Abstract
Anisakid nematodes are the most infamous parasites occurring in seafood with ability to infect humans. In the present study, the infective stages of five anisakid larval types, including Anisakis types I and III, Terranova types I and II and Contracaecum larval type, as well as adult Anisakis paggiae are reported from 16 host species from New Caledonian waters. The specific identity of the larval types was investigated using ITS sequence data. Anisakis larval types I and III were identified as Anisakis typica and Anisakis brevispiculata, respectively, based on identical ITS sequences. However, the specific identity of the Terranova larval types and Contracaecum larval type remains unknown until a matching ITS sequence from a well-identified adult is available. Several fish host species are reported for the first time for anisakid larval types found in this study. Considering that third-stage larvae of anisakids are known to be the infective stage of the parasite for humans and the popularity of seafood in New Caledonia, presence of these parasites in New Caledonian fish is of high importance in terms of public health and raising awareness among various stakeholders. Although adult nematodes in the present study were identified as Anisakis paggiae, the spicule length is shorter in our specimens and falls within the range reported for Anisakis oceanicus previously reported in Pacific waters from black fish (genus Globicephala) and later synonymised with Anisakis physeteris. However, our specimens are different from A. physeteris in morphology of ventriculus. Anisakis paggiae has been reported from whales in southern hemisphere and this is the first report from the Pacific regions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Seafood products account for more than 40% of the protein intake of people in New Caledonia (Gontard and de Coudenhove 2013). Although there have been studies on marine parasites from this area, most of these studies cover a range of parasitic phyla (Justine 2007, 2010; Justine et al. 2010a, b, 2012a; Myers et al. 2000) and the focus is rarely just on nematodes of zoonotic importance. Identification of nematodes, particularly for zoonotic marine ascaridoids, is often not completed down to the species level in these studies and molecular analyses are often not used to specifically identify larvae. Identification of potentially zoonotic parasites is especially important due to the high consumption of fish and seafood in this area, including raw fish. A series of lists of fish parasites from New Caledonia have been published, mainly for the fish families Epinephelidae, Lethrinidae and Lutjanidae (Justine et al. 2010a, b, 2012a) and other families (Justine 2010), including mainly coral reef fish and also some deep-sea fish from the external slope of the coral reef. However, the authors pointed out that the results were only about a fraction of the true biodiversity of an ecosystem as many species were yet unidentified. It was outlined that many records simply labelled as ‘unidentified anisakid larvae’ were certainly hiding a higher diversity. Our previous works have aimed at elucidating this hidden biodiversity of anisakid larvae (Shamsi et al. 2015, 2017).
The aim of the present study was to identify nematode parasites found in marine fish and selected aquatic animals in New Caledonian waters using combined molecular and morphological approaches. This is an essential step if any future work is to be done on areas such as risk assessment, control, prevention and seafood safety. The focus of this study is on anisakid nematode parasites only, due to their zoonotic significance.
Materials and methods
Hosts
Fish were mostly purchased from the fish market and were dead at the time of purchase. Fish from other places were collected under permit from Province Sud, New Caledonia, given to Institut de Recherche pour le Développement (IRD), Nouméa, and were killed according to local and French laws and humane rules. Stranded mammals were dead at the time of the parasitological inspection. Host morphological identification was performed by Jean-Lou Justine using current literature (Froese and Pauly 2018) and then confirmed by ichthyologists (see acknowledgements). In some instances, the precise identification of fish hosts was problematic; these fish did not perfectly match descriptions of existing species and therefore have only been identified to the genus level.
Parasite collection
Parasites (larvae and adults) were generally collected via a variation of the wash method (Justine et al. 2012b) from the abdominal cavities of the fish. Most were found in the stomach and intestinal lumen but some were found in encapsulations on surfaces of the abdominal organs. Specimens were washed in saline and fixed in 70% ethanol. Specimens from marine mammals were collected by other scientists and given to Jean-Lou Justine years after their collection. All parasites were deposited in Museum National d’Histoire Naturelle, Paris.
Morphology
A small cross-section of the mid body from each parasite was excised and kept frozen for molecular studies. The anterior and posterior ends were then mounted on microscope slides and cleared in lactophenol for morphological examination. For larger, thicker worms, the anterior and posterior ends were soaked in Berland’s fluid for 48 h prior to being mounted for additional clearing. Specimens were assigned into distinct groups based on the morphology of lips (labia), tail, reproductive, digestive and excretory systems (Cannon 1977; Jabbar et al. 2012; Murata et al. 2011; Shamsi et al. 2011, 2012; Shamsi and Suthar 2016). A number of representatives from each group were selected for detailed measurement of important bodily features. A drawing of each parasite was made using a microscope equipped with a drawing tube. Images were captured via a light microscope equipped with a digital camera.
PCR and sequencing
For the groups that were subject to molecular study, one representative from each parasite group per host species was selected for molecular examination. Genomic DNA (gDNA) was isolated from each individual specimen via sodium dodecyl-sulphate/proteinase K treatment, column-purified (Wizard™ DNA Clean-Up, Promega) and eluted into 45 μl of water. PCR was used to amplify the ITS-1 and ITS-2 regions using primer sets SS1: 50-GTTTCCGTAGGTGAACCTGCG-30 (forward) and NC13R: 50-GCTGCGTTCTTCATCGAT-30 (reverse) for the former and SS2: 50-TTGCAGACACATTGAGCACT-30 (forward) and NC2: 50-TTAGTTTCTTTTCCTCCGCT-30 (reverse) for the latter region (Shamsi and Suthar 2016). Cycling conditions: initial 94 °C/5′, then 94 °C/30″, 55 °C/40″, 72 °C/40″ × 30 cycles, 72 °C/5 extension and 4 °C. A 4 μl aliquot of each amplicon was examined on a 1.5% w/v agarose gel. Amplicons were purified over mini-columns (Wizard™ PCR Prep, Promega, WI, USA), eluted in 35 μl H2O and then subjected to automated sequencing using the same primers as for PCR (Table 1).
Phylogenetic analyses
ITS-1 and ITS-2 sequences were either generated in our current study, or were obtained from GenBank (Table 2). If obtained from GenBank, only sequences from well-identified adults for which museum vouchers are available were considered. ITS-1 and ITS-2 sequence data were concatenated by using Geneious version 11.0.5 (Kearse et al. 2012). Combined sequences were aligned by Geneious alignment algorithm, and then were double checked with all variable sites in the original trace files for confirmation. Alignments were then truncated to 768, 794 and 711 characters, based on the shortest sequence of the alignment, for Contracaecum spp., Anisakis spp. and Terranova spp., respectively. Same gene region from Heterakis gallinarum was used as an outgroup. Phylogenetic relationship among species was calculated by MEGA7.0.26 (Kumar et al. 2016) using neighbour-joining method with p-distances model.
Results
Twenty-three host individuals from 16 species were examined. Five larval types belonging to three genera, Anisakis, Contracaecum and Terranova and one adult, Anisakis paggiae were found. Table 1 shows occurrence of anisakids in various hosts. Below detailed characteristics for each anisakid nematode found in this study is provided followed by a phylogenetic tree showing their genetic relationships. For the following results sections, all measurements are in millimetres unless otherwise stated. Mean measurements are given followed by range and number of specimens measured in parentheses.
Morphological and genetic characterisation
Anisakis larval type I of Cannon 1977
Materials examined: Two from Echeneis naucrates (Museum accession number: JNC209A), one from Epinephelus areolatus (Museum accession number: JNC204H), one from Nemipterus furcosus (Museum accession number: JNC252E), two from Upeneus vittatus (Museum accession numbers: JNC3348A and 3347), one from Carangoides fulvoguttatus (Museum accession number: JNC3298), one from Scomberoides sp. (Museum accession number: JNC3378A) and one from Decapterus macarellus (Museum accession number: JNC2018).
Morphometrics: Third-stage larvae (Fig. 1a, b). Body length 19.38 (15.1–23.925, n = 9), maximum body width 0.46 (0.34–0.6, n = 9). Cuticle with fine transverse annulations, most apparent on tail. Poorly developed labia. Boring tooth present. Excretory pore immediately below tooth. Nerve ring 0.29 (0.24–0.4, n = 9) from anterior end. Muscular oesophagus, length 1.79 (1.425–2.05, n = 9), 9.27% (7.35–10.32%, n = 9) of total body length, ending in glandular ventriculus. Ventriculus 0.76 (0.475–1.05, n = 9) in length, 3.92% (2.45–5.42%, n = 9) of total body length and joined obliquely to intestine (Fig. 1a). No caeca or diverticula present. Rectum short and oblique to anus. Tail conical, short, with rounded tip, decorated with a single mucron. Anus 0.13 (0.09–0.23, n = 9) from posterior end.
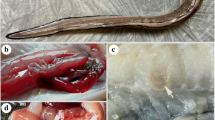
Nematodes found in the present study. a, b Anisakis larval type I from Echeneis naucrates (specimen number: 206–10); a anterior end, scale bar 0.5 mm, b posterior end, scale bar 0.25 mm. c, d Anisakis larval type III from Beryx splendens (specimen number: 274–9); c anterior end, scale bar 0.5 mm; d posterior end, scale bar 0.25 mm. e, f Terranova larval type I from Epinephelus ongus, specimen number 266-1; e anterior end, scale bar 0.5 mm; f posterior end, scale bar 0.25 mm. g, h Terranova larval type II from Echeneis naucrates, specimen number 206-13; g anterior end, scale bar 0.25 mm; h posterior end, scale bar 0.25 mm. i, j Contracaecum larval type from Periopthalmus argentilineatus, specimen number 261-5; i anterior end, scale bar 0.25 mm; j posterior end, scale bar 0.25 mm. k–m Adult Anisakis paggiae males from Kogia breviceps; k anterior end, scale bar 1 mm; l posterior end lateral view, scale bar 0.25 mm; m posterior end ventral view, scale bar 0.25 mm
Genetic characterisation: Nine specimens, one from each fish species, were selected to obtain their ITS-1and ITS-2 sequences. ITS-1 (accession numbers MH190354-62) to and ITS-2 (accession numbers MH190312-20) were 348 and 355 bp in length, respectively. No nucleotide differences were observed within ITS-1 or ITS-2 sequences among specimens in the present study. Sequences were identical to those of Anisakis typica (GenBank accession number: MF642334 and MF642335) and were grouped phylogenetically with previously characterised Anisakis typica (Fig. 2).
Phylogenetic analysis of the concatenated ITS-1 and ITS-2 sequence data for Contracaecum spp., Anisakis spp. and Terranova spp., with Heterakis gallinarum as an outgroup, using the using neighbour-joining method with p-distances model. Bootstrap (1000 replicates) support values are indicated above the branches. Numbers after taxa refer to ITS-1 and ITS-2, respectively. If only one number is presented, it indicates that ITS sequence data were extracted from a bigger region of rDNA
Anisakis larval type III (Murata et al. 2011 )
Materials examined: Ten specimens from Beryx splendens (Museum accession number: JNC1576).
Morphometrics: Third-stage larvae (Fig. 1c, d). Body length 23.32 (15.95–27.9, n = 10). Maximum body width 0.83 (0.63–1.05, n = 10). Thick, transversely annulated cuticle. Poorly developed labia. Boring tooth present. Excretory pore immediately below tooth. Nerve ring 0.33 (0.25–0.38, n = 10) from anterior end. Muscular oesophagus, 1.46 (0.425–2.18, n = 10) in length, 6.26% (1.82–9.35%, n = 10) of total body length, ending in glandular ventriculus. Ventriculus 0.65 (0.3–1.53, n = 10) long, 2.79% (1.29–6.54%, n = 10) of total body length. No caeca or diverticula present. Rectum short and oblique to anus. Tail conical, short with rounded tip, decorated with a single mucron. Anus 0.11 (0.05–0.18, n = 10) from posterior end.
Genetic characterisation: Four specimens were subjected to PCR for amplification and obtaining sequences of the ITS-1 and ITS-2. ITS-1 and ITS-2 were 375 and 272 bp long respectively (accession numbers MH190363-6 and MH190321-4). Apart from one specimen (GenBank accession number MH190365) which had different nucleotide at alignment position 117 (in the ITS-1 region), ITS sequences were identical among our specimens. A search in GenBank for matching sequences belonging to adult Anisakis spp. showed that adult A. brevispiculata (KC342887 and FN891881) from the Philippines (Quiazon et al. 2013) and Australian waters (Shamsi et al. 2012), respectively, are almost identical with our specimens.
Terranova larval type I of Cannon ( 1977 )
Materials examined: Eight specimens from Saurida undosquamis (numbers: JNC571B), Sufflamen fraenatum (JNC297B), Scomberoides sp. (JNC3378B), Atule mate (JNC3365), Epinephelus ongus (JNC3275), Decapterus macarellus (JNC2019A) and Lutjanus rivulatus (JNC1864).
Morphometrics: Third-stage larvae (Fig. 1e, f). Body length 9.92 (7.5–12.55, n = 8). Maximum body width 0.25 (0.19–0.32, n = 8). Delicately annulated cuticle. Poorly developed labia. Boring tooth present. Excretory pore immediately below tooth. Nerve ring 0.27 (0.2–0.33, n = 8) from anterior end. Muscular oesophagus, 1 (0.7–1.28, n = 8) long, 10.08% (7.06–12.9%, n = 8) of total body length. Oesophagus ending in elongated glandular ventriculus, 1.09 (0.35–1.45, n = 8) long, 10.99% (3.53–14.62%, n = 8) of total body length. Intestinal caecum present, 1.23 (0.6–1.7, n = 8) long. Ratio of intestinal caecum to ventriculus 1.13:1 (1.23, 1.09, n = 8). Tail strongly annulated, conical, smoothly tapered. Anus 0.13 (0.11–0.17, n = 8) from posterior end.
Genetic characterisation: Eight specimens including one from each above mentioned host (accession numbers MH190367-74) plus one from Variola albimarginata (accession number MH190375) were sequenced in both ITS-1 and ITS-2. The length of ITS-1 was 437 bp. ITS-1 was identical among all sequences as well as with GenBank accession number KC437344, belonging to Terranova sp. type I from Western Australian waters (Jabbar et al. 2013). The length of ITS-2 was 266 bp, identical among sequences (MH190325-33) including two nucleotide ambiguities at alignment positions 43 (Y) and 147 (R), as well as identical to JX848681 belonging to Terranova larval type I from Lizard Island in Australia (Jabbar et al. 2012). Terranova type I in the present study was grouped with previously known Terranova type I from southern hemisphere.
Terranova larval type II of Cannon ( 1977 )
Materials examined: Ten specimens from Carangoides cf. orthogrammus (JNC569C), Scomberomorus commerson (JNC898D), Epinephelus areolatus (JNC205E), Echeneis naucrates (JNC209A), Nemipterus furcosus (JNC279B), Sphyraena qunie (JNC394A), Scomberoides sp. (JNC3378B) and Atule mate (JNC3365).
Morphometrics: Third-stage larvae (Fig. 1g, h). Body length 6.63 (5.42–8.3, n = 10). Maximum body width 0.23 (0.18–0.28, n = 10). Delicately annulated cuticle. Poorly developed labia. Boring tooth present. Excretory pore immediately below tooth. Nerve ring 0.25 (0.22–0.32, n = 10) from anterior end. Muscular oesophagus, 0.85 (0.73–1.03, n = 10) long, 12.82% (11.01–15.54%, n = 10) of total body length, ending in oval-shaped glandular ventriculus. Ventriculus 0.34 (0.29–0.38, n = 10) long, 5.13% (4.37–5.73%, n = 10) of total body length. Intestinal caecum present, 0.68 (0.61–0.85, n = 10) long. Ratio of intestinal caecum to ventriculus 2:1 (0.68, 0.34, n = 10). Tail strongly annulated, conical, smoothly tapered. Anus 0.13 (0.09–0.15, n = 10) from posterior end.
Genetic characterisation: One specimen from each host mentioned above, except for Nemipterus furcosus, plus one from Sufflamen frenatum each were subjected to amplification of the ITS-1 and ITS-2 regions and subsequent sequencing.
ITS-1 was 437 bp, except for specimen from Saurida undosquamis (accession number MH190376); all had identical sequence (accession numbers MH190377-84). The difference was for alignment position 99 for which it was CC in MH190376 instead of CT for others. Blast in GenBank resulted showed our specimens have identical ITS-1 sequence as LN795828 previously reported in Australian waters. ITS-2 was 252 bp and identical among all sequences in the present study (MH190334-42). Blast in GenBank resulted showed our specimens have identical ITS-2 sequence as LN651110 previously reported in New Caledonian waters. Phylogenetic analyses showed that all Terranova type II are grouped together (Fig. 2).
Contracaecum larval type
Materials examined: Eight specimens from Periophthalmus argentilineatus (JNC 3320).
Morphometrics: Third-stage larvae (Fig. 1i, j). Body length 3.43 (2.67–4.25, n = 8). Maximum body width 0.14 (0.11–0.2, n = 8). Cuticle strongly annulated. Poorly developed labia. Boring tooth present. Excretory pore located ventrally, immediately below boring tooth. Nerve ring 0.19 (0.17–0.24, n = 8) from anterior end. Muscular oesophagus 0.45 (0.38–0.53, n = 8) in length, 13.12% (11.08–15.45%, n = 8) of total body length. Oesophagus ending in glandular, subglobular ventriculus. Ventriculus length 0.04 (0.03–0.05, n = 8), 1.17% (0.87–1.46%, n = 8) of total body length. Intestinal caecum length 0.27 (0.2–0.33, n = 8). Ventricular appendix 0.35 (0.25–0.42, n = 8) long. Ratio of intestinal caecum to ventricular appendix 0.77:1 (0.27, 0.35, n = 8). Tail conical and without mucron. Anus 0.1 (0.09–1.1, n = 6) from posterior end.
Genetic characterisation: All specimens above plus four additional specimens from the same fish species were subjected to amplification of the ITS-1 and ITS-2 region followed by the sequences. ITS-1 was at least 443 bp (accession numbers MH190385-95) with two polymorphic sites including alignment positions 53 and 415 which were A and G, respectively in MH190392, MH190393, MH190395, and G and A in the remaining sequences. ITS-2 was 260 bp, identical among all specimens (accession numbers MH190343-53) except for alignment position 63 which was T in four specimens (MH190353, MH190343, MH190344 and MH190348) and C in the remaining specimens. No highly similar sequence was found in GenBank.
Anisakis paggiae adults
Materials examined: Ten specimens; five male, five female (museum accession numbers: JNC 1735 and JNC 1531 from hosts Kogia sima and Kogia breviceps.
Morphometrics:
Males (Fig. 1k–m) with fine transverse annulated cuticle. Body length 35.12 (15.98–49.25, n = 5). Three protruding labia, 0.1 (0.07–0.15, n = 5) in height, 0.13 (0.12–0.16, n = 5) in width. Dorsal and ventro-lateral labia with low anterior projection with dentigerous ridges on inner surface. Dorsal lip with two double papillae. Ventro-lateral lips with one double papilla, one single papilla and one amphid. Deirids small and papillate, located around level of first third of oesophagus, posterior to nerve ring, 0.74 (0.52–0.9, n = 8) from anterior end. Nerve ring 0.55 (0.29–1.15, n = 5) from anterior end. Muscular oesophagus 2.63 (2.08–3.48, n = 5) in length, 7.49% (5.92–9.91%, n = 5) of total body length, ending in glandular ventriculus. Ventriculus violin-shaped with a distinct constriction in the middle, 0.53 (0.43–0.62, n = 5) in length, 1.51% (1.22–1.77%, n = 5) of total body length. No caeca or diverticula present. Two short, stout and slightly subequal spicules present. Right spicule 0.2 (0.14–0.31, n = 5), left spicule 0.22 (0.15–0.32, n = 5). Ratio between right and left spicule = 1:1.1. Tail 0.2 (0.16–0.25, n = 5) in length. Three narrow denticulate caudal plates (plectanes) present, posterior to cloaca. Pre-cloacal papillae pairs numerous and arranged in single rows. One median papilla and one pair of proximal papillae lateral to cloaca. One pair of double paracloacal papillae. Four pairs of distal papillae.
Females with fine transverse annulated cuticle. Body length 39.22 (25.43–46.83, n = 5). Maximum body width 0.82 (0.37–1.15, n = 5). Three protruding labia, 0.1 (0.08–0.12, n = 5) in height, 0.12 (0.1–0.15, n = 5) in width. Dorsal and ventro-lateral labia with low anterior projection with dentigerous ridges on inner surface. Dorsal lip with two double papillae. Ventro-lateral lips with 1 double papilla, one single papilla and one amphid. Deirids small and papillate, located around level of first third of oesophagus, posterior to nerve ring, 0.89 (0.58–1.13, n = 8) from anterior end. Nerve ring 0.54 (0.36–0.83, n = 5) from anterior end. Muscular oesophagus 3.03 (2.23–3.83, n = 5) in length, 7.73% (5.69–9.77%, n = 5) of total body length, ending in glandular ventriculus. Ventriculus violin-shaped with a distinct constriction in the middle, 0.57 (0.42–0.65, n = 5) long, 1.45% (1.07–1.66%, n = 5) of total body length. No caeca or diverticula present. Vulva small, 11.13 (8.75–13.38, n = 5) from anterior end. Tail conical with rounded tip. No mucron present. Anus 0.28 (0.18–0.35, n = 5) from posterior end.
Genetic characterisation: These specimens were fixed in formalin and therefore molecular work could not be carried out.
Phylogenetic analyses of taxa found in this study and comparison with congeners
Phylogenetic analysis of the ITS-1 and ITS-2 sequence data was performed using neighbour-joining method. Phylogenetic analyses showed that the five larval types found in the present study were genetically distinct (Fig. 2), divided into three distinct clades. Clade I (green highlighted) included members of Terranova larval types found in the resent study, revealing clear distinction between types I and II supported by a strong bootstrap value. Clade II (pink highlighted) included members of Contracaecum larva, all resolved in one group. The group was distinct from other Contracaecum spp. found in Australia and/or for which comparable data were available in GenBank. Clade III (blue highlighted) represented Anisakis spp. Two Anisakis larval types found in the present study were resolved separately. Anisakis type I was grouped with members of Anisakis typica and Anisakis type III was clustered with members of A. brevispiculata.
Discussion
Five morphotypes belonging to three genera of anisakid nematodes, including Anisakis, Terranova and Contracaecum, were found in New Caledonian waters during this study. The morphological distinction between larvae was supported by ITS sequence data. Phylogenetic analysis revealed presence of three distinct clades in which sequences from these three genera clustered. Clade I included the most common anisakid larvae in the present study, Terranova larval types, including larval types I and II which could be morphologically differentiated based on the ratio of intestinal caecum length to ventriculus length. They were considerably different in their ITS sequence data and resolved separately in the phylogenetic tree (Fig. 2). However, not much is known about the specific identity of Terranova larval types yet. Larvae referred to as ‘Terranova sp.’ have the potential to belong to a number of different genera including Pseudoterranova, Terranova and Pulchrascaris (Shamsi and Suthar, 2016). Although adults of Terranova scoliodontis have been reported from the tiger shark, Galeocerdo cuvier, off New Caledonia (Moravec and Justine 2006), there is no comparable molecular data available to investigate its relationship with larvae found in the present study.
Terranova larval type I has not been previously reported in New Caledonian fish, therefore the present study is a new geographical record; however, previous publications have listed specimens found simply as ‘Terranova sp.’ larvae with no further clarification as to type. Terranova larval type II was reported from New Caledonia previously (Shamsi et al. 2015) in 11 host species. Except for Atule mate and Carangoides cf. orthogrammus, the remaining six host species; Echeneis naucrates (live sharksucker), Epinephelus areolatus (areolate grouper), Nemipterus furcosus (fork-tailed threadfin bream), Scomberoides sp., Scomberomorus commerson (narrow-barred Spanish mackerel) and Sphyraena qunie (great barracuda) are new host records. Hosts A. mate and Scomberoides sp. were infected with both larval types I and II. ITS region sequences were a match for those of specimens obtained from larvae from 13 species of Australian fishes (Shamsi and Suthar 2016). ITS-1 and ITS-2 regions in specimens examined by Shamsi and Suthar (2016) showed a high degree of similarity, therefore suggesting they all belong to one species. However, there is a lack of comparable sequence data from well-described adults in GenBank, so the specific identity of specimens from the present study and from Shamsi and Suthar (2016) remain unknown.
Finding of Contracaecum larval type is another interesting finding of the present study. Human infection due to Contracaecum larval type can occur (Shamsi and Butcher 2011); however, Periophthalmus argentilineatus is not an edible fish. In the phylogenetic tree, all Contracaecum larvae found in the present study were grouped separately from all other known Contracaecum spp. reported in Australia and also from those with the highest similar ITS sequence (Fig. 2), suggesting it may belong to a new species or to a known species for which ITS sequence data is not available. There is no report of adult Contracaecum from New Caledonia but presence of the infective stage of the parasite in fish suggests that adults must be present in birds or marine mammals nearby. Periophthalmus argentilineatus is a mangrove fish and the parasite cycle may involve other hosts. Our specimens of P. argentilineatus were also infected with acanthocephalan larvae (Bray and Justine 2013) another parasite group that have marine mammals or birds as their definitive hosts.
Clade III included two different Anisakis larval types in New Caledonian fish, including types I and III. Although Anisakis larval types I and III could easily be differentiated morphologically based on the morphology of the ventriculus and tail, specific identification is not possible due to lack of characteristic features with taxonomic values, such as spicules. Based on morphology, Anisakis type I larva could be belong to A. simplex, A. pegreffii, A. berlandi or A. typica. Similarly, Anisakis type III could be A. brevispiculata, A. paggiae or A. physeteris. The grouping of taxa clade III of our phylogenetic tree suggests Anisakis larval type I to be A. typica and Anisakis larval type III to be A. brevispiculata. Previously, Anisakis type I larvae were reported from 16 species of fish in New Caledonia (Shamsi et al. 2015). There is also a report of Anisakis sp. larva in this geographical region from E. areolatus (Justine et al., 2010a); however, the larval type was not stated and specific identification was unknown as molecular work was not performed. Except for C. fulvoguttatus, the remaining five fish species are new host records.
In contrast, Anisakis larval type III was found only in one fish, Beryx splendens (splendid alfonsino), in the present study. In opposition to all other fish mentioned in the present study, which are coral reef-associated fish, this species is a deep-sea fish, collected from a Seamount off New Caledonia. This fish host was not included in previous bibliographic works of parasites from fish from New Caledonia. The present study is a new parasite-host record and first report of Anisakis type III larvae in New Caledonia. Beryx splendens has a circumglobal distribution and Anisakis parasites of this host species have been recorded in other geographical locations. ITS molecular work on Anisakis larval types II, III and IV from Beryx splendens from Japan revealed type II larvae were a 100% match to A. physeteris, type III larvae had a high level of similarity to A. brevispiculata and type IV larvae were 100% identical to A. paggiae (Murata et al., 2011). Third-stage larvae of A. physeteris have been found in B. splendens from the Mediterranean Sea (Psomadakis et al. 2012). In our study, Anisakis type III larvae had identical sequences with A. brevispiculata reported from whales in Australia and the Philippine waters. Alignment of ITS sequences of A. brevispiculata from southern hemisphere and those in the Philippine waters shows presence of 1 bp difference in both ITS-1 and ITS-2. For some Anisakis spp. (members of Anisakis simplex sensu lato), one base pair difference in either ITS-1 or ITS-2 region means a distinct species (Shamsi et al. 2012). It cannot be concluded at this stage that the base pair difference observed between A. brevispiculata from different parts of the world is indicative of the presence of distinct species. More studies using other genes are necessary to investigate this.
Although adult nematodes collected from whales in the present study were placed under Anisakis paggiae, a closely related species to A. brevispiculata (Mattiucci et al., 2018), the spicule length in our specimens is shorter than both A. paggiae and A. brevispiculata, fitting within the range reported for Anisakis oceanicus, previously reported in pacific waters from black fish (genus Globicephala) (Johnston and Mawson, 1951) and currently considered as A. physeteris (Davey, 1971). For the time being due to the lack of molecular data, the specimens in the present study are placed under A. paggiae and if this is correct then this is the first report of the species from the Pacific region.
In conclusion, this study shows that new and known anisakid nematodes are commonly found in New Caledonian waters. All of the specimens from fish were in third larval stage and adults were only found in whales. Third-stage larvae of anisakids are known to be the infective stage of the parasite for humans, therefore presence of these parasites in New Caledonian fish, where seafood consumption is high (Charlton et al. 2016), is very important. Similar to some of the neighbouring countries, e.g. in Australia (Shamsi and Shorey 2018), the absence of report from humans in this region could be due to the lack of awareness among health practitioners. Considering the high level of seafood consumption in New Caledonia, it is crucial to raise awareness among various stake holders about these parasites and risk they may pose to public health.
References
Bray RA, Justine J-L (2013) A digenean parasite in a mudskipper: Opegaster ouemoensis sp n. (Digenea: Opecoelidae) in Periophthalmus argentilineatus Valenciennes (Perciformes: Gobiidae) in the mangroves of New Caledonia. Folia Parasitol 60:7–16
Cannon LRG (1977) Some larval ascaridoids from south-eastern Queensland marine fishes. Int J Parasitol 7:233–243
Charlton KE, Russell J, Gorman E, Hanich Q, Delisle A, Campbell B, Bell J (2016) Fish, food security and health in Pacific Island countries and territories: a systematic literature review. BMC Public Health 16:285
D'Amelio S, Cavallero S, Dronen NO, Barros NB, Paggi L (2012) Two new species of Contracaecum Railliet & Henry, 1912 (Nematoda: Anisakidae), C. fagerholmi n. sp and C. rudolphii F from the brown pelican Pelecanus occidentalis in the northern Gulf of Mexico. Syst Parasitol 81:1–16
Davey JT (1971) A revision of the genus Anisakis Dujardin, 1845 (Nematoda: Ascaridata). J Helminthol 45:51–72
dos Reis Sardella CJ, Luque JL (2016) Morphological and molecular diagnostic of Anisakis typica and Anisakis brevispiculata of southeastern coast of Brazil. Braz J Vet Med 38(Supl. 3):124–130
Froese, R., Pauly, D., (Eds) (2018) FishBase. World Wide Web electronic publication. www.fishbase.org, version (02/2018)
Gontard T, de Coudenhove G (2013) Economic monitoring study: the fisheries sector in New Caledonia. SPC Fish Newsletter 141:29–36
Jabbar A, Asnoussi A, Norbury LJ, Eisenbarth A, Shamsi S, Gasser RB, Lopata AL, Beveridge I (2012) Larval anisakid nematodes in teleost fishes from Lizard Island, northern Great Barrier Reef, Australia. Mar Freshw Res 63:1283–1299
Gu X, Zhu J-Y, Jian K-L, Wang B-J, Peng X-R, Yang G-Y, Wang T, Zhong Z-J, Peng K-Y (2016) Absence of population genetic structure in Heterakis gallinarum of chicken from Sichuan, inferred from mitochondrial cytochrome c oxidase subunit I gene. Mitochondrial DNA Part A 27:3612–3617
Jabbar A, Fong RWJ, Kok KX, Lopata AL, Gasser RB, Beveridge I (2013) Molecular characterization of anisakid nematode larvae from 13 species of fish from Western Australia. Int J Food Microbiol 161:247–253
Johnston TH, Mawson PM (1951) Report on some parasitic nematodes from the Australian Museum. Rec Aust Mus 22:289–297
Justine J-L (2007) Parasite biodiversity in a coral reef fish: twelve species of monogeneans on the gills of the grouper Epinephelus maculatus (Perciformes: Serranidae) off New Caledonia, with a description of eight new species of Pseudorhabdosynochus (Monogenea: Diplectanidae). Syst Parasitol 66:81–129
Justine JL (2010) Parasites of coral reef fish: how much do we know? With a bibliography of fish parasites in New Caledonia. Belg J Zool 140(Suppl):155–190
Justine J-L, Beveridge I, Boxshall GA, Bray RA, Miller TL, Moravec F, Trilles J-P, Whittington ID (2010a) An annotated list of parasites (Isopoda, Copepoda, Monogenea, Digenea, Cestoda and Nematoda) collected in groupers (Serranidae, Epinephelinae) in New Caledonia emphasizes parasite biodiversity in coral reef fish. Folia Parasitol 57:237–262
Justine J-L, Beveridge I, Boxshall GA, Bray RA, Moravec F, Whittington ID (2010b) An annotated list of fish parasites (Copepoda, Monogenea, Digenea, Cestoda and Nematoda) collected from emperors and emperor bream (Lethrinidae) in New Caledonia further highlights parasite biodiversity estimates on coral reef fish. Zootaxa 2691:1–40
Justine J-L, Beveridge I, Boxshall GA, Bray RA, Miller TL, Moravec F, Trilles J-P, Whittington ID (2012a) An annotated list of fish parasites (isopoda, Copepoda, Monogenea, Digenea, Cestoda, Nematoda) collected from snappers and bream (Lutjanidae, Nemipteridae, Caesionidae) in New Caledonia confirms high parasite biodiversity on coral reef fish. Aquatic Biosystems 8:22
Justine J-L, Briand MJ, Bray RA (2012b) A quick and simple method, usable in the field, for collecting parasites in suitable condition for both morphological and molecular studies. Parasitol Res 111:341–351
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649
Kijewska A, Rokicki J, Sitko J, Węgrzyn G (2002) Ascaridoidea: a simple DNA assay for identification of 11 species infecting marine and freshwater fish, mammals, and fish-eating birds. Exp Parasitol 101(1):35–39
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Mattiucci S, Cipriani P, Webb SC, Paoletti M, Marcer F, Bellisario B, Gibson DI, Nascetti G (2014) Genetic and morphological approaches distinguish the three sibling species of the Anisakis simplex species complex with a species designation as Anisakis berlandi n. sp. for A. simplex sp C (Nematoda: Anisakidae). J Parasitol 100:199–214. https://doi.org/10.1645/12-120.1
Mattiucci S, Cipriani P, Levsen A, Paoletti M, Nascetti G (2018) Molecular epidemiology of Anisakis and Anisakiasis: an ecological and evolutionary road map. Adv Parasitol 99:93–263
Moravec F, Justine J-L (2006) Three nematode species from elasmobranchs off New Caledonia. Syst Parasitol 64:131–145
Murata R, Suzuki J, Sadamasu K, Kai A (2011) Morphological and molecular characterization of Anisakis larvae (Nematoda: Anisakidae) in Beryx splendens from Japanese waters. Parasitol Int 60:193–198
Myers N, Mittermeier RA, Mittermeier CG, da Fonesca GAB, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 403:853–858
Nadler SA, D'Amelio S, Dailey MD, Paggi L, Siu S, Sakanari JA (2005) Molecular phylogenetics and diagnosis of Anisakis, Pseudoterranova, and Contracaecum from northern Pacific marine mammals. J Parasitol 91(6):1413–1429
Psomadakis PN, Stefanni S, Merella P, Ferrando S, Amato A, Vacchi M (2012) Additional records of Beryx splendens (Osteichthyes: Berycidae) from the Mediterranean Sea, with notes on molecular phylogeny and parasites. Ital J Zool 79:111–119
Quiazon KMA, Yoshinaga T, Santos MD, Ogawa K (2009) Identification of Larval Anisakis spp. (Nematoda: Anisakidae) in Alaska Pollock (Theragra chalcogramma) in Northern Japan Using Morphological and Molecular Markers. J Parasitol 95(5):1227–1232
Quiazon KMA, Santos MD, Yoshinaga T (2013) Anisakis species (Nematoda: Anisakidae) of dwarf sperm whale Kogia sima (Owen, 1866) stranded off the Pacific coast of southern Philippine archipelago. Vet Prasitol 197:221–230
Shamsi S, Butcher AR (2011) First report of human anisakidosis in Australia. Med J Aust 194:199–200
Shamsi S, Shorey H (2018) Seafood-borne parasitic diseases in Australia: are they rare or underdiagnosed? Int J Med 48:591–596
Shamsi S, Suthar J (2016) Occurrence of Terranova larval types (Nematoda: Anisakidae) in Australian marine fish with comments on their specific identities. PeerJ 4:e1722–e1722
Shamsi S, Norman R, Gasser R, Beveridge I (2009) Redescription and genetic characterization of selected Contracaecum spp. (Nematoda: Anisakidae) from various hosts in Australia. Parasitol Res 104(6):1507–1525
Shamsi S, Gasser RB, Beveridge I (2011) Mutation scanning-coupled sequencing of nuclear ribosomal DNA spacers (as a taxonomic tool) for the specific identification of different Contracaecum (Nematoda: Anisakidae) larval types. Mol Cell Probes 25, 13–18
Shamsi S, Gasser R, Beveridge I (2012) Genetic characterisation and taxonomy of species of Anisakis (Nematoda:Anisakidae) parasitic in Australian marine mammals. Invertebr Syst 26:204–212
Shamsi S, Poupa AM, Justine J-L (2015) Characterisation of Ascaridoid larvae from marine fish off New Caledonia, with description of new Hysterothylacium larval types XIII and XIV. Parasitol Int 64:397–404
Shamsi S, Briand MJ, Justine J-L (2017) Occurrence of Anisakis (Nematoda: Anisakidae) larvae in unusual hosts in southern hemisphere. Parasitol Int 66:837–840
Shamsi S, Steller E, Chen Y (2018) New and known zoonotic nematode larvae within selected fish species from Queensland waters in Australia. Int J Food Microbiol 272:73–82
Acknowledgements
The following colleagues and students participated in the parasitologal survey over years: Bernard Marchand, Eva Řehulkova, František Moravec, Pierre Legendre, Ian Beveridge, Aude Sigura, Marine Briand, Julie Mounier, Charlotte Evenat, Charles Beaufrère, Audrey Guérin and Anaïs Guillou. Some fish were provided by Claude Chauvet. Fish were identified, generally from photographs, by Bernard Séret, Ronald Fricke and/or Jack Randall. Parasites from Beryx splendens, collected by Manina Tehei and Philippe Borsa, were donated by Philippe Borsa. Parasites from stranded mammals were collected and donated by Claire Garrigue.
Author information
Authors and Affiliations
Corresponding author
Additional information
Section Editor: Christopher Whipps
Rights and permissions
About this article
Cite this article
Shamsi, S., Chen, Y., Poupa, A. et al. Occurrence of anisakid parasites in marine fishes and whales off New Caledonia. Parasitol Res 117, 3195–3204 (2018). https://doi.org/10.1007/s00436-018-6018-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-018-6018-0