Abstract
The sheep body louse, Bovicola ovis (B. ovis), is one of the most significant ectoparasites affecting Australia’s sheep flocks. Despite this, detection methods for B. ovis infestation are limited to visual inspection and ELISA. A colourimetric loop-mediated isothermal amplification (LAMP) method was developed and evaluated for the detection of B. ovis DNA. Diagnostic sensitivity and specificity of LAMP were compared with those of visual inspection and PCR and validated using field samples collected from 22 farms. Two different DNA extraction methods using a commercial kit and a boiling method were also compared. The highest sensitivity and specificity were observed when PCR was used and DNA was extracted using a commercial kit. Compared with PCR, the LAMP assay demonstrated a sensitivity and specificity of 90% and 92% when DNA was extracted by a commercial kit and 100% and 75% when DNA was extracted by the boiling method, respectively. The LAMP test developed in this study could potentially serve as a point-of-care diagnostic tool for monitoring of sheep flocks as well as surveillance of B. ovis populations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wool production is an integral component of the Australian economy, generating $2.6 billion in revenue, making wool one of Australia’s most valuable export commodities (DAWR 2018). Sheep lice can affect wool quality and quantity, as well as leading to additional treatment costs for producers with infested sheep. There are several species of sheep lice found in Australia. The most significant sheep lice species is Bovicola ovis (B. ovis, formerly Damalinia ovis), commonly referred to as the ‘sheep body louse’ (James 2013). The percentage of lousy flocks in Australia will differ from state to state, based on environmental and flock variables that influence lice transmission rates. It is estimated that approximately 25% of Australia’s sheep flocks are afflicted by a form of sheep louse (Wojtek et al. 2001). Total eradication of sheep lice from the national flock is also considered unlikely, due to inadequate biosecurity, in addition to the increased prevalence of treatment-resistant lice as a result of frequent or other improper chemical usage (Joshua et al. 2010). Thus, there is demand for an accurate, efficient and sensitive detection method of sheep lice infestation, to prevent unnecessary chemical usage, reduce treatment costs and support efficient sustainable overall sheep lice control (Popp et al. 2012). Fleece parting and observation of lice is an inexpensive, quick and common technique used for identifying lice infestations (James et al. 2002). While the method is very convenient, the technique lacks the sensitivity required for accurate detection of small infestations (Morcombe et al. 1996). Therefore, enzyme-linked immunosorbent assay (ELISA) was developed for detection of B. ovis. This test has high sensitivity and specificity (Popp et al. 2012) but was not widely accepted by producers, due to cost and an inconvenient 3-day testing period required before the results could be determined (Wojtek et al. 2001). Molecular diagnostics offer some advantages over other detection assays for parasite detection. Molecular techniques can be more sensitive and specific and thus could be particularly useful when parasitic infestations are low in numbers or require differentiation from morphologically similar or identical species (Pritt 2015).
The aim of this study was to evaluate a colourimetric LAMP diagnostic method for detection of B. ovis that could potentially be carried out on farm, as a rapid point-of-care test.
Materials and methods
Samples
The B. ovis isolates used in this study included parasites that were available in the collection of the parasitology group in the School of Animal and Veterinary Sciences at Charles Sturt University and field samples collected from 22 different flocks of sheep. The field samples were collected from flocks during wool shearing. Samples were collected in the form of residues off cutters and combs that were used for shearing. Samples were collected when the cutters or combs were changed during shearing, which happened with a frequency of once every 20 sheep. Cutters of shearing handpieces were rinsed with approximately 100 ml of 70% ethanol or distilled water and brushed to collect the accumulated debris. Within each batch of roughly 20 sheep, two fleeces, freshly shorn off sheep that demonstrated evidence of damage by lice, were visually inspected. Alternatively, when no evidence of fleece damage was obvious, two fleeces were randomly selected to be visually examined, in order to confirm the presence or absence of any lice infestation. The visual detection of sheep lice followed the protocol outlined before (Joshua et al. 2010). The samples were stored in the PC2 laboratory at − 20 °C until use.
DNA extraction
DNA was extracted from the washings (collected in 70% ethanol and dH2O) using a Wizard® SV Genomic DNA Purification Kit (Promega, Australia) with slight modifications. Samples stored in ethanol were precipitated (2000×g, 5 min) and washed two times with dH2O before DNA extraction. Positive control DNA sample was prepared from B. ovis isolates homogenised in a pestle and mortar before DNA extraction.
A second method of DNA extraction (boiling method) was also evaluated. A fraction of prepared samples (500-μl subsample of each prepared washing) was incubated at 100 °C for 10 min, and the supernatant was used as the DNA template. The NanoDrop 2000 (Thermo Scientific, Australia) was used for measuring the quantity and quality of DNA samples.
LAMP and PCR primers
LAMP primers were designed using PrimerExplorerV5 (Eiken). The LAMP primers were designed based on the B. ovis 18S small subunit ribosomal RNA (rRNA) gene (GenBank accession no. AY077769) sequence (Table 1). Primers were ordered from Bioneer Pacific (Australia) and were HPLC purified. The position of LAMP primers in the target DNA is illustrated in Fig. 1.
PCR primers (cox1-F 5′-CCGAGAGGGGAAAGAAGGAG-3′ and cox1-R 5′-CAGTGGGCACAGCAATGAT-3′) were designed from B. ovis cytochrome oxidase subunit 1 (cox1) gene (GenBank accession number GU569309) using DNASTAR software.
Detection of B. ovis using PCR-cox1
Extracted genomic DNA (using a commercial kit) from B. ovis was used as the template for optimisation of PCR. PCR amplification was performed in 25 μl reaction volume on an iCycler thermal cycler (Bio-Rad). The reaction mixture contained 2 μl extracted genomic DNA, 25 μM of each primer, 1.5 mM MgCl2, 1250 μM of each dNTP, 5 × GoTaq Green Flexi Reaction Buffer and 1 U of GoTaq DNA Polymerase (Promega, Australia). PCR cycling was one cycle of 95 °C for 4 min; 35 cycles of 94 °C for 30 s, 55 °C for 30 s and 72 °C for 30 s; and a final cycle of 72 °C for 2 min. All field samples were tested and amplified PCR products were analysed by agarose gel electrophoresis.
LAMP reaction
The LAMP reaction mix contained outer primers (F3 and B3) at 2 μM each, inner primers (FIP and BIP) at 16 μM each, loop primers (LF and LB) at 4 μM each, WarmStart colourimetric 2X Master Mix (New England Biolabs, Australia) and DNA template in a total volume of 25 μl. LAMP reactions were incubated for 60 min at 65 °C in a heat block. Results were determined by visual observation of the colour of the LAMP reaction solutions, with successful amplification from samples containing B. ovis DNA exhibiting a change of the colour from red to yellow.
Sensitivity test for PCR-cox1 and LAMP-18S reaction
The sensitivity of PCR-cox1 and LAMP-18S was determined using serial 10-fold dilutions of DNA extracted from B. ovis specimens. DNA extracted from the B. ovis was serially diluted 10-fold from 5.6 to 5.6 × 10−7 ng/μl. Each DNA dilution was tested using both PCR-cox1 and LAMP-18S.
Sequencing and nucleotide sequence analysis of PCR amplicons
The PCR products were purified using the Wizard® SV Gel and PCR Clean-Up System (Promega, Australia) according to the manufacturer’s instructions. Selected PCR amplicons were sequenced in both directions using cox1 primers by Australian Genome Research Facility Ltd. (AGRF Ltd., Brisbane). Sequence data were analysed using multiple sequence alignments using ClustalW software (Thompson et al. 1994) and BioEdit Sequence Alignment Editor (version 6.0.9.0). GenBank accession numbers were assigned to the nucleotide sequences of the B. ovis isolates.
Comparison of clinical sensitivity and specificity of the evaluated detection methods
New detection tests are usually evaluated against a gold standard test. Therefore, the sensitivity and specificity of the visual inspection, LAMP and PCR were measured using a 2 × 2 contingency table, assuming PCR as the gold standard test. Given that PCR is not necessarily a gold standard diagnostic tool for B. ovis, we also performed latent class analysis, which combines results from different diagnostic tools, and produce sensitivities and specificities via a statistical model in the absence of any reference gold standard (Beath 2017). In this analysis, two latent classes (presence or absence of sheep lice), and a random effect to model the conditional dependence amongst the diagnostic tests, were used. Latent class analysis was performed using the randomLCA package (Beath 2017) in the computing environment R version 3.5.3 (https://www.R-project.org). Based on latent class analysis, PCR using DNA extracted by a commercially available kit was determined to have 100% sensitivity and specificity. Therefore, PCR on DNA extracted by a commercial kit was determined to be the gold standard diagnostic test, and subsequently, sensitivity and specificity of each diagnostic assay were calculated using the MedCalc 2 × 2 contingency table (www.medcalc.org/calc/diagnostic_test.php).
Results
Detection of sheep lice using visual assessment
Wool derangement is one potential sign of lice infestation; however, in this study, the presence of sheep lice was only considered confirmed when adult lice or lice eggs were actually observed. The presence of sheep lice was confirmed in eight out of the 22 samples from shearing sheds when assessed visually (Table 2).
Detection of sheep lice using PCR
DNA samples extracted from samples collected/preserved in dH2O did not produce consistent or satisfactory results in PCR (results are not shown). However, samples collected in 70% ethanol produced an expected size of amplicon (171 bp), and therefore, for the rest of study, only samples collected in ethanol were processed and tested.
Out of the two DNA extraction methods (commercial kit and boiling method), an expected size of DNA fragment was amplified in 10 out of 22 samples when DNA was extracted using the commercial kit. However, specific and non-specific DNA fragments were amplified in 16 out of 22 samples when a boiling method was utilised. Results from PCR are summarised in Table 2.
Detection of sheep lice using LAMP-18S
LAMP assay successfully amplified DNA from B. ovis–positive field samples collected from the 22 farms.
A change of colour from red to yellow was observed in the positive control, while the negative control remained red.
Out of 22 samples collected at shearing, 10 samples were recorded positive when DNA was extracted using a commercial kit. However, 13 samples out of 22 were positive when DNA was extracted using a boiling method (Table 2).
Laboratory sensitivity of PCR and LAMP using DNA extracted by a commercial kit
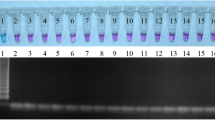
The laboratory sensitivity of PCR and LAMP was assessed by testing serial dilutions of extracted DNA using a commercial kit. Sheep lice DNA was detected up to 5.6 × 10−3 ng in the PCR and up to 5.6 × 10−2 ng in LAMP (Fig. 2a).
Sensitivity of PCR and LAMP using DNA extracted by (a) a commercial kit and (b) a boiling method at different dilutions of B. ovis DNA. M, molecular marker (Sigma-Aldrich, Australia); lanes 1–8 corresponding with the DNA concentration 5.6 ng, 5.6 × 10−1 ng, 5.6 × 10−2 ng, 5.6 × 10−3 ng, 5.6 × 10−4 ng, 5.6 × 10−5 ng, 5.6 × 10−6 ng and 5.6 × 10−7 ng, respectively
The test sensitivity when DNA was extracted by the boiling method was 10 times higher than that of the commercial kit for the PCR and LAMP at 5.6 × 10−4 ng and 5.6 × 10−3 ng, respectively (Fig. 2b).
Clinical sensitivity and specificity of the evaluated diagnostic methods
Performance of the diagnostic tests in this study was evaluated based on a DNA extraction method using a statistical 2 × 2 contingency table assuming PCR as the gold standard test and latent class analysis assuming there is no gold standard test available. The sensitivity and specificity of visual assessment were 80% and 100%, respectively.
When the DNA was extracted by the commercial kit, PCR and LAMP showed sensitivities of 100% and 90%, respectively, and specificities of 100% and 91.67%, respectively. However, when the DNA was extracted using the boiling method, PCR and LAMP showed sensitivities of 80% and 100%, respectively, and specificities of 33.33% and 75%, respectively (Table 3).
The lowest negative likelihood ratio (LR−) was achieved by the PCR (DNA extracted using the commercial kit) and LAMP (DNA extracted using the boiling method) with 0.0 (a LR− below 0.1 essentially eliminates the chance that a patient has the disease) (Chaouch et al. 2019) (Table 3).
In addition, results from the 2 × 2 contingency table were compared with latent class analysis. The sensitivity and specificity of visual assessment, PCR and LAMP were found similar in both statistical models.
Discussion
It is estimated that 23.3% of Australian sheep flocks are infested with lice (Reeve and Walkden-Brown 2014). Lice infestation of sheep flocks is an important economic loss to the Australian wool industry and cost $81 million per year in 2014 (Lane et al. 2015). With a trend towards increased lice infestation (Cotter 2019; Joshua et al. 2010; Popp et al. 2012) and in the absence of a rapid detection tool, a reliable, rapid and cost-effective diagnostic tool is required for efficient and sustainable lice control and management.
Wool parting to observe lice is considered a primary method of evaluation; however, lice can be very difficult to detect, especially with light infestations (< 400 lice bodies) (Joshua et al. 2010), and effectiveness relies on the observation and other skills of the assessor (Morcombe et al. 1996). Assessment can also be labour-intensive and time-consuming. ELISA testing is generally more sensitive than visual inspections (Popp et al. 2012); however, this approach requires samples to be sent to a suitably equipped laboratory and hence delays results when producers would prefer more timely results to decide on the need for off-shears treatment and minimise losses associated with sheep lice. For effective control of sheep lice, timely detection of B. ovis is crucial.
To our knowledge, there is no commercially available molecular test for the detection of sheep lice. This might be due to the fact that molecular diagnostics usually require samples to be sent to a laboratory which may not be convenient for farmers. This study compared visual assessment and two different molecular diagnostic methods (PCR and LAMP) for the detection of sheep lice using samples collected at shearing time. Two different DNA extraction methods were also evaluated, a commercial kit–based approach (Wizard® SV Genomic DNA Purification Kit, Promega, Australia) and a simple, crude boiling method, as previously used for samples containing bacteria or parasites (Sepahvand et al. 2017; Zhu et al. 2006).
The LAMP reaction is a novel approach to DNA amplification that is based on autocycling strand displacement DNA amplification using the Bst DNA polymerase enzyme under isothermal conditions (Notomi et al. 2000). The four specific primers will anneal to six different regions of the target DNA sequence and amplify the DNA at constant temperature of 65 °C without requiring a thermal cycler. Therefore, compared with PCR, LAMP does not need expensive laboratory equipment, and this makes this method potentially suitable for point-of-care testing (Niessen et al. 2013).
PCR showed greater sensitivity and specificity (100% and 100%) compared with LAMP (90% and 91.7%) when DNA template was extracted using a commercial kit. However, LAMP showed higher sensitivity and specificity (100% and 75%) compared with PCR (80% and 33.3%) when DNA template was extracted using a boiling method. This could be due to the fact that LAMP is less susceptible to contaminants such as crude DNA preparation when compared with the PCR (Martzy et al. 2017).
When DNA was extracted by the boiling method, the positive likelihood ratio (LR+) indicated that the samples detected as positive by LAMP are potentially true positives. However, the negative likelihood ratio (LR−) of PCR was greater than 0.1 (0.6) and lower than 1, which indicates the possibility of lower false negative results.
Compared with the DNA extraction using a commercial kit, more positive results were produced by PCR following the boiling extraction method which could be due to the presence of cellular and environmental contamination (Abdelhai et al. 2016). Therefore, false positive results seen in the PCR following the boiling method might be due to non-specific amplification. Since the shearing debris have been used for DNA extraction, contamination of 18S rRNA and cox1 gene from other possible insects present in the wool at the time of shearing might have also contributed to the false positive results in molecular tests.
Extraction of DNA using a commercial kit can be very effective in removing cell and environmental contaminants, resulting in a purified DNA from field samples and preventing non-specific bands in the PCR. However, this method of DNA extraction requires laboratory equipment, such as a centrifuge, and technical expertise. In order to use PCR as a diagnostic test and extract DNA with a commercial kit, samples should be sent to the diagnostic laboratory where all required instruments and expertise are available. This approach, however, might be inconvenient for farmers, as they need rapid results to allow them to make more timely treatment decisions.
On a sheep farm, a crude DNA preparation method, such as the boiling method, followed by a colourimetric LAMP-based detection assay could provide a convenient and applicable point-of-care test for detection of sheep lice during shearing. Performing a LAMP does not require sophisticated laboratory instruments, and gel electrophoresis can be avoided when a colourimetric technique is used for visualisation of results; hence, testing and interpretation of results would be simplified. This study provided an initial suggestion that LAMP using DNA extracted by boiling of collected samples could be superior in terms of practicality and still can provide satisfactory detection capability.
One of the primary advantages of LAMP over PCR could be high specificity and sensitivity, due to the use of six primer binding sites and low susceptibility to sample inhibitors (Niessen et al. 2013). Generally, when a LAMP assay is used for diagnostic purposes, sensitivity of the test could be increased if clinical samples such as blood or body fluids are utilised. This is due to the relatively clean samples which are less contaminated with dirt or environmental impurities. However, one challenging finding from this study is that LAMP proved to be less sensitive than PCR. This could be due to a high degree of environmental contaminants like wool and dirt in the collected samples. Therefore, further study might be required to improve the DNA extraction method to eliminate environmental contaminants.
This is the first study that evaluated a LAMP assay for the detection of B. ovis as a point-of-care diagnostic test. The developed LAMP assay was compared with PCR using two different methods of DNA extraction and visual assessment of sheep lice. The LAMP assay developed in this study was found to be more convenient than PCR for the detection of B. ovis in clinical samples and has the potential to be used as a point-of-care diagnostic test for better management and control of sheep lice.
References
Abdelhai M, Hassanin HM, Sun X (2016) Comparative study of rapid DNA extraction methods of pathogenic bacteria. Am J Biosci Bioeng 4:1–8
Beath K (2017) randomLCA : an R package for latent class with random effects analysis. J Stat Softw 81. https://doi.org/10.18637/jss.v081.i13
Chaouch M, Aoun K, Ben Othman S, Ben Abid M, Ben Sghaier I, Bouratbine A, Ben Abderrazak S (2019) Development and assessment of Leishmania major- and Leishmania tropica-specific loop-mediated isothermal amplification assays for the diagnosis of cutaneous leishmaniasis in Tunisia. Am J Trop Med Hyg. https://doi.org/10.4269/ajtmh.19-0097
Cotter J (2019) Sheep lice, Biosecurity can prevent introduction. http://www.liceboss.com.au/sheep-goats/prevention/sheep-lice-biosecurity-can-prevent-introduction.php. Accessed Aug 2019
DAWR (2018) Wool (FACT Sheet) Department of Agriculture and Water Resources, Australian Government. http://www.agriculture.gov.au/about/commitment/portfolio-facts/wool. Accessed Aug 2019
James PJ (2013) Biology of sheep lice (Bovicola ovis). LiceBoss. http://www.liceboss.com.au/files/pages/notes/Biology_of_sheep_lice_Bovicola_ovis.pdf. Accessed Aug 2019
James PJ, Garrett JA, Moon RD (2002) Sensitivity of two-stage sampling to detect sheep biting lice (Bovicola ovis) in infested flocks. Vet Parasitol 103:157–166. https://doi.org/10.1016/S0304-4017(01)00585-4
Joshua E, Junk G, Levot G (2010) Sheep lice (primefacts). Department of Primary Industries, NSW. https://www.dpi.nsw.gov.au/__data/assets/pdf_file/0005/318704/Sheep-lice.pdf. Accessed Aug 2019
Lane J, Jubb T, Shephard R, Webb-Ware J, Fordyce G (2015) Priority list of endemic diseases for the red meat industries. Meat and Livestock Australia Ltd, Sydney
Martzy R et al (2017) A loop-mediated isothermal amplification (LAMP) assay for the rapid detection of Enterococcus spp. in water. Water Res 122:62–69. https://doi.org/10.1016/j.watres.2017.05.023
Morcombe P, Young G, Ball M, Dunlop R (1996) The detection of lice (Bovicola ovis) in mobs of sheep: a comparison of fleece parting, the lamp test and the table locks test. Aust Vet J 73:170–173. https://doi.org/10.1111/j.1751-0813.1996.tb10020.x
Niessen L, Luo J, Denschlag C, Vogel RF (2013) The application of loop-mediated isothermal amplification (LAMP) in food testing for bacterial pathogens and fungal contaminants. Food Microbiol 36:191–206. https://doi.org/10.1016/j.fm.2013.04.017
Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T (2000) Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28:e63–e63. https://doi.org/10.1093/nar/28.12.e63
Popp S, Eppleston J, Watt BR, Mansfield S, Bush RD (2012) The prevalence of lice (Bovicola ovis) in sheep flocks on the central and southern Tablelands of New South Wales. Ani Prod Sci 52:659–664. https://doi.org/10.1071/AN11240
Pritt BS (2015) Chapter 4 - Molecular diagnostics in the diagnosis of parasitic infection. In: Sails A, Tang Y-W (eds) Methods in Microbiology, vol 42. Academic Press, pp 111–160. https://doi.org/10.1016/bs.mim.2015.05.001
Reeve I, Walkden-Brown, S. (2014) Benchmarking Australian sheep parasite control: cross-sectional survey report Meat and Livestock Australia Limited Sydney
Sepahvand A, Pestehchian N, Yousefi HA, Gharehbaba RP (2017) Comparison and evaluation of four methods for extracting DNA from Giardia duodenalis cysts for PCR targeting the tpi gene. J Parasit Dis 41:263–267. https://doi.org/10.1007/s12639-016-0790-5
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. https://doi.org/10.1093/nar/22.22.4673
Wojtek PM, Paul Y, Shiell B, Levot G (2001) Development of a lice detection test for ‘on-farm’. Paper presented at the FLICS Conference Launceston,
Zhu K, Jin H, He Z, Zhu Q, Wang B (2006) A continuous method for the large-scale extraction of plasmid DNA by modified boiling lysis. Nat Protoc 1:3088–3093. https://doi.org/10.1038/nprot.2006.452
Acknowledgements
The authors would like to thank Ag resellers and the farmers from the Riverina in NSW, who participated in this study by providing samples.
Funding
This work was supported by the Graham Centre for Agricultural Innovation (grant number 50714) and the School of Animal and Veterinary Sciences (grant number 40702) at Charles Sturt University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
All the experiments complied with the current policies of Charles Sturt University.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Section Editor: Douglas D. Colwell
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wong, S.A., Woodgate, R.G., Pant, S.D. et al. Rapid detection of Bovicola ovis using colourimetric loop-mediated isothermal amplification (LAMP): a potential tool for the detection of sheep lice infestation on farm. Parasitol Res 119, 395–401 (2020). https://doi.org/10.1007/s00436-019-06552-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-019-06552-y