Abstract
Water is considered an important vehicle for the spread of human toxoplasmosis in several countries. Toxoplasma gondii oocysts can persist in the environment for long periods, being highly resistant to the various chemical inactivation processes commonly used by water supply systems, distinctly from simple filtration and flocculation that are efficient in removing oocysts from drinking water. The existing methodologies for identification and quantification of this parasite in water samples are not standardized and have limitations. This study aimed to evaluate the presence of T. gondii oocysts in surface water samples used as a source for the production of drinking water in the State of São Paulo, through the implementation of a specific methodology using real-time PCR technique (qPCR). Volumes of 20 L of the sample were concentrated by filtration in Envirocheck® HV capsules. For DNA extraction, the PowerSoil DNA isolation® kit (currently DNeasy PowerSoil®) was used. The target sequence selected for qPCR was a 62-base-pair fragment of the B1 gene. In the initial recovery evaluation of the method in four replicates of reverse osmosis water, the mean recovery was 48.5% (SD ± 11.5), while the mean recovery for method performance in matrices was 3.2% (SD ± 3.2) (rainy season) and 62.0% (SD ± 6.2) (dry period), suggesting that the characteristics of the samples and the climatic conditions interfere in the recovery efficiency. Of the 39 samples analyzed (May to December 2015), 7.7% (3/39) were positive for T. gondii, and among the ten sources studied; the occurrence of the oocysts was detected in 30% (3/10).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Waterborne diseases occur predominantly in developing countries, but constitute a serious global challenge (Ramírez-Castillo et al. 2015). Globally, about 3.4 million people, mostly children, die every year due to the occurrence of these diseases. According to the WHO, targeted investments in improving water quality could reduce the global burden of waterborne diseases by approximately 4% (WHO/UNICEF 2010).
Among the waterborne pathogens, Toxoplasma gondii has been prominent due to the spread of oocysts through the water, causing outbreaks of toxoplasmosis in different parts of the world (Karanis et al. 2013). Studies compiled by Karanis et al. (2013) suggest that the epidemiological impact of the dissemination of the T. gondii oocysts by water is underestimated. The water delivery of the oocysts is favored due to the long period; they can remain viable in the environment, the low infectious dose and their resistance to the processes of water disinfection such as chlorination, ozonation, and ultraviolet irradiation (Jones and Dubey 2010).
Despite the low prevalence in feces and short duration of T. gondii oocyst shedding by cats, they could lead to substantial environmental contamination (Torrey and Yolken 2013). Furthermore, in some areas, serological prevalence to T. gondii can reach up to 50% in cats, especially in free-living ones (Hartmann et al. 2013). In Brazil, Pena and Soares (2006) studied the serologic and parasitological prevalence of T. gondii in 237 cats from 15 municipalities in the São Paulo State and found a seroprevalence of 35.4% (84/237) by the modified agglutination test (MAT).
From 1979 to 2015, 16 outbreaks due to waterborne parasites were reported in Latin American countries. Giardia spp. and Cryptosporidium spp. were the parasites most frequently found in water samples, while T. gondii and Cyclospora cayetanensis were the parasites that caused the most outbreaks in Latin American countries (Rosado-García et al. 2017).
According to estimates by Dubey et al. (2012), Brazil is one of the regions with the highest prevalence of T. gondii in humans, with about 50% of children and 50 to 80% of women of childbearing age having antibodies to this parasite. The high prevalence of parasite in humans in Brazil was also evidenced by Garcia Bahia-Oliveira et al. (2003), Sobral et al. (2005), and De Moura et al. (2006).
In Campos dos Goytacazes (Rio de Janeiro, Brazil), toxoplasmosis is highly endemic among the low-income population and the lack of access to treated water is considered as the potential source of transmission of T. gondii oocysts among the population (Garcia Bahia-Oliveira et al. 2003); however, in this endemic setting, an important issue is the continuous contamination of drinking water with T. gondii oocysts thus causing asymptomatic infections (Vieira et al. 2015).
In Brazil, the first and also the largest worldwide outbreak of toxoplasmosis by water transmission occurred in the city of Santa Isabel do Ivaí, Paraná State. During the period from November 2001 to January 2002, about 600 people sought health services with symptoms characteristic of toxoplasmosis and 426 presented serology suggestive of acute toxoplasmosis. The outbreak was associated with contamination of the city’s water reservoir by cat feces. Additionally, removal of oocysts in water treatment was not efficient due to the absence of filtration and flocculation processes, as well as inadequate chlorination process (Garcia Bahia-Oliveira et al. 2003; De Moura et al. 2006).
Reports of the occurrence of waterborne toxoplasmosis outbreaks have reinforced the importance of developing parasite detection methods in these samples. Although there is interest of the scientific community in the development of methods for the recovery of T. gondii in aquatic matrices, still no methodology is standardized and universally used (Villena et al. 2004; Karanis et al. 2013; Palos Ladeiro et al. 2015).
In the present study, we have used qPCR for the detection and quantification of T. gondii DNA in water samples because it is a highly sensitive, specific, and fast molecular approach for this purpose (Yang et al. 2009). We chose a 62-base fragment of the B1 gene (Kompalic-Cristo et al. 2007) as a target sequence because it was repeated in 35 copies in the genome of T. gondii (Burg et al. 1989) and was conserved among the different parasite genotypes, taking into account the high genotypic diversity of T. gondii parasites circulating in Brazil (Shwab et al. 2014).
Considering the relevance of T. gondii presence in water sources utilized for public supplies, the present study aimed to evaluate the presence of this parasite in surface water samples through the implementation of a specific qPCR methodology using the B1 gene as a target with a view to contribute to the development method for quantification of T. gondii in surface waters.
Materials and methods
Study area
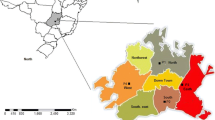
Ten points of surface water collection in catchment areas for human consumption (Table 1 and Fig. 1) were selected because they were impacted by the discharge of crude and treated sewage and because they presented results with high concentrations of Giardia spp. cysts and Cryptosporidium spp. oocysts according to the data from the Report of the São Paulo State Inland Water Quality Monitoring Network conducted by the Environmental Company of the State of São Paulo during the year 2014 (CETESB 2015).
The collection points evaluated in this study are located in industrial areas, where five of the densest Brazilian metropolitan regions are concentrated: metropolitan region of São Paulo, metropolitan region of Campinas, metropolitan region of South Coast, metropolitan region of the Paraíba Valley, and Sorocaba metropolitan region, with a high industrial activity and that together adds up to 32.7 million inhabitants.
Sampling
In the period from May to December 2015, the CETESB’s sampling division collected 40 samples of surface water in the ten selected points. The sampling frequency was carried out every 2 months, and a volume of 20 L of surface water was collected at each point, according to the collection procedure described in the National Guide for Collection and Preservation of Water, Sediment, Water Communities and Liquid Effluents (CETESB/ANA 2011). Samples from Ribeirão dos Cristais River were collected in June of 2018 in order to evaluate the reproducibility and detection limit of the method. The samples were transported under refrigeration (2 to 8 °C) to the laboratory and analyzed within 24 h after collection.
Oocysts of T. gondii
Purified T. gondii oocyst suspensions were provided from the Laboratory of the Department of Preventive Medicine and Animal Health—Faculty of Veterinary Medicine and Animal Science—University of São Paulo. Oocysts were obtained from the feces of cats fed with experimentally infected mice by T. gondii genotype BrIII. The suspensions of oocysts were counted microscopically in a hemacytometer under a bright field, using a Leica Microscope (DMLB), and a 90% sporulation rate was observed.
Evaluation of method performance
Initial recovery
The initial recovery evaluation of the method was performed by analyzing four replicate samples of 20 L of reverse osmosis water that was contaminated with oocysts of T. gondii genotype BrIII (6.68 × 104 oocysts inoculated in 20 L of reverse osmosis water sample). The reverse osmosis water used was from the Laboratory of the Division of Microbiology and Parasitology of CETESB.
Recovery and reproducibility in matrices
The recovery evaluation tests of the method in matrices were performed in nine surface water points of collection from the ten points selected for this study in the wet period (November and December 2015). The samples were processed in two aliquots of 20 L: one contaminated with the suspension of T. gondii oocyst genotype BrIII containing 6.68 × 104 oocysts (spiked) and another without contamination (blank).
The reproducibility of the method in matrices was performed in four aliquots of 20 L from Ribeirão dos Cristais River collected during the dry period (June 2018): three aliquots contaminated with the suspension of T. gondii oocysts genotype BrIII containing 5.13 × 104 oocysts (spiked) and one aliquot without contamination (blank).
The recovery percentage of the T. gondii DNA method in reverse osmosis water and surface water samples was calculated according to Eq. (1):
Where:
- C1:
-
Number of DNA copy T. gondii of the contaminated sample (spike)
- C2:
-
Number of DNA copy T. gondii of the uncontaminated sample (blank)
- C3:
-
Number of DNA copy T. gondii of the suspension added to the contaminated samples
Detection limit of the method
A hemocytometer-counted suspension of 5.13 × 105 oocysts in 1 mL of distilled water was used as initial dilution of T. gondii oocysts. Suspensions of 5.13 × 104, 5.13 × 103, 5.13 × 102, 5.13 × 101, and 5.13 × 100 oocysts were made by serial dilution and then were inoculated in 20 L of each analyzed sample to estimate detection limit of the method for samples of surface water and reverse osmosis water.
Concentration of the samples by filtration
The concentration of the samples was performed according to method 1623.1 (USEPA 2012). Twenty-liter volumes of each sample were concentrated by filtration using Envirocheck® HV (Pall Gelman Laboratory) capsules as specified by the manufacturer. After filtration, the capsules were treated with sufficient amount of the dispersing solution (5% sodium hexametaphosphate solution) until the entire area of the inner membrane was in contact with this solution (about 125 mL) and the particulate matter was eluted from the capsules by means of elution buffer containing detergent (Laureth-12, Pall Gelman Laboratory). The eluates were centrifuged for 15 min at 1500g. All pellet yielded was analyzed.
DNA extraction of T. gondii
The sediments obtained after the centrifugation of the samples were submitted to DNA extraction, using the PowerSoil DNA isolation®, MO BIO extraction kit (currently DNeasy PowerSoil®, QIAGEN), according to the procedures recommended by the manufacturer. The samples were added to a bead beating tube for homogenization and cell lysis occurred by mechanical and chemical methods. Total genomic DNA was captured on a silica membrane in a spin column format and was washed and eluted from the membrane. After the extraction, the purity of the extracted DNA was verified by spectrophotometry (A260/A280), using Biodrop equipment (Denville Scientific Inc.). The DNA was stored at − 80 °C until use.
Quantification of T. gondii (qPCR)
The quantification of T. gondii DNA was performed by the qPCR technique using the TaqMAn® Minor Groove Binder detection system to amplify the target sequence of the 62-bp fragment of the B1 gene. The primers and the respective probe used in this study are described in Table 2 (Kompalic-Cristo et al. 2007).
For the qPCR reaction, a final volume containing 25 μL was used: 9.5 μL of ultra-pure water, 5.0 μL of TaqMan®Environmental Master Mix 2.0 (Applied Biosystem®), 3.5 μL of primer/probe solution (Applied Biosystem®) (0.9 μM of each primer and 0.25 μM TaqMan-MGB probe), 2.0 μL of MgCl2 (5 mM), and 5.0 μL of DNA extracted from the sample. Amplification was performed by means of an initial preincubation at 50 °C for 2 min, followed by 40 cycles at 95 °C for 10 min, 95 °C for 15 s, and 60 °C for 1 min (Kompalic-Cristo et al. 2007).
All qPCR reactions included positive controls (T. gondii oocysts of BrIII genotype) and negative controls (non-template controls, NTC). Samples and positive and negative controls were tested in duplicate in each reaction, and the same procedure was performed to evaluate the efficiency of the standard curve.
Amplification data were obtained using the StepOnePlusTM Real-Time PCR System (Applied Biosystem®) from the Laboratory of Molecular Biology of the Microbiology and Parasitology Division of CETESB. The results were generated through the software StepOne™ (V2.2.2) where the values generated by the quantification are plotted with the cycle number of the qPCR as a function of the cycle of this reaction in which the fluorescence was detected (Cq) for each of the different samples. The data generated by this graph was used to calculate the values of the efficiency of the reaction (E) and of the correlation between the values obtained by the quantification during the qPCR and the values previously known (R2).
Standard curve preparation
The Biological Sequence Alignment Editor, BioEdit program (Hall 1999), was used for the generation of the alignment of the gene fragment chosen for the construction of the standard DNA curve with the product of qPCR generated by the primers used in this study. After alignment analysis, a 466-bp sequence (GenBank DQ789361.1) was chosen, which was commercially synthesized by Integrated DNA Technologies (IDT, Lowa, USA) as gBlocks Gene Fragments® and named B1 Partial. This sequence was made available, lyophilized and quantified (200 ng), and resuspended according to the manufacturer’s recommendations. Serial dilutions on the order of 10-fold of the 466-bp synthetic fragment were used to generate the standard curve of concentrations expressed in log units (log10) versus the values obtained in amplification cycles.
The values of mean, standard deviation, and coefficient of variation were calculated for the reproducibility tests by comparison of the standard curves amplified on alternate days (Pfaffl 2004).
Evaluation of possible false negative results in qPCR
Serial dilutions of DNAs extracted from artificially contaminated surface water samples (recovery evaluation tests of the method in matrices) on the order of 10-fold (pure DNA, 1:10 and 1:100) were performed to evaluate possible false negative results from the presence of inhibitors in the surface water samples analyzed and were subjected to qPCR under the same amplification conditions as described in “Materials and methods” section.
Results
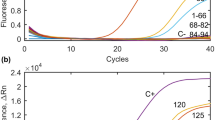
The results obtained for the evaluation of the initial recovery for four replicates of reverse osmosis water samples were 55.5%, 31.8%, 49.9%, and 56.7% (mean 48.5%; SD ± 11.5) (Fig. 2). For the surface water matrices, the mean recovery for method performance was 3.2% (SD ± 3.2) in the rainy period and 62% (SD ± 6.2) in the dry period. The maximum method recovery result in the rainy period (November to December 2015) was 8.9% in the Ribeirão dos Cristais River. We did not obtain any recovery at the Cotia River in the rainy period (Fig. 3).
The reproducibility in matrices was performed in the dry period (June 2018) for samples from the Ribeirão dos Cristais River. The recovery results were 55.4%, 62.9%, and 67.6% (mean 62.0%; SD ± 6.2) (Fig. 4). The methodology used was able to quantify the minimum 25.6 oocysts/L (5.13 × 102 oocysts/20 L) for both surface water and reverse osmosis water. It is, however, important to keep in mind that the detection limit was based on the serial dilution of a hemocytometer-counted suspension and could be inaccurate.
Thirty-nine surface water samples were analyzed to investigate the presence of T. gondii DNA. Positive results were observed in three (7.7%) samples collected in three different water sources, representing 30% (3/10) of positivity in the sampling areas. All positive samples were collected during November and December 2015 (Table 3).
Among the sources with positive results for T. gondii, the most critical was the Jaguari River, in the municipality of Pedreira (1.79 × 104 genomic copies of T. gondii/L, Cq 25.4), followed by the Corumbataí River in the municipality of Piracicaba (3.15 × 102 genomic copies of T. gondii/L, Cq 32.4), and by the Paraíba River in the municipality of Aparecida (1.28 × 102 genomic copies of T. gondii/L, Cq 32.7).
In the qPCR reaction validation assays, primers and probe revealed positive amplification results only for samples containing T. gondii DNA. The best performance was obtained with the addition of 2.0 μL of MgCl2 (5 mM) to the total mix of qPCR reactions. The detection capacity of the primers and probe at different DNA concentrations was evaluated by means of a 10-fold serial dilution of 5.0 × 107 genomic copies/μL to 5 genomic copies/μL of the synthetic fragment containing 466 bp, which was used for the construction of the standard curve. The best amplification reactions obtained in the validation assays with the respective primers and probe were between logs from 5.0 × 106 to 5.0 × 101 genomic copies/μL, considering as detection limit Cq equal to 36.5. The limit of detection (LOD) and the limit of quantification (LOQ) were 5.0 × 100 and 5.0 × 101 genomic copies/μL, respectively. Standard curve validation for T. gondii showed ranges of correlation coefficient (R2) from 0.999 to 0.998, efficiency from 91.6 to 95.9%, and slope from − 3.541 to − 3.423. The coefficient of variation showed good performance in reproducibility (≤ 0.03) for the standard curve used in this study. The results obtained in the linear regression curve for this assay were efficiency = 95.9%, R2 = 0.998, and slope = − 3.423.
Discussion
Because there are no standardized methods for the detection and quantification of T. gondii in water samples, it is not possible to compare the performance of our test. But it is recognized that several researchers are putting efforts to contribute for reaching a method for T. gondii from environmental samples (Isaac-Renton et al. 1998; Kourenti et al. 2003; Villena et al. 2004; Sotiriadou and Karanis 2008; Aubert and Villena 2009; Yang et al. 2009; Gallas-Lindemann et al. 2013; Mahmoudi et al. 2015; Wells et al. 2015; Lora-Suarez et al. 2016).
In our study, the initial recovery rates were higher than 31.8% indicating that the proposed method is promising. In the recovery evaluation tests using the method in surface water matrices performed in the rainy season, the mean recovery was 3.2% (SD ± 3.2) and the maximum recovery was 8.9% (Ribeirão dos Cristais River). However, the results of reproducibility evaluation tests in the dry period for samples from Ribeirão dos Cristais River (n = 3) were 55.4%, 62.9%, and 67.6% with mean of 62.0% (SD ± 6.2), suggesting that the characteristics of the samples and also the climatic conditions (rainfall) interfere in the recovery efficiency of the method.
The T. gondii DNA recovery variation observed in this study can be mainly related to the water quality conditions in the rainy season due to the resuspension of particles from the sediment and from rainwater runoff. The turbidity results of the analyzed samples, although it was variable between samples from different points, were not conclusive to affirm that the turbidity interfered in the recovery of the method. Further studies should be performed using a larger number of samples to better evaluate the impact of turbidity values on recovery rates during dry and wet seasons. Kuhn and Oshima (2002), using Envirocheck® capsule for concentration of Cryptosporidium parvum oocysts in Rio Grande surface water samples (Las Cruces, New Mexico), obtained low recoveries (0.4%) in samples with high turbidity (159 NTU, Nephelometric Turbidity Units) and better recoveries (71.9%) in samples with low turbidity (3.9 NTU).
It is also necessary to consider the steps of own method as limiting factors. The one of critical step is the water sample concentration that involves large volumes of water, and so can occur losses of the target organism as reported by Kerambrun et al. (2015).
One of the advantages of using Envirocheck® HV filtration technique for water samples concentration is that large volumes can be filtered (Villena et al. 2004; Aubert and Villena 2009; Yang et al. 2009). Another one is the pretreatment of the capsule with sodium hexametaphosphate solution (Mccuin and Clancy 2005). This reagent decreases the impact of turbidity on the recovery of the target organism (USEPA 2012), dissolving the material collected on the membrane surface (McCuin and Clancy 2005). Some authors suggest that the recovery efficiency of cysts and oocysts of parasites in turbid waters is low regardless of the method used (Nieminski and Iii 1995; Digiorgio et al. 2002; Borchardt et al. 2009). DNA extraction from the sporulated oocysts requires effective rupture of the walls of the oocysts and sporocysts, as well as the membranes of the sporozoites to access the genetic material (Bushkin et al. 2013; Dumetre et al. 2013; Samuelson et al. 2013; Hohweyer et al. 2016). Methods combining chemical, enzymatic, and/or mechanical lysis procedures at temperatures of at least 56 °C should be preferred for the efficient release of DNA from oocysts (Paulos et al. 2016).
Although there is a standardized and universally used method for the detection and quantification of Cryptosporidium spp. and Giardia spp. in water samples (USEPA 2012), there is no equivalent method for T. gondii, mainly due to the absence of an IMS technique available for the parasite. (Harito et al. 2017) evaluated the use of the lectin-magnetic separation (LMS) technique to isolate T. gondii from water sample concentrates. The results of this study indicated that LMS with wheat germ agglutinin (WGA) coupled to magnetic beads could be an efficient step in the analysis of water sample concentrates, with subsequent detection and quantification by microscopy or by qPCR.
There are divergences in the literature among which is the best target sequence for the detection of T. gondii. Some authors choose to use B1 gene markers, since in addition to being repeated 35 times in the parasite genome (Burg et al. 1989), it has been shown to be very sensitive and conserved among the different genotypes of T. gondii (Contini et al. 2005). However, with the description of the sequence of 529 base pairs (Homan et al. 2000), repeated 200 to 300 times in the genome of the respective parasite, several studies appeared to compare markers from this new sequence with the gene B1. Edvinsson et al. (2006) suggest that new work should ensure that the sequence of 529 base pairs is actually conserved among the different T. gondii genotypes.
Synthetic DNA oligonucleotides were utilized (gBlocks TM, IDT) due to the difficulty in obtaining a standardized suspension containing sufficient amount of T. gondii oocysts to build the standard curve of the qPCR reaction. This strategy was considered interesting and advantageous because it provided simplicity in obtaining the synthesized, purified, and ready-to-use DNA fragments in addition to cost reduction compared to vectors constructed for cloning. The use of synthetic DNA oligonucleotides has presented advantages for the construction of standard curve in the absolute quantification of virus by qPCR reaction (Tourinho et al. 2015). Fumian et al. (2016) standardized a standard curve for the simultaneous detection of norovirus and rotavirus by multiplex RT-qPCR using synthetic oligonucleotides, obtaining satisfactory results.
In our study, we detected the presence of T. gondii DNA in 7.7% of the surface water samples analyzed, and these results corroborate with results reported in other countries in different studies and conditions of T. gondii detection in water samples (Villena et al. 2004; Gallas-Lindemann et al. 2013; Mahmoudi et al. 2015; Wells et al. 2015).
The samples with positive results for T. gondii obtained in our study were collected in the rainy season, indicating a greater introduction of contaminants in the body of water due to the rainwater that carries them. In CA, Shapiro et al. (2015) applied an alternative surveillance approach using mussels to evaluate the contamination of T. gondii marine habitats and also showed a peak of parasite-positive samples during the period of high precipitation.
Considering the important role of T. gondii in several waterborne outbreaks that waterborne oocysts are responsible for most cases of asymptomatic toxoplasmosis, as well as the fact that the world’s largest outbreak of toxoplasmosis by water occurred in our country (Garcia Bahia-Oliveira et al. 2003; De Moura et al. 2006) and also that the parasite was evidenced in three of the ten water bodies evaluated in this study, it is important to carry out periodic and more extensive monitoring of T. gondii in the water bodies used for public supply in the State of São Paulo, using an optimized and standardized analysis method.
Conclusions
The present study was the first to demonstrate the presence of T. gondii in surface water samples in the State of São Paulo and opens perspectives for other studies that may guide future investigations and support policymakers in guiding new regulations in relation to water quality.
The experience obtained in this study in the evaluation of the performance of the method for the recovery of T. gondii in surface water matrices and also in the quantification of the parasite by qPCR provided important subsidies for the continuity of research in this area. In order to elucidate the questions generated in relation to our recovery results obtained in surface water samples, we believe that conducting new research is key to verify the percentages of loss in each step of the method, in order to achieve efficiency in the recovery of this parasite in environmental samples due to their relevance and impact on human health.
We considered the qPCR test implemented in this study as a rapid, sensitive, and specific tool for the detection and quantification of T. gondii in surface water samples, thus contributing to the risk management of public water supply systems.
References
Aubert D, Villena I (2009) Detection of Toxoplasma gondii oocysts in water: proposition of a strategy and evaluation in Champagne-Ardenne region, France. Mem Inst Oswaldo Cruz 104:290–295. https://doi.org/10.1590/S0074-02762009000200023
Borchardt MA, Spencer SK, Bertz PD, Ware MW, Dubey JP, Alan Lindquist HD (2009) Concentrating Toxoplasma gondii and Cyclospora cayetanensis from surface water and drinking water by continuous separation channel centrifugation. J Appl Microbiol 107:1089–1097. https://doi.org/10.1111/j.1365-2672.2009.04316.x
Burg JL, Grover CM, Pouletty P, Boothroyd JC (1989) Direct and sensitive detection of a pathogenic protozoan, Toxoplasma gondii, by polymerase chain reaction. J Clin Microbiol 27:1787–1792
Bushkin GG, Motari E, Carpentieri A (2013) Evidence for a structural role for acid-fast lipids in oocyst walls of Cryptosporidium, Toxoplasma, and Eimeria. mBio 4:1–8. https://doi.org/10.1128/mBio.00387-13.Editor
CETESB (2015) Qualidade das águas superficias no Estado de São Paulo 2014 (Quality of superficialwaters in Sao Paulo State 2014)
CETESB/ANA (2011) Guia Nacional de Coleta e Preservação de Amostras - Água, Sedimento, Comunidades Aquáticas e Efluentes Líquidos (National guide for sampling and preservation of samples:water, sediment, aquatic communities and wastewater). Cia Ambiental do Estado São Paulo, 326p
Contini C, Seraceni S, Cultrera R, Incorvaia C, Sebastiani A, Picot S (2005) Evaluation of a real-time PCR-based assay using the lightcycler system for detection of Toxoplasma gondii bradyzoite genes in blood specimens from patients with toxoplasmic retinochoroiditis. Int J Parasitol 35:275–283. https://doi.org/10.1016/j.ijpara.2004.11.016
De Moura L, Garcia Bahia-Oliveira LM, Wada MY et al (2006) Waterborne toxoplasmosis, Brazil, from field to gene. Emerg Infect Dis 12:326–329. https://doi.org/10.3201/eid1202.041115
Digiorgio CL, Gonzalez DA, Huitt CC (2002) Cryptosporidium and Giardia recoveries in natural waters by using Environmental Protection Agency method 1623. Appl Environ Microbiol 68:5952–5955. https://doi.org/10.1128/aem.68.12.5952-5955.2002
Dubey JP, Lago EG, Gennari SM et al (2012) Toxoplasmosis in humans and animals in Brazil: high prevalence, high burden of disease, and epidemiology. Parasitology 139:1375–1424. https://doi.org/10.1017/S0031182012000765
Dumetre A, Dubey JP, Ferguson DJ et al (2013) Mechanics of the Toxoplasma gondii oocyst wall. Proc Natl Acad Sci U S A 110:11535–11540. https://doi.org/10.1073/pnas.1308425110
Edvinsson B, Lappalainen M, Evengard B et al (2006) Real-time PCR targeting a 529-bp repeat element for diagnosis of toxoplasmosis. Clin Microbiol Infect 12:131–136. https://doi.org/10.1111/j.1469-0691.2005.01332.x
Fumian TM, Leite JPG, Rocha MS, de Andrade JSR, Fioretti JM, de Assis RMS, Assis MRS, Fialho AM, Miagostovich MP (2016) Performance of a one-step quantitative duplex RT-PCR for detection of rotavirus A and noroviruses GII during two periods of high viral circulation. J Virol Methods 228:123–129. https://doi.org/10.1016/j.jviromet.2015.11.008
Gallas-Lindemann C, Sotiriadou I, Mahmoodi MR, Karanis P (2013) Detection of Toxoplasma gondii oocysts in different water resources by loop mediated isothermal amplification (LAMP). Acta Trop 125:231–236. https://doi.org/10.1016/j.actatropica.2012.10.007
Garcia Bahia-Oliveira LM, Jones JL, Azevedo-Silva J et al (2003) Highly endemic, waterborne toxoplasmosis in North Rio de Janeiro State, Brazil. Emerg Infect Dis 9:55–62. https://doi.org/10.3201/eid0901.020160
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Harito JB, Campbell AT, Tysnes KR, Dubey JP, Robertson LJ (2017) Lectin-magnetic separation (LMS) for isolation of Toxoplasma gondii oocysts from concentrated water samples prior to detection by microscopy or qPCR. Water Res 114:228–236. https://doi.org/10.1016/j.watres.2017.02.044
Hartmann K, Addie D, Belák S et al (2013) Toxoplasma Gondii infection in cats: ABCD guidelines on prevention and management. https://doi.org/10.1177/1098612X13489228
Hohweyer J, Cazeaux C, Travaillé E, Languet E, Dumètre A, Aubert D, Terryn C, Dubey JP, Azas N, Houssin M, Loïc F, Villena I, la Carbona S (2016) Simultaneous detection of the protozoan parasites Toxoplasma, Cryptosporidium and Giardia in food matrices and their persistence on basil leaves. Food Microbiol 57:36–44. https://doi.org/10.1016/j.fm.2016.01.002
Homan WL, Vercammen M, De Braekeleer J, Verschueren H (2000) Identification of a 200- to 300-fold repetitive 529 bp DNA fragment in Toxoplasma gondii, and its use for diagnostic and quantitative PCR. Int J Parasitol 30:69–75. https://doi.org/10.1016/S0020-7519(99)00170-8
Isaac-Renton J, Bowie WR, King A et al (1998) Detection of Toxoplasma gondii oocysts in drinking water. Appl Environ Microbiol 64:2278–2280
Jones JL, Dubey JP (2010) Waterborne toxoplasmosis - recent developments. Exp Parasitol 124:10–25. https://doi.org/10.1016/j.exppara.2009.03.013
Karanis P, Aldeyarbi HM, Mirhashemi ME, Khalil KM (2013) The impact of the waterborne transmission of Toxoplasma gondii and analysis efforts for water detection: an overview and update. Environ Sci Pollut Res 20:86–99. https://doi.org/10.1007/s11356-012-1177-5
Kerambrun E, Ladeiro MP, Dupuis E, Villena I (2015) Zebra mussel as a new tool to show evidence of freshwater contamination by waterborne Toxoplasma gondii. J Appl Microbiol 120:498–508. https://doi.org/10.1111/jam.12999
Kompalic-Cristo A, Frotta C, Suárez-Mutis M, Fernandes O, Britto C (2007) Evaluation of a real-time PCR assay based on the repetitive B1 gene for the detection of Toxoplasma gondii in human peripheral blood. Parasitol Res 101:619–625. https://doi.org/10.1007/s00436-007-0524-9
Kourenti C, Heckeroth A, Tenter A, Karanis P (2003) Development and application of different methods for the detection of Toxoplasma gondii in water. Society 69:102–106. https://doi.org/10.1128/aem.69.1.102-106.2003
Kuhn RC, Oshima KH (2002) Hollow-fiber ultrafiltration of Cryptosporidium parvum oocysts from a wide variety of 10-L surface water samples. NRC Research Press 549:542–549. https://doi.org/10.1139/W02-049
Lora-Suarez F, Rivera R, Triviño-Valencia J, Gomez-Marin JE (2016) Detection of protozoa in water samples by formalin/ether concentration method. Water Res 100:377–381. https://doi.org/10.1016/j.watres.2016.05.038
Mahmoudi MR, Kazemi B, Haghighi A, Karanis P (2015) Detection of Acanthamoeba and Toxoplasma in river water samples by molecular methods in Iran. Iran J Parasitol 10:250–257
Mccuin RM, Clancy JL (2005) Methods for the recovery, isolation and detection of Cryptosporidium oocysts in wastewaters. J Microbiol Methods 63:73–88. https://doi.org/10.1016/j.mimet.2005.02.020
Nieminski EVAC, Iii FWS (1995) Comparison of two methods for detection of Giardia cysts and Cryptosporidium oocysts in water. Appl Environ Microbiol 61:1714–1719
Palos Ladeiro M, Bigot-Clivot A, Aubert D et al (2015) Assessment of Toxoplasma gondii levels in zebra mussel (Dreissena polymorpha) by real-time PCR: an organotropism study. Environ Sci Pollut Res 2. https://doi.org/10.1007/s11356-015-4296-y
Paulos S, Mateo M, de Lucio A, Hernández-de Mingo M, Bailo B, Saugar JM, Cardona GA, Fuentes I, Mateo M, Carmena D (2016) Evaluation of five commercial methods for the extraction and purification of DNA from human faecal samples for downstream molecular detection of the enteric protozoan parasites Cryptosporidium spp., Giardia duodenalis, and Entamoeba spp. NU SC. J Microbiol Methods 127:68–73. https://doi.org/10.1016/j.mimet.2016.05.020
Pena HFJ, Soares RM (2006) Toxoplasma gondii infection in cats from São Paulo state, Brazil: seroprevalence, oocyst shedding, isolation in mice, and biologic and molecular characterization. Res Vet Sci 81:58–67. https://doi.org/10.1016/j.rvsc.2005.09.007
Pfaffl MW (2004) Quantification strategies in real-time PCR. In: A-Z of Quantitative PCR, Bustin SA editor, 87–112p., FIVEphoton Biochemicals
Ramírez-Castillo F, Loera-Muro A, Jacques M, Garneau P, Avelar-González F, Harel J, Guerrero-Barrera A (2015) Waterborne pathogens: detection methods and challenges. Pathogens 4:307–334. https://doi.org/10.3390/pathogens4020307
Rosado-García FM, Guerrero-Flórez M, Karanis G, Hinojosa MDC, Karanis P (2017) Water-borne protozoa parasites: the Latin American perspective. Int J Hyg Environ Health 220:783–798. https://doi.org/10.1016/j.ijheh.2017.03.008
Samuelson J, Bushkin GG, Chatterjee A, Robbins PW (2013) Strategies to discover the structural components of cyst and oocyst walls. Eukaryot Cell 12:1578–1587. https://doi.org/10.1128/EC.00213-13
Shapiro K, Vanwormer E, Aguilar B, Conrad PA (2015) Surveillance for Toxoplasma gondii in California mussels (Mytilus californianus) reveals transmission of atypical genotypes from land to sea. Environ Microbiol 17:4177–4188. https://doi.org/10.1111/1462-2920.12685
Shwab EK, Zhu X-Q, Majumdar D et al (2014) Geographical patterns of Toxoplasma gondii genetic diversity revealed by multilocus PCR-RFLP genotyping. Parasitology 141:453–461. https://doi.org/10.1017/S0031182013001844
Sobral CA, Amendoeira MRR, Teva A et al (2005) Seroprevalence of infection with Toxoplasma gondii in indigenous Brazilian populations. Am J Trop Med Hyg 72:37–41
Sotiriadou I, Karanis P (2008) Evaluation of loop-mediated isothermal amplification for detection of Toxoplasma gondii in water samples and comparative findings by polymerase chain reaction and immunofluorescence test (IFT). Diagn Microbiol Infect Dis 62:357–365. https://doi.org/10.1016/j.diagmicrobio.2008.07.009
Torrey EF, Yolken RH (2013) Toxoplasma oocysts as a public health problem. Trends Parasitol 29:380–384. https://doi.org/10.1016/j.pt.2013.06.001
Tourinho RS, Almeida CR, De Lemos AS (2015) Application of synthetic standard curves for absolute quantification of hepatitis A and E by real-time PCR. J Genet Genome Res 2:1
USEPA (2012) USEPA method 1623.1: Cryptosporidium and Giardia in water by filtration/IMS/FA. EPA 816-R-12
Vieira FP, Alves G, Martins LM et al (2015) Waterborne toxoplasmosis investigated and analysed under hydrogeological assessment: new data and perspectives for further research. Mem Inst Oswaldo Cruz 110:929–935. https://doi.org/10.1590/0074-02760150262
Villena I, Aubert D, Gomis P, Ferte H, Inglard JC, Denis-Bisiaux H, Dondon JM, Pisano E, Ortis N, Pinon JM (2004) Evaluation of a strategy for Toxoplasma gondii oocyst detection in water. Appl Environ Microbiol 70:4035–4039. https://doi.org/10.1128/AEM.70.7.4035-4039.2004
Wells B, Shaw H, Innocent G, Guido S, Hotchkiss E, Parigi M, Opsteegh M, Green J, Gillespie S, Innes EA, Katzer F (2015) Molecular detection of Toxoplasma gondii in water samples from Scotland and a comparison between the 529bp real-time PCR and ITS1 nested PCR. Water Res 87:175–181. https://doi.org/10.1016/j.watres.2015.09.015
WHO/UNICEF (2010) Progress on sanitation and drinking water: 2010 Update. WHO Libr 60
Yang W, Lindquist HDA, Cama V, Schaefer FW, Villegas E, Fayer R, Lewis EJ, Feng Y, Xiao L (2009) Detection of Toxoplasma gondii oocysts in water sample concentrates by real-time PCR. Appl Environ Microbiol 75:3477–3483. https://doi.org/10.1128/AEM.00285-09
Acknowledgements
To Professor Dr. Hilda Fátima de Jesus Pena, Professor Dr. Solange Maria Gennari, and Professor Dr. Rodrigo Martins Soares, from the Laboratory of the Department of Preventive Medicine and Animal Health, School of Veterinary and Zootechnic Medicine, University of Sao Paulo, for their key contributions and donations (Toxoplasma gondi oocysts). Additionally, we are grateful to Professor Dr. Luciana Regina Meireles Jaguaribe Ekman from the Tropical Medicine Institute, University of Sao Paulo, for their substantial contributions in the development of the present study and Dr. Elayse Maria Hachich from the Division of Microbiology and Parasitology of São Paulo State Environmental Company for their support at the beginning of this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Section Editor: Panagiotis Karanis
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Galvani, A.T., Christ, A.P.G., Padula, J.A. et al. Real-time PCR detection of Toxoplasma gondii in surface water samples in São Paulo, Brazil. Parasitol Res 118, 631–640 (2019). https://doi.org/10.1007/s00436-018-6185-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-018-6185-z