. miyakoensis Sakai, Kawai, Zhang et Sato, 2019 (Sakai et al. 2019).
Postharvest myoliquefaction caused by multivalvulidan infection in commercially important fish has a significant economic impact on natural water marine fisheries and aquaculture (Egusa 1986; Moran et al. 1999). The known causative species of this phenomenon include Unicapsula seriolae Lester 1982, K. thyrsites (Gilchrist, 1924), K. lateolabracis, K. megacapsula Yokoyama et Itoh, 2005, K. musculoliquefaciens (Matsumoto 1954), K. neothunni, K. paniformis Kabata et Whitaker, 1981, K. pleurogrammi Kasai, Li, Mafie et Sato, 2016, K. rosenbuschi (Gelormini, 1944), and K. aburakarei Li, Inoue, Tanaka, Zhang et Sato, 2020 (Matsumoto 1954; Kabata and Whitaker 1981; Lester 1982; Moran et al. 1999; Yokoyama et al. 2004, 2006; Yokoyama and Itoh 2005; Whipps and Kent 2006; Li et al. 2013, 2020b; Kasai et al. 2016a). The parasitism of these species is not directly related to postharvest myoliquefaction, as enzymatic digestion of host muscle can be affected by multiple factors (Konagaya 1984; Dawson-Coates et al. 2003; Funk et al. 2007, 2008; Zhou and Li-Chan 2009). Originally, K. lateolabracis was described as a cause of postharvest myoliquefaction in Chinese sea bass Lateolabrax sp. cultured in western Japan (Yokoyama et al. 2004). However, all Japanese parrotfish parasitized with K. lateolabracis did not exhibit this phenomenon when Shirakashi et al. (2014) examined 17 infected fish samples.
In this study, we examined eight Japanese parrotfish specimens fished in the Philippine Sea, distant from the previous study on Japanese parrotfish kudoid infection (Shirakashi et al. 2014), and found two Kudoa spp. in the brain and trunk muscles. We attempted to identify the isolated kudoid species based on morphological criteria and molecular-genetic characterization.
Materials and methods
Fish samples and parasitological examination
Four whole Japanese parrotfish each were purchased on January 10 and February 4, 2019, from a local fish market in Kochi, southwestern Japan. They were fished in the Philippine Sea (Northwest Pacific Ocean), off Kochi, Japan. The samples were transported on ice to the laboratory in Yamaguchi University within 1 day of purchase. Parasitological examinations of the fish samples were performed as previously described (Inoue et al. 2021). Briefly, the gills, viscera, and brain were removed and examined under a dissection microscope. Thin slices of the trunk muscle were placed in physiological saline, pressed between glass plates, and examined under a dissection microscope.
Fresh myxospores were measured according to Lom and Arthur (1989). All measurements are expressed in µm unless otherwise stated, and the ranges are presented with the means in parentheses. The tissue-embedded myxosporean plasmodia were divided into two groups and fixed in either 10% neutral-buffered formalin solution or 70% ethanol for further analyses. The specimens in this study were deposited in the Meguro Parasitological Museum, Tokyo, Japan, under collection numbers 21774 and 21775.
DNA extraction, amplification, and sequencing
Parasite DNA was extracted from the myxosporean plasmodia frozen after isolation at − 20 °C. The methods for DNA extraction, amplification of rDNA fragments by polymerase chain reaction (PCR), and purification of the PCR products were performed as previously described (Li et al. 2013; Kasai et al. 2015). Further molecular-genetic characterization of kudoid isolates was conducted on the mitochondrial DNA (mtDNA), i.e., cytochrome c oxidase subunit 1 (cox-1) and small and large ribosomal RNA subunits (rns-rnl), according to our previous study (Sakai et al. 2018). When direct sequencing results were not satisfactory, purified PCR products were cloned into the pTA2 plasmid vector (TArget Clone™; TOYOBO, Dojima Hama, Osaka, Japan) according to the manufacturer’s instructions. Following propagation, the plasmid DNA was extracted using a FastGene Plasmid Mini Kit (NIPPON Genetics Co., Tokyo, Japan), and inserts from multiple independent clones, at least three, were sequenced using universal M13 forward and reverse primers. The nucleotide sequences obtained in this study are available from the DDBJ/EMBL/GenBank databases under accession numbers LC640102–LC640108.
Phylogenetic analyses
Fragments of the newly obtained rDNA sequences were analyzed to identify highly similar nucleotide sequences using the Basic Local Alignment Search Tool (BLAST) of the National Center for Biotechnology Information website (NCBI; https://www.ncbi.nlm.nih.gov/). For phylogenetic analysis, the newly obtained 18S and 28S rDNA sequences of Kudoa spp. from this study and related sequences retrieved from the GenBank database (NCBI) were aligned using the MEGA7 software (Kumar et al. 2016), with subsequent manual adjustments. The accession numbers of the analyzed sequences are provided in the figures of the phylogenetic trees. Regions judged to be poorly aligned and characters with a gap in any sequences were excluded from subsequent analyses; 1415 characters, of which 194 were variable, remained for subsequent analysis of the 18S rDNA, and 591 characters, of which 216 were variable, remained for subsequent analysis of the 28S rDNA. Similarly, 437 characters, of which 172 were variable, remained for subsequent analysis of the cox-1 mtDNA, and 961 characters, of which 471 were variable, remained for subsequent analysis of the rns-rnl mtDNA. Maximum likelihood (ML) analysis was performed with the program, PhyML (Guindon and Gascuel 2003; Dereeper et al. 2008) provided on the “phylogeny.fr” website (http://www.phylogeny.fr/). The probability of inferred branching was assessed by the approximate likelihood-ratio test (aLRT), an alternative to the non-parametric bootstrap estimation of branch support (Anisimova and Gascuel 2006). Outlier Kudoa spp. forming cysts, such as K. bora (Fujita, 1930), K. iwatai Egusa et Schiomitsu, 1983, and K. lutjanus Wang, Huang, Tsai, Cheng, Tsai, Chen, Chen, Chu, Liaw, Chang et Chen, 2005, to a majority of Kudoa spp. were used as an outgroup for construction of the ML phylogenetic trees.
Results
Parasitological examination
Two batches of the Japanese parrotfish samples, with four samples in each batch, were purchased in January and February 2019, and kudoid plasmodia were found in the cranial cavity and trunk muscles of all eight samples. Myxospores isolated from plasmodia located in different organs exhibited distinct morphology.
Kudoa prunusi Meng, Yokoyama, Shirakashi, Grabner, Ogawa, Ishimaru, Sawada et Murata, 2011
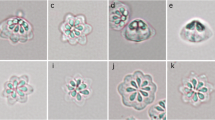
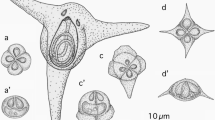
Kudoa prunusi was identified as round to oval plasmodia floating in the subarachnoid space (Fig. 1), measuring 0.22–1.93 (0.73) mm by 0.21–1.65 (0.64) mm (n = 80). There were 3–57 (17.9) plasmodia of varying dimensions per fish. The myxospores were stellate in apical view, with six or five SVs and a corresponding number of PCs (Fig. 2). The ratio of myxospores with six SVs/PCs to those with five SVs/PCs was 75%:25% (n = 40). The lateral view of the myxospores was rounded-pyramidal. The dimensions of the K. prunusi myxospores isolated in this study are shown in Table 1.
BLAST searches using the 18S and partial 28S rDNA nucleotide sequences, 1719-bp and 800-bp in length, respectively, of the K. prunusi isolate (DDBJ/EMBL/GenBank accession no. LC640106) showed the highest nucleotide identity with an isolate of a K. prunusi isolate from the brain of cultured Thunnus orientalis (Temminck et Schlegel, 1844) in Wakayama, western Japan, followed by various Kudoa spp. isolated from brain tissue (Table 2). Two partial fragments of K. prunusi mitochondrial genes, cox-1 and rns-rnl, were sequenced for the first time (LC640102–LC640104). The cox-1 sequences of the two K. prunusi isolates exhibited a high affinity with K. yasunagai (LC382003), exhibiting 91.08% (398/437) and 91.30% (399/437) identity, followed by K. miyakoensis (LC3820004) with 82.84% (362/437) and 83.07% (363/437) identity.
Remarks
Kudoa prunusi was originally described from the brain of juvenile Pacific bluefin tunas cultured in Wakayama Prefecture, Japan, at the edge of the Philippine Sea (Meng et al. 2011). The myxospores of the species were characterized as penta-radiate in apical view, with five (rarely six) SVs/PCs per myxospore. The ratio of these two morphotypes in a plasmodium was 80:20. As in the original description (Meng et al. 2011), we also found two myxospore morphotypes in the same plasmodium in the present isolate; however, the ratio was approximately opposite, 25:75 for five and six SVs/PCs, respectively. The morphometric values of the present isolate were comparable with those of K. prunusi, K. yasunagai, and K. neurophila (Table 1). The nucleotide sequences of the 18S and 28S rDNA of the present isolate identified it as K. prunusi. The K. prunusi parasitism in Japanese parrotfish brain tissue established a new host record and expanded its geographical distribution to the open sea area of the Philippine Sea.
Taxonomic summary
Host: Calotomus japonicus (Valenciennes, 1840): Japanese parrotfish (Actinopteryi: Eupercaria/misc: Scaridaedae: Sparisomatinae).
Locality: Philippine Sea (Northwest Pacific Ocean), off Kochi, western Japan.
Site of infection: Subarachnoid space.
Materials deposited: Specimen no. 21775, Meguro Parasitological Museum, Tokyo, Japan.
Deposited rDNA sequences: DDBJ/EMBL/GenBank accession nos. LC640106 (rDNA) and LC640102–LC640104 (mtDNA).
Prevalence: 100% (8/8).
Kudoa lateolabracis Yokoyama, Whipps, Kent, Mizuno et Kawakami, 2004
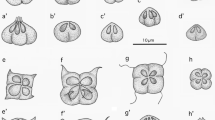
Kudoa lateolabracis was frequently found in the trunk muscle of all eight Japanese parrotfish examined in this study. The plasmodia, developed in pseudocysts in the myofibers, measured 0.98–3.79 (2.01) mm by 0.09–0.33 (0.16) mm. The myxospores were cruciform in apical view, with four SVs and a corresponding number of PCs. The sizes of the SVs and pyriform PCs were unequal, with one large PC/SV opposite one small PC/SV and two intermediate PCs/SVs between the former two (Fig. 3). The lateral view of the myxospores was asymmetric-pyramidal. The myxospore dimensions of K. lateolabracis isolated in this study are shown in Table 3.
BLAST searches using 18S and partial 28S rDNA nucleotide sequences, 1721-bp and 800-bp in length, respectively, of the present K. lateolabracis isolate (LC640107 and LC640108), showed an absolute or close to absolute similarity to K. lateolabracis from Lateolabrax sp. (AY382606) and Calotomus japonicus (AB844442). A partial fragment of K. lateolabracis mitochondrial rns-rnl was sequenced for the first time (LC640105).
Remarks
Kudoa lateolabracis was originally described from the liquefied muscle of Chinese sea bass farmed in the Inland Sea of Japan, off Ehime Prefecture (Yokoyama et al. 2004), from fish seeds imported from China. Additional records of the species came from the muscle of the Japanese parrotfish at the edge of the Philippine Sea, off Wakayama, Japan (Shirakashi et al. 2014), and the olive flounder farmed in Wakayama (Shin et al. 2016). The two previous reports suggested that K. lateolabracis might be endemic in the seawater around western Japan, and farmed Chinese sea bass might be infected in the farming sea, the Inland Sea of Japan. The current record of K. lateolabracis in Japanese parrotfish from the Philippine Sea, off Kochi, further supports the hypothesis that K. lateolabracis is an endemic kudoid species in the sea around southwestern Japan.
Taxonomic summary
Host: Calotomus japonicus (Valenciennes, 1840): Japanese parrotfish (Actinopteryi: Eupercaria/misc: Scaridaedae: Sparisomatinae).
Locality: Philippine Sea (Northwest Pacific Ocean), off Kochi, western Japan.
Site of infection: Pseudocysts in the myofiber of trunk muscles.
Materials deposited: Specimen no. 21774, Meguro Parasitological Museum, Tokyo, Japan.
Deposited rDNA sequences: DDBJ/EMBL/GenBank accession nos. LC640107 (18S rDNA), LC640108 (28S rDNA), and LC640105 (mtDNA).
Prevalence: 100% (8/8).
Phylogenetic analyses
Phylogenetic trees based on the 18S and 28S rDNA of Kudoa spp. with cruciform myxospores (including K. lateolabracis) and Kudoa spp. with brain tropism (including K. prunusi) were constructed using the cyst-forming kudoid species (K. iwatai, K. lutjanus, and K. bora) as an outgroup. The two aforementioned kudoid groups formed separate robust clades (Figs. 4 and 5). Phylogenetically, K. prunusi, K. yasunagai, K. lemniscati, K. chaetodoni, and K. miyakoensis were closely related to each other, and K. neurophila (Grossel, Dyková, Handlinger et Munday, 2003) and K. lethrini Burger, Cribb et Adlard, 2007 were positioned relatively distantly at the root of the five Kudoa spp. Similarly, Kudoa spp. with cruciform myxospores, including K. lateolabracis, formed a robust clade in the 18S or 28S rDNA phylogenetic trees (Figs. 4 and 5). The topological position of K. lateolabracis in the tree differed using different rDNA regions.
Maximum likelihood phylogenetic tree of Kudoa spp. based on 28S rDNA sequences (591 characters). Labeling of each isolate is similar to Fig. 4 legend. New sequences from this study are marked with gray backgrounds
Phylogenetic trees based on the mitochondrial DNA genes cox-1 and rns-rnl were constructed using the newly obtained sequences of K. prunusi and K. lateolabracis (Fig. 6). The available kudoid species were limited to eight Kudoa spp. for cox-1 and seven for rns-rnl. The phylogenetic relationships between different species were approximately similar to phylogenetic relationships based on the rDNA regions.
Unrooted maximum likelihood phylogenetic trees based on partial mitochondrial gene sequences (A, cox-1 and B, rns-rnl) of representative Kudoa spp. with five or more SVs/PCs per myxospore, and K. lateolabracis with cruciform myxospores comprised four SVs and PCs. Labeling of each isolate is similar to Fig. 4. New sequences from this study are marked with gray backgrounds
Discussion
This study detected K. prunusi plasmodia in the brain and K. lateolabracis plasmodia in the trunk muscle of all eight Japanese parrotfish samples examined, which originated from the northwestern Philippine Sea, off southwestern Japan (Kochi). Kudoa prunusi was characterized by myxospores with predominately six SVs/PCs, in contrast to its original description from the brain of cultured Pacific bluefin tuna, in which the species was characterized as having predominately five SVs/PCs (Meng et al. 2011). Molecular-genetic characterization of the 18S and 28S rDNA and mitochondrial DNA genes (cox-1) clearly differentiated this K. prunusi isolate from K. yasunagai, characterized by a myxospore with six or seven, rarely five, SVs/PCs. Accordingly, the Japanese parrotfish has set a new host record for K. prunusi and is speculated as an important reservoir host of the species in its natural waters. Shirakashi et al. (2014) recorded K. yasunagai prevalence of 94.1% (16/17) in the brain of Japanese parrotfish fished in the Philippine Sea, off Wakayama. Furthermore, our previous study (Sakai et al. 2019) detected K. yasunagai in the brain of one of three Japanese parrotfish collected from the same area as this study.
As discussed by Meng et al. (2011), reliable species differentiation of K. prunusi from related species with brain tropism, such as K. yasunagai, K. miyakoensis, and K. chaetodoni, is not feasible due to the high morphological variations of myxospores, e.g., the number of SVs/PCs, overlapping measurements, and low molecular-genetic variations (few nucleotide substitutions) in the 18S and 28S rDNA (Shin et al. 2016; Sakai et al. 2019; Inoue et al. 2021). This study (Fig. 6) suggests the possibility of assessing cox-1 mtDNA sequencing as an alternative for specific identification. However, more isolates need to be sequenced to clarify the intra- and inter-specific variations in Kudoa spp. with brain tropism.
Shirakashi et al. (2014) reported K. lateolabracis plasmodia, which formed pseudocyst in the myofibers, in the trunk muscle of the Japanese parrotfish at a prevalence of 41.5% (17/41). Their report was the second host record for the species after its original description in the liquefied muscles of Chinese sea bass farmed in the Inland Sea of Japan, off Ehime (Yokoyama et al. 2004). As mentioned above, the common occurrence of K. lateolabracis in Japanese parrotfish in its natural waters, in the Philippine Sea, off southwestern Japan, indicates the endemicity of the species in the waters around Japan. In this study, a partial rns-rnl mtDNA sequence of K. lateolabracis characterized by cruciform myxospores with four SVs/PCs was obtained for the first time (Takeuchi et al. 2016; Sakai et al. 2018, 2019; Li et al. 2020a). It might be possible to use mtDNA genes to identify the geographical origin of an isolate, as postulated for K. septempunctata (Takeuchi et al. 2016; Yokoyama et al. 2017).
Several Kudoa spp. with cruciform myxospores have been differentiated from the well-known K. thyrsites (Gilchrist, 1924) in the last two decades using rDNA molecular-genetic characterization: e.g., K. mirabilis Naidenova et Gaevskaya, 1991; K. minithyrsites Whipps, Adlard, Bryant, Lester, Findlay et Kent, 2003; K. lateolabracis; K. whippsi Burger et Adlard, 2010; K. gunterae Burger et Adlard, 2010; K. cheilodipteri Heiniger, Cribb et Adlard, 2013; K. parathyrsites Kasai, Li, Mafie et Sato, 2016; K. akihitoi Kasai, Setsuda et Sato, 2017; and K. aburakarei Li, Inoue, Tanaka, Zhang et Sato, 2020 (Whipps et al. 2003; Yokoyama et al. 2004; Burger and Adlard 2010a; Heiniger et al. 2013; Kasai et al. 2016b, 2017a; Li et al. 2020b; Giulietti et al. 2020). For any kudoid species, reliable species identification and disclosure of substantial biodiversity in multivalvulidan myxosporeans could be achieved by integrated taxonomic approaches with morphological observation, intense molecular-genetic characterization, and ecological investigation (Atkinson et al. 2015). This study contributes to the understanding of the biogeography and epidemiological status of multivalvulidans, which is a particularly important issue for commercial edible fish farming and wild fishes living in aquaculture areas (Egusa and Nakajima 1980; Sugiyama et al. 1999; Burger et al. 2008).
References
Anisimova M, Gascuel O (2006) Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol 55:539–552
Atkinson SD, Bartošová-Sojková P, Whipps CM, Bartholomew JL (2015) Approaches for characterising myxozoan species. In: Okamura B, Gruhl A, Bartholomew JL (eds) Myxozoan evolution, ecology and development. Springer International Publishing, Switzerland, pp 111–123
Burger MAA, Adlard RD (2010a) Four new species of Kudoa Meglitsch, 1947 (Myxosporea: Multivalvulida) from Australia with recommendations for species descriptions in the Kudoidae. Parasitology 137:793–814
Burger MAA, Adlard RD (2010b) Phenotypic variation in a significant spore character in Kudoa (Myxosporea: Multivalvulida) species infecting brain tissue. Parasitology 137:1759–1772
Burger MAA, Adlard RD (2011) Low host specificity in the Kudoidae (Myxosporea: Multivalvulida) including seventeen new host records for Kudoa thalassomi. Folia Parasitol 58:1–16
Burger MAA, Cribb TH, Adlard RD (2007) Patterns of relatedness in the Kudoidae with descriptions of Kudoa chaetodoni n. sp. and K lethrini n. sp. (Myxosporea: Multivalvulida). Parasitology 134:669–681
Burger MAA, Barnes AC, Adlard RD (2008) Wildlife as reservoirs for parasites infecting commercial species: host specificity and a redescription of Kudoa amamiensis from teleost fish in Australia. J Fish Dis 31:835–844
Dawson-Coates JA, Chase JC, Funk V, Booy MH, Haines LR, Falkenberg CL, Whitaker DJ, Olafson RW, Pearson TW (2003) The relationship between flesh quality and numbers of Kudoa thyrsites plasmodia and spores in farmed Atlantic salmon, Salmo salar L. J Fish Dis 26:451–459
Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie J-M, Gascuel O (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:465–469
Egusa S (1986) The order Multivalvulida Shulman, 1959 (Myxozoa: Myxosporea): a review. Fish Pathol 21:261–274 ((in Japanese with English summary))
Egusa S, Nakajima K (1980) Kudoa amamiensis n. sp. (Myxosporea: Multivalvulida) found in cultured yellowtails and wild damselfishes from Amami-Ohshima and Okinawa, Japan. Bull Japan Soc Sci Fish 46:1193–1198
Funk VA, Raap M, Sojonky K, Jones S, Robinson J, Falkenberg C, Miller KM (2007) Development and validation of an RNA- and DNA-based quantitative PCR assay for determination of Kudoa thyrsites infection levels in Atlantic salmon Salmo salar. Dis Aquat Org 75:239–249
Funk VA, Olafson RW, Raap M, Smith D, Aitken L, Haddow JD, Wang D, Dawson-Coates JA, Burke RD, Miller KM (2008) Identification, characterization and deduced amino acid sequence of the dominant protease from Kudoa paniformis and K. thyrsites; a unique cytoplasmic cysterine protease. Comp Biochem Physiol B 149:477–489
Giulietti L, Karlsbakk E, Cipriani P, Shayo SD, Storesund JE, Levsen A (2020) Molecular characterization of the myoliquefactive fish parasite Kudoa mirabilis (Cnidaria, Kudoidae) from SW Indian Ocean and its phylogenetic relationship with the Kudoa thyrsites species complex. Microorganisms 8:1352. https://doi.org/10.3390/microorganisms8091352
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Heiniger H, Cribb TH, Adlard RD (2013) Intra-specific variation of Kudoa spp. (Myxosporea: Multivalvulida) from apogonid fishes (Perciformes), including the description of two new species, K. cheilodipteri n. sp. and K. cookii n. sp., from Australian waters. Syst Parasitol 84:193–215
Inoue K, Li Y-C, Ghosh S, Yunus M, Zhang J-Y, Sato H (2021) Identification of a new species, Unicapsula aequilobata n. sp., and Unicapsula seriolae (Myxozoa: Myxosporea: Multivalvulida) in carangid fish from the South China Sea. Parasitol Res 120:2379–2389
Kabata Z, Whitaker DJ (1981) Two species of Kudoa (Myxosporea: Multivalvulida) parasitic in the flesh of Merluccius productus (Ayres, 1855) (Pisces: Teleostei) in the Canadian Pacific. Can J Zool 59:2085–2091
Kasai A, Li Y-C, Setsuda A, Mafie E, Sato H (2015) Genetic characterization of Kudoa iwatai and Kudoa trachuri in commercial marine fish (Platycephalus sp. and Trachurus japonicus) for human consumption. Jpn J Vet Parasitol 14:22–30
Kasai A, Li Y-C, Mafie E, Sato H (2016a) Morphological and molecular genetic characterization of two Kudoa spp., K. musculoliquefaciens, and K. pleurogrammi n. sp. (Myxosporea: Multivalvulida), causing myoliquefaction of commercial marine fish. Parasitol Res 115:1883–1892
Kasai A, Li Y-C, Mafie E, Sato H (2016b) New host records of monacanthid fish for three Kudoa spp. (K. septempunctata, K. thyrsites, and K. shiomitsui) prevalent in the olive flounder (Paralichthys olivaceus), with the description of K. parathyrsites n. sp. from a black scraper (Thamnaconus modestus). Parasitol Res 115:2741–2755
Kasai A, Setsuda A, Sato H (2017a) Morphological and genetic characterization of Kudoa whippsi (Myxosporea: Multivalvulida) from Cheilodactylus zonatus in the western Pacific Ocean off Japan, and two new Kudoa spp. (K. akihitoi n. sp. and K. empressmichikoae n. sp.) from Acanthogobius hasta in the Sea of Ariake. Japan Parasitol Res 116:647–659
Kasai A, Tsuduki H, Jimenez LA, Li Y-C, Tanaka S, Sato H (2017b) Incidence of three Kudoa spp., K. neothunni, K. hexapunctata, and K. thunni (Myxosporea: Multivalvulida), in Thunnus tunas distributed in the western Pacific Ocean. Parasitol Res 116:1137–1150
Konagaya S (1984) Studies on the jellied meat of fish, with special reference to that of yellowfin tuna. Bull Tokai Reg Fish Res Lab 114:1–101 ((in Japanese with English summary))
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Lester RJG (1982) Unicapsula seriolae n. sp. (Myxosporea, Multivalvulida) from Australian yellowtail Kingfish Seriola lalandi. J Protozool 29:584–587
Li Y-C, Sato H, Tanaka S, Ohnishi T, Kamata Y, Sugita-Konishi Y (2013) Characterization of the ribosomal RNA gene of Kudoa neothunni (Myxosporea: Multivalvulida) in tunas (Thunnus spp.) and Kudoa scomberi n. sp. in a chub mackerel (Scomber japonicus). Parasitol Res 112:1991–2003
Li Y-C, Tamemasa S, Zhang J-Y, Sato H (2020a) Phylogenetic relationships of three Kudoa spp. with morphologically similar myxospores (K. iwatai, K. lutjanus, and K. bora), with the redescription of K. uncinata and K. petala and description of a new species (K. fujitai n. sp.) in fishes in the South China Sea. Parasitol Res 119:1221–1236
Li Y-C, Inoue K, Tanaka S, Zhang J-Y, Sato H (2020b) Identification of four new Kudoa spp. (Myxozoa: Myxosporea: Multivalvulida) in commercial fishes collected from South China Sea, Atlantic Ocean, and Bering Sea by integrated taxonomic approach. Parasitol Res 119:2113–2128
Lom J, Arthur JR (1989) A guideline for the preparation of species descriptions in Myxosporea. J Fish Dis 12:151–156
Matsukane Y, Sato H, Tanaka S, Kamata Y, Sugita-Konishi Y (2010) Kudoa septempunctata n. sp. (Myxosporea: Multivalvulida) from an aquacultured olive flounder (Paralichthys olivaceus) imported from Korea. Parasitol Res 107:865–872
Matsumoto K (1954) On the two new Myxospridia, Chloromyxum musculoliquefaciens sp. nov. and Neochloromyxum cruciformum gen. et sp. nov., from the jellied muscle of swordfish, Xiphias glandius Linné, and Lateolabrax japonicus (Temmink et Schlegel). Bull Japan Soc Sci Fish 20:469–478
Meng F, Yokoyama H, Shirakashi S, Grabner D, Ogawa K, Ishimaru K, Sawada Y, Murata O (2011) Kudoa prunusi n. sp. (Myxozoa: Myxosporea) from the brain of Pacific bluefin tuna Thunnus orientalis (Temminck & Schlegel, 1844) cultured in Japan. Parasitol Int 60:90–96
Miller TL, Adlard RD (2012) Brain infecting kudoids of Australia’s coral reefs, including a description of Kudoa lemniscati n. sp. (Myxosporea: Kudoidae) from Lutjanus lemniscatus (Perciformes: Lutjanidae) off Ningaloo Reef, Western Australia. Parasitol Int 61:333–342
Moran JDW, Whitaker DJ, Kent ML (1999) A review of the myxosporean genus Kudoa Meglitsch, 1947, and its impact on the international aquaculture industry and commercial fisheries. Aquaculture 172:163–196
Sakai H, Kato E, Sakaguchi S, Setsuda A, Sato H (2018) Morphological and molecular genetic characterization of Kudoa konishiae n. sp. (Myxosporea: Multivalvulida) in the muscle of Japanese Spanish mackerel (Scomberomorus niphonius). Parasitol Res 117:893–904
Sakai H, Kawai T, Zhang J, Sato H (2019) New host records of three Kudoa spp. (K. yasunagai, K. thalassomi, and K. igami) with notable variation in the number of shell valves and polar capsules in spores. Parasitol Res 118:143–157
Shin SP, Ishitani H, Shirakashi S (2016) Development of a multiplex PCR to detect Kudoa spp. and to distinguish Kudoa septempunctata in olive flounder Paralichthys olivaceus. Aquaculture 464:37–41
Shirakashi S, Yamane IH, Yanagida T, Yokoyama H (2014) First report of Kudoa species in the somatic muscle of the Japanese parrotfish Calotomus japonicus (Scaridae) and a description of Kudoa igami, n. sp. (Myxozoa: Multivalvulida). Parasitol Res 113:2515–2524
Sugiyama A, Yokoyama H, Ogawa K (1999) Epizootiological investigation on kudoosis amami caused by Kudoa amamiensis (Multivalvulida: Myxozoa) in Okinawa Prefecture, Japan. Fish Pathol 34:39–43 ((in Japanese with English summary))
Takeuchi F, Ogasawara Y, Kato K, Sekizuka T, Nozaki T, Sugita-Konishi Y, Ohnishi T, Kuroda M (2016) Genetic variants of Kudoa septempunctata (Myxozoa: Multivalvulida), a flounder parasite causing foodborne disease. J Fish Dis 39:667–672
Whipps CM, Kent ML (2006) Phylogeography of the cosmopolitan marine parasite Kudoa thyrsites (Myxozoa: Myxosporea). J Eukaryot Microbiol 53:364–373
Whipps CM, Adlard RD, Bryant MS, Lester RJG, Findlay V, Kent ML (2003) First report of three Kudoa species from eastern Australia: Kudoa thyrsites from mahi mahi (Coryphaena hippurus), Kudoa amamiensis and Kudoa minithyrsites n. sp. from sweeper (Pempheris ypsilychnus). J Eukaryot Microbiol 50:215–219
Yokoyama H, Itoh N (2005) Two multivalvulid myxozoans causing postmortem myoliquefaction: Kudoa megacapsula n. sp. from red barracuda (Sphyraena pinguis) and Kudoa thyrsites from splendid alfonso (Beryx splendens). J Parasitol 91:1132–1137
Yokoyama H, Whipps CM, Kent ML, Mizuno K, Kawakami H (2004) Kudoa thyrsites from Japanese flounder and Kudoa lateolabracis n. sp. from Chinese Sea bass: causative myxozoans of post-mortem myoliquefaction. Fish Pathol 39:79–85
Yokoyama H, Yanagida T, Takemaru I (2006) The first record of Kudoa megacapsula (Myxozoa: Multivalvulida) from farmed yellowtail Seriola quinqueradiata originating from wild seedlings in south Korea. Fish Pathol 41:159–163
Yokoyama H, Mekata T, Satoh J, Nishioka T, Mori K (2017) Morphological and molecular comparisons between Japanese and Korean Isolates of Kudoa septempunctata (Myxozoa: Multivalvulida) in the olive flounder Paralichthys olivaceus. Fish Pathol 52:152–157
Zhou LS, Li-Chan ECY (2009) Effect of Kudoa spores, endogenous protease activity and frozen storage on cooked texture of minced Pacific hake (Merluccius productus). Food Chem 11:1076–1082
Funding
This study was partially supported by a Grant-in-Aid for Food Science and Research 2019 from The Toyo Suisan Foundation (HS) and the Japan Society for the Promotion of Science (JSPS) KAKENHI grant number 18K05995 (HS).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Section Editor: Christopher Whipps
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Inoue, K., Kasai, A., Rosyadi, I. et al. Occurrence of Kudoa prunusi and K. lateolabracis (Myxozoa: Myxosporea: Multivalvulida) in Philippine-Sea Japanese parrotfish (Calotomus japonicus). Parasitol Res 121, 601–612 (2022). https://doi.org/10.1007/s00436-021-07418-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-021-07418-y