Abstract
Cattle and buffaloes, popular protein sources worldwide, are intermediate hosts for several Sarcocystis species. These coccidian protozoans cause sarcocystosis resulting in subclinical and chronic infections in striated muscles by forming macrocysts or microcysts. In Thailand, Lao People’s Democratic Republic, and Cambodia, Sarcocystis species have been reported, but molecular identification has been lacking. This study investigates the prevalence of infection, histo-morphology, and molecular identification of Sarcocystis species in hearts of cattle and buffalo sold in local markets. A phylogenetic tree inferred from a portion of the 18S ribosomal (r) RNA gene was used to identify the genus and species of Sarcocystis. The mitochondrial cytochrome c oxidase subunit 1 (cox-1 gene) was sequenced to confirm the species of host tissue. In Thailand, Sarcocystis was detected in 66.7% (14/21) of samples. In Lao People’s Democratic Republic, 90% (9/10) of samples were infected and in Cambodia 100% (8/8). For the first time from these countries, we report Sarcocystis cruzi, Sarcocystis heydorni, and Sarcocystis levinei found in taurine cattle (Bos taurus) and water buffalo (Bubalus bubalis). Zoonotic protozoan transmission needs to be controlled by inspection activities by local health inspectors, and appropriate action is required at all points in the food chain by competent authorities to protect consumer health and prevent sarcocystosis in cattle and water buffaloes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sarcocystis is a foodborne zoonotic pathogen classified in the phylum Apicomplexa and can infect animals and humans (Dubey 2015; Fayer et al. 2015). These parasites require two hosts in a simple predator–prey relationship to complete their life cycle (Fayer 2004). The intermediate host is infected by ingesting sporocysts, and then merogony occurs in the endothelial cells. Merozoites are released into the blood circulation and develop into sarcocysts in the muscle cells (Fayer et al. 2015; Lindsay et al. 1995). Cattle are intermediate hosts of five Sarcocystis species (Sarcocystis cruzi, S. hirsuta, S. hominis, S. rommeli, and S. heydorni), with the first three of these causing disease (Decker Franco et al. 2018; Dubey 2015). Sarcocystis hominis (syn. S. bovihominis) and S. heydorni are zoonotic: Humans can act as definitive hosts of these species (Fayer et al. 2015; Dubey 2015). Human infections are generally asymptomatic, but intestinal symptoms, such as nausea, abdominal discomfort, watery diarrhea, and dizziness, can occur (Fayer 2004; Dubey 2015). Four species of Sarcocystis are known from water buffalo (Bubalus bubalis): S. fusiformis, S. buffalonis, S. levinei, and S. dubeyi (Dissanaike and Kan 1978; Claveria and Cruz 2000; Dubey et al. 2014; Gjerde et al. 2016).
Infected livestock, such as cattle, buffaloes, sheep, and horses, can experience fever, lethargy, poor growth, low feed use, reduced milk production, lameness, wool and hair loss, abortion, and death (Fayer et al. 2015). Sarcocysts in muscle samples can be detected by microscopy via histological sections or by transmission electron microscopy (TEM). Species are differentiated by morphological features of sarcocysts such as size, presence or absence of septa, and cyst-wall thickness (thick > 3 µm and thin < 1 µm). A problem for morphological identification is that fresh meat samples with a high parasite burden are required (Fayer 2004; Xue et al. 2019). Nowadays, molecular methods have become popular. These are more efficient and sensitive than TEM for species identification of Sarcocystis in meat samples (Xue et al. 2019). The 18S ribosomal (r) RNA gene (18S rRNA) is an appropriate genetic marker able to distinguish between the five known species infecting cattle (Xue et al. 2019).
In Thailand, Sarcocystis screening has been reported during meat inspection (Nateeworanart et al. 2004; Dokmaikaw et al. 2017); however, the results to date have been limited to only morphological identification: There have been no molecular studies in Thailand. Sarcocystis has never been reported from Lao People’s Democratic Republic or Cambodia.
This study aimed to investigate the prevalence, histo-morphology, molecular characterization, and genetic diversity of Sarcocystis in hearts of cattle and buffaloes sold as meat products in local markets in Thailand, Lao PDR, and Cambodia.
Materials and methods
Sample collection
Heart muscle samples were collected from retail meat booths at local markets in Thailand, Lao PDR, and Cambodia from June 2018 to Dec 2019. A total of 39 samples were examined using a modified muscle-squashing method (Latif et al. 1999). Briefly, about 1 g of tissue cut into small pieces, approximately 3–5 mm thick, was crushed firmly between two glass slides, washed with 0.85% normal saline, and examined under a stereomicroscope for the presence of sarcocysts. The sarcocyst-positive samples were preserved with RNAlater® (QIAGEN, Germantown, MD) and then kept at -70 °C for molecular analysis. Samples found to be negative by the squashing method were subjected to a squeezing process (Latif et al. 1999), and the resulting fluid was observed microscopically for the presence of bradyzoites. The presence of the bradyzoites, and also absence was collect using for molecular analysis. Each individual sample was tested using the molecular method.
Histological examination
For the morphological study, all tissue samples that were positive by the squashing method were fixed in 10% buffered formalin to pause autolysis. After fixation, each sample was trimmed as appropriate, pre-embedded to infiltrate tissue samples with paraffin and to replace water content of tissue with wax, followed by embedding. The paraffin blocks were stored at room temperature until sectioned on a microtome to yield 5-µm-thick sections. These were stained with hematoxylin and eosin. After staining, a coverslip was placed on the tissue section, mounting medium was added, and the section was microscopically examined at × 400 magnification (Slaoui and Fiette 2011).
DNA extraction and amplification
Genomic DNA was extracted from individual samples using a NucleoSpin® tissue kit (Macherey–Nagel GmbH & Co., Duren, Germany), according to the manufacturer’s instructions. Resultant DNA samples were stored at -20 °C until use. Polymerase chain reaction (PCR) was performed using a primer pair specific for the 18S rRNA gene of Sarcocystis (Table 1). Genus and species of host tissue was identified using sequences of mitochondrial DNA, amplified with primers (spanning tRNA-PRO and part of the control region) listed in Table 1. For both primer pairs, a 50 µL reaction volume contained 1 × FastStart High Fidelity Reaction Buffer with 1.8 mM MgCl2 (Roche Applied Science, Mannheim, Germany), 0.2 mM dNTP (Vivantis, Malaysia), 0.2 µM of each primer, 1.25 U of FastStart High Fidelity Enzyme Blend (Roche Applied Science), and 5 µL of DNA template. PCR was done using a SimpliAmp™ Thermal Cycler (Applied Biosystems (ABI), Singapore). The PCR product was loaded on 1% agarose gel, electrophoresed, and stained with ethidium bromide for visualization, and its molecular weight estimated against a 100-bp DNA ladder RTU (GeneDireX®, Taiwan). The amplified products were sequenced at First BASE Laboratories Sdn Bhd (Selangor, Malaysia) using the PCR primers as sequencing primers by the BigDye terminator v3.1 cycle sequencing kit (ABI).
Sequence alignment and phylogenetic analysis
A nucleotide BLAST search was done using each sequence through the National Center for Biotechnology Information (NCBI). Reference sequences were downloaded from NCBI and aligned with our new sequences using the ClustalW program (Thompson et al. 1994) in Bioedit (Hall 1999: http://www.mbio.ncsu.edu/bioedit/bioedit.html).
Phylogenetic analyses of partial 18S rRNA sequences of Sarcocystis were conducted using MEGA7 (Kumar et al. 2016). The phylogenetic tree was constructed using the maximum likelihood method with the Tamura 3-parameter model of sequence evolution (Tamura 1992) and tested using 1000 bootstrap resamplings. Toxoplasma gondii was the outgroup for rooting the tree.
Results
Prevalence and morphology of sarcocysts
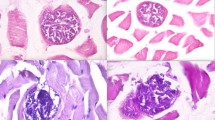
The origin of each host sample (24 from taurine cattle Bos taurus and 15 from water buffalo Bubalus bubalis) and the associated mitochondrial sequence from each host is shown in Table S1. Out of 39 samples, 31 (79.5%) were positive by microscopic examination which detected sarcocysts or bradyzoites. Positive samples included 8/8 (100%) from Cambodia, 9/10 (90%) from Lao PDR, and 14/21 (66.7%) from Thailand. Light microscopy showed fusiform sarcocysts varying in length:width ratios. Histological sections showed thin cyst walls (0.61 to 0.92-µm thick) (Fig. 1). Sarcocysts ranged from 183.67 to 329.21 µm in length and 64.27 to 80.83 µm in width. Bradyzoites were packed tightly into compartments of irregular shape separated by thin septa. Bradyzoites found by use of the squeezing method are not figured here.
Histological sections (longitudinal and cross-sections) of sarcocysts in cattle and buffalo heart muscle from Cambodia (a1–2 and b1–2), Lao PDR (c1–c2 and d1–2), and Thailand (g1–2 and f1–2) stained with hematoxylin and eosin (HE), (a1 at 200 magnification; b1, c1, d1, e1, and f1 at 600 magnification; whereas a2, b2, c2, d2, e2, and f2 at 1000 magnification). Panels a and e are from taurine cattle (Bos taurus) specimens, while panels b, c, d, and f show sarcocysts from water buffalo (Bubalus bubalis)
Molecular analyses
All 31 samples positive by microscopic examination were also positive by PCR, while the remaining eight microscopically negative samples were negative by PCR. Each unique partial 18S rRNA Sarcocystis sequence has been submitted to GenBank, as shown in Table 2. Sarcocystis heydorni was found in taurine cattle, and S. cruzi was found in both in taurine cattle and water buffalo. Sarcocystis levinei was found in water buffalo.
Sarcocystis 18S sequences from Cambodia (KHM 5–KHM 11) and Thailand (THA 1–THA 14) shared 97.7–100% similarity with S. cruzi from China, Germany, the USA, Australia, and Argentina (Fig. 2). All sequences from Lao PDR (LAO 1–LAO 9) were identical to that of S. levinei from Egypt (Fig. 2). In the phylogenetic tree, sequences of S. cruzi from taurine cattle and water buffalo in Thailand and Cambodia were intermingled in the tree with sequences of S. levinei from water buffalo in Lao PDR (Fig. 2). Sarcocystis cruzi exhibits some variation in its 18S rRNA gene. The KHM 1 sequence from Cambodia was 99.8% similar to that of S. heydorni from China and the Netherlands (Fig. 2), indicating the presence of S heydorni in Cambodia.
Phylogenetic tree of Sarcocystis spp. based on 18S rRNA sequences obtained during this study and reference sequences from GenBank. The tree was inferred using the maximum likelihood (ML) method and the Tamura 3-parameter model of sequence evolution. Bootstrap resampling (1000) was used to assess support for individual nodes. Toxoplasma gondii was selected as the outgroup. Symbols designate whether samples were derived from taurine cattle (Bos taurus) (animal symbol with small horns)
 or water buffalo (Bubalus bubalis) (animal symbol with large horns)
or water buffalo (Bubalus bubalis) (animal symbol with large horns)

Discussion
Sarcocystis is a globally important foodborne zoonotic pathogen (Dubey 2015). Consuming raw beef infected with Sarcocystis can cause sickness in humans. This study investigated the prevalence of Sarcocystis infection in hearts of cattle and buffaloes sold in local markets of three countries Thailand, Lao PDR, and Cambodia using microscopic observation and molecular identification. Histological examination showed that all sarcocyst walls were less than 1-μm thick, which means that all belonged within the thin-walled group (i.e. S. cruzi-like, S. heydorni-like and S. levinei-like) (Dubey et al. 2015; JyothiSree et al. 2017). Molecular identification is therefore essential for distinguishing among these species. Our molecular work identified S. cruzi, S. levinei, and S. heydorni (Decker Franco et al. 2018; Dubey et al. 2015; Dissanaike and Kan 1978).
Based on the 18S rRNA gene, we identified that S. cruzi from taurine cattle and water buffalo in Thailand and Cambodia and S. levinei from water buffalo in Lao PDR. Sarcocystis levinei and S. cruzi are very closely related (Gjerde et al. 2016) and our analysis also demonstrates that sequences of S. cruzi fall into the same clade as those of S. levinei (Fig. 2) (Gjerde et al. 2016). Although the 18S rRNA gene sequence is said to be a suitable marker for identification of genus and species (Xue et al. 2019), it cannot discriminate between S. cruzi and S. levinei. The relationship between these species and their taxonomic status requires further investigation, including use of full-length 18S rRNA sequences and other genetic markers (i.e. the mitochondrial cytochrome c oxidase subunit I, nuclear 28S ribosomal (r) RNA genes, and the internal transcribed spacer regions).
Sarcocystis cruzi is global in distribution and is transmitted by canids (Dissanaike and Kan 1978). It most generally causes subclinical infection in cattle, but can lead to fatal systemic disease with fatal myocarditis (Aráoz et al. 2019). This was the most common species found in the myocardium tissue of domestic cattle (B. taurus) in Cambodia and Thailand, in line with other reports that it is the most common parasite in cattle worldwide (Dubey et al. 2016). We also found this species in water buffalo (B. bubalis), which reflects the fact that both bovid species often live in the same area. We noted that two main groups of 18S rRNA variants were represented among S. cruzi isolates from our study region (Fig. 2), similar to those previously reported in infected Malaysian cattle (Ng et al. 2015). This indicates a degree of intra-specific variation within S. cruzi.
Sarcocystis levinei was described by Dissanaike and Kan (1978) from water buffaloes in Malaysia. We found it in water buffalo in Lao PDR. Ultrastructure of S. levinei sarcocysts was described from water buffaloes in the Philippines (Claveria and Cruz 2000). This species is evidently typically found in water buffaloes from Southeast Asian countries.
Sarcocystis heydorni was described as a new species of Sarcocystis with thin-walled sarcocysts (< 1 µm) in cattle (Dubey et al. 2015). Its sarcocysts differ from those of other thin-walled species in having distinctive villar protrusions (Dubey et al. 2015). We found S. heydoni in a sample of cattle heart tissue from Cambodia. This species was experimentally shown to infect a human volunteer (Dr. Heydorn) who ate raw minced beef from Turkey (Dubey et al. 1988). There is a report of S. heydorni from cattle in China: Ingestion by a human volunteer did not produce a patent infection in that case (Hu et al. 2016).
One limitation of our study is that we only examined heart tissue. We may therefore have failed to find other species of Sarcocystis. For example, felid-transmitted species parasitize other tissues and are not found in the heart (Dubey et al. 2015). Examination of other parts of the body, such as tongue, esophagus, heart tissue and diaphragm, would increase the chances of finding additional Sarcocystis species (Dubey et al. 2016).
Conclusions
We have presented the first molecular characterization of Sarcocystis infections within the heart samples of cattle and buffalo in three countries, Thailand, Cambodia, and Lao PDR. Phylogenetic relationships among several Sarcocystis species were analyzed. Sarcocystis cruzi appears to be the most prevalent Sarcocystis species in cattle in Cambodia and Thailand. Sarcocystis levinei was commonly found in water buffalo in Lao PDR, whereas S. heydorni, a known human pathogen, S. heydorni, was found in cattle from Cambodia. It is important to investigate the prevalence and molecular characterization of Sarcocystis infections in meat products more widely to fully evaluate the potential public health risk from bovine Sarcocystis infections in Asian countries.
References
Aráoz V, da Silva SC, Moré G, Banchero G, Riet-Correa F, Giannitti F (2019) Fatal Sarcocystis cruzi induced eosinophilic myocarditis in a heifer in Uruguay. J Vet Diagn Invest 31:656–660
Claveria FG, Cruz MJ (2000) Sarcocystis levinei infection in Philippine water buffaloes (Bubalus bubalis). Parasitol Int 48:243–247
Dissanaike AS, Kan SP (1978) Studies on Sarcocystis in Malaysia. I. Sarcocystis levinei n. sp. from the water buffalo Bubalus bubalis. Z Parasitenkd Parasitol Res 55:127–138
Decker Franco C, Schnittger L, Florin-Christensen M (2018) Sarcocystis. In: Florin Christensen M, Schnittger L (eds) Parasitic protozoa of farm animals and pets. Springer, Cham, pp 103–124
Dokmaikaw O, Suntharawitoon P, Singsuk C (2017) Prevalence of zoonotic Sarcocystis spp. isolated from cardiac muscle of cattle in Muang Chachoengsao District. Chachoengsao Province KKU Sci J 45:354–359 (in Thai with English abstract)
Dubey JP (2015) Foodborne and waterborne zoonotic sarcocystosis. Food Waterborne Parasitol 1:2–11
Dubey JP, Fayer R, Speer CA (1988) Experimental Sarcocystis hominis infection in cattle: lesions and ultrastructure of sarcocysts. J Parasitol 74:875–879
Dubey JP, Calero-Bernal R, Rosenthal BM, Speer CA, Fayer R (2016) Sarcocystosis of animals and humans. CRC Press Taylor & Francis group, Boca Raton, FL
Dubey JP, van Wilpe E, Calero-Bernal R, Verma SK, Fayer R (2015) Sarcocystis heydorni, n. sp. (Apicomplexa: Protozoa) with cattle (Bos taurus) and human (Homo sapiens) cycle. Parasitol Res 114:4143–4147
Fayer R (2004) Sarcocystis spp. Human Infections Clin Microbiol Rev 17:894–902
Fayer R, Esposito DH, Dubey JP (2015) Human infections with Sarcocystis species. Clin Microbiol Rev 28:295–311
Gjerde B, Hilali M, Abbas IE (2016) Molecular differentiation of Sarcocystis buffalonis and Sarcocystis levinei in water buffaloes (Bubalus bubalis) from Sarcocystis hirsuta and Sarcocystis cruzi in cattle (Bos taurus). Parasitol Res 115:2459–2471
Guan F, Jin YT, Zhao J, Xu AC, Luo YY (2018) A PCR method that can be further developed into PCR-RFLP assay for eight animal species identification. J Anal Methods Chem. https://doi.org/10.1155/2018/5890140
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Hu JJ, Wen T, Chen XW, Liu TT, Esch GW, Huang S (2016) Prevalence, morphology, and molecular characterization of Sarcocystis heydorni sarcocysts from cattle (Bos Taurus) in China. J Parasitol 102:545–548
JyothiSree C, Venu R, Samatha V, Malakondaiah P, Rayulu VC (2017) Prevalence and microscopic studies of Sarcocystis infection in naturally infected water buffaloes (Bubalus bubalis) of Andhra Pradesh. J Parasit Dis 41:476–482
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Latif BMA, Al-Delemi JK, Mohammed BS, Al-Bayati SM, Al-Amiry AM (1999) Prevalence of Sarcocystis spp. in meat-producing animals in Iraq. Vet Parasitol 84:85–90
Lindsay DS, Blagburn BL, Braund KG (1995) Sarcocystis spp. and sarcocystosis. Br Med J 5:249–254
Moré G, Abrahamovich P, Jurado S, Bacigalupe D, Marin JC, Rambeaud M, Venturini L, Venturini MC (2011) Prevalence of Sarcocystis spp. in Argentinean cattle. Vet Parasitol 177:162–165
Nateeworanart S, Chanetmahun U, Samreuy S, Uteam W, Kambenmad U (2004) Prevalence of Sarcocystis spp. in cardiac muscle of swine in Samut Prakan Province Thailand. J Southeast Asian Educ 35:82–83
Ng YH, Fong MY, Subramaniam V, Shahari S, Lau YL (2015) Genetic variants of Sarcocystis cruzi in infected Malaysian cattle based on 18S rDNA. Res Vet Sci 103:201–204
Slaoui M, Fiette L (2011) Histopathology procedures: from tissue sampling to histopathological evaluation. In: Gautier JC (ed) Drug safety evaluation. Methods in Molecular Biology (Methods and Protocols), vol 691. Humana Press, Clifton, pp 69–82
Tamura K (1992) Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G + C-content biases. Mol Biol Evol 9:678–687
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Xue R, Yan W, Qian W, Wang T, Zhang M, Wei Z, Han L, He B, Dou J (2019) Prevalence and molecular characterization of Sarcocystis infections of retail beef products from central China. Acta Trop 190:339–343
Acknowledgements
We would like to thank Professor David Blair for the valuable suggestions and assistance with this paper’s presentation through Khon Kaen University Publication Clinic. We thank Mr. Mesa Un and Dr. Sakhone Laymanivong for collecting the sample from Cambodia and Lao PDR, respectively.
Funding
This study was supported by the Faculty of Medicine, Khon Kaen University (grant number IN62128), a Thailand Research Fund’s Distinguished Research Professor Grant (PMI and WM, grant number DPG 6280002) and Research Program (RP64010), Khon Kaen University. LS was supported by a scholarship under the Post-Doctoral Training Program at the Khon Kaen University Research Affairs and Graduate School (grant number 60164). RR received a scholarship under the Post-Doctoral Training Program at the Khon Kaen University, Thailand.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Disclaimer
The funders had no role in study design, data collection, interpretation, or the decision to submit the work for publication.
Additional information
Section Editor: Leonhard Schnittger
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hongchuta, S., Intapan, P.M., Thanchomnang, T. et al. Preliminary findings and molecular characterization of thin-walled Sarcocystis species in hearts of cattle and buffaloes in Thailand, Lao PDR, and Cambodia. Parasitol Res 120, 2819–2825 (2021). https://doi.org/10.1007/s00436-021-07241-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-021-07241-5