Abstract
Water buffalo (Bubalus bubalis) is the intermediate host for at least four species of Sarcocystis: S. fusiformis, S. buffalonis, S. levinei, and S. sinensis/S. dubeyi. Here, a new species, Sarcocystis dehongensis, is reported in 51 of 756 (6.7%) water buffaloes in China. By light microscopy, the cysts of S. dehongensis were macroscopic, up to 18.5 mm long and 95 μm in diameter; 4.9–11.9 μm villous protrusions extended beyond the sarcocyst wall. Using transmission electron microscopy, the sarcocyst wall had lancet- or leaf-like protrusions in longitudinal section, but the cross section showed that the protrusions appeared as mushroom-like in shape with a core of tightly packed microtubules, similar to “type 24.” BLAST searches revealed that S. dehongensis shared the most similarities with the 18S rDNA sequence of S. hardangeri (92.4%) and mitochondrial cox1 gene sequence of S. ovalis (81.0%), whereas no sequences in GenBank were found to be significantly similar to the ITS-1 region of S. dehongensis. A phylogenetic analysis based on 18S rDNA and mitochondrial cox1 gene sequences suggested that S. dehongensis was closely related to Sarcocystis species from cervids that employ corvids as definitive hosts.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sarcocystis species are among the most common and widespread protozoan parasites of livestock. Almost all these hosts harbor more than one species of Sarcocystis found as microscopic or, less frequently, as macroscopically visible sarcocysts in striated muscular tissues. The species of Sarcocystis within a given host can be distinguished by the structure of their cyst walls and the characterization of genetic markers (Dubey et al. 2016). Most of available evidence indicates that Sarcocystis species of livestock should be host-specific (Dubey et al. 2016).
The water buffalo (Bubalus bubalis) has two macroscopic sarcocysts (S. fusiformis and S. buffalonis) and at least two microscopic sarcocysts (S. levinei and S. sinensis/S. dubeyi) (Chen et al., 2011; Dubey et al., 2016). Here, we report a new bubaline Sarcocystis species on the basis of epidemiology, morphology, and molecular characterization. Additionally, the phylogenetic relationships of the new species with other known Sarcocystis spp. were investigated using the sequences of 18S ribosomal DNA (rDNA) and mitochondrial cytochrome c oxidase subunit 1 (cox1) gene.
Materials and methods
During 2014–2016, samples from the esophagus, tongue, heart, neck, diaphragm, and skeletal and abdominal muscles were collected from six farmers’ markets in Dehong Autonomous Prefecture of China, near the boundaries of Myanmar and China. Most of the water buffalo were obtained from local farmers, whereas the rest were imported from the neighborhood of Myanmar.
In the laboratory, 0.5-mm pieces of muscle from each tissue were squeezed between two glass slides and examined for sarcocysts using a dissecting microscope. Sarcocysts were separated manually from the muscle and processed for light microscopy (LM), transmission electron microscopy (TEM), and DNA analysis.
For TEM, five sarcocysts from a water buffalo were fixed in 2.5% glutaraldehyde in cacodylate buffer (0.1 M, pH 7.4) at 4 °C and post-fixed in 1% osmium tetroxide in the same buffer, dehydrated in a graded alcohol series, and embedded in Epon-Araldite mixture. Ultrathin sections were stained with uranyl acetate and lead citrate and examined using a JEM100-CX transmission electron microscope (JEOL Ltd., Tokyo, Japan) at 80 kV.
For DNA analysis, three individual sarcocysts from three buffaloes were subjected to genomic DNA extraction using the phenol/chloroform method after 0.01% proteinase K and 0.25% trypsin digestion. Three primer pairs, S1/B (Fischer and Odening 1998; Medlin et al. 1988), SF1/SR9 (Gjerde 2013, 2014a), and SU1F/ 5.8SR2 (Gjerde 2014b) were used to amplify the three genetic markers, 18S rDNA, mitochondrial cox1 gene, and ITS-1 gene, respectively. PCR products were gel purified, cloned, sequenced, and characterized using the method detailed in a previous paper (Hu et al. 2016).
Phylogenetic analyses were conducted separately on nucleotide sequences of 18S rDNA and mitochondrial cox1 gene by means of MEGA6 software (Tamura et al. 2013). The maximum parsimony (MP) trees for the two genes were created with a Tree-Bisection-Regrafting (TBR) algorithm. The reliability of the MP phylograms was tested with the bootstrap method using 1000 replications. The 18S rDNA analysis involved 51 nucleotide sequences, and 2086 aligned positions of 49 taxa; Hammondia heydorni (GQ984224), H. hammondi (AF096498), and Toxoplasma gondii (L37415) were chosen as outgroups. The mitochondrial cox1 gene analysis involved 30 nucleotide sequences, and 999 aligned positions of 29 taxa; T. gondii (JX473253), H. triffittae (JX473247), and H. heydorni (JX473251) were used as outgroup species to root the tree.
Results
LM and TEM observations on sarcocysts
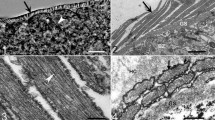
Sarcocysts of the new species were found in 51 of 756 (6.7%) water buffalo. The highest prevalence was 5.2% (39/756) found in skeletal muscles, followed by 4.4% (34/756) in abdominal muscles, 3.9% (29/750) in neck muscles, 0.5% (4/756) in the esophagus, but none in the hearts, tongues, or diaphragms. Using LM, the sarcocysts were macroscopic, 450–18,500 × 40–95 μm (n = 63), and had a thick wall with numerous, 4.9–11.9 μm (n = 30) long, villar protrusions (Fig. 1a). They were septate and contained bradyzoites, measuring 11.6–15.6 × 1.6–2.5 μm, (n = 33).
Morphological characteristics of Sarcocystis dehongensis n. sp. isolated from skeletal muscles of water buffalo. a Sarcocyst (unstained, light microscopy) bounded by villar protrusions (vp). b Longitudinal section of a sarcocyst (under transmission electron microscopy, TEM). The sarcocyst is surrounded by host cell (hc), and a bundle of microtubules (mt) exists in the core of the vp. c Cross section of a sarcocyst under TEM. Note the mushroom-like vp, the bundle of microtubules (mt), and a layer of ground substances (gs) beneath the vp
By TEM, the sarcocyst wall contained numerous protrusions, the morphology of which changed according to the cut section. In longitudinal section, the villar protrusions measured 10.1–14.8 × 2.5–3.8 μm (n = 12), appearing as elongated lancet- or leaf-like shape. In the core of the protrusions, microtubules were condensed, while microtubules were not detected within the ground substance layer (Fig. 1b). In the cross section, the cyst wall formed numerous mushroom-like protrusions with a slender neck stood on the ground substance; in the central area of the protrusions, the microtubules bundled together and appeared as unrooted umbrella-like shapes (Fig. 1c). The cyst wall had minute undulations over the entire surface. A layer of ground substances measuring 3.4–5.0 μm (n = 8) in thickness was located immediately beneath the primary sarcocyst wall.
Molecular characterization of the 18S rDNA, mitochondrial cox1 gene, and ITS-1 gene
Three 18S rDNA sequences (KY711373–KY711375) from three clones, each from an individual sarcocyst, were 1870, 1871, and 1873 bp in length, respectively. The identities between the three clones were 98.7, 98.9, and 99.6%, respectively. The differences included both insertions/deletions and nucleotide substitutions. For clone 1, the most similar 18S rDNA sequence in GenBank was that of S. hardangeri (EF056013) from reindeer (Rangifer tarandus), with 92.4% identity, followed by a sequence of S. ovalis from sika deer (Cervus nippon) (LC184602; 92.2% identity). The same was true for clones 2 and 3, with identities of 92.5 and 92.4% to S. hardangeri (EF056013), and 92.4 and 92.3% to S. ovalis (LC184602), respectively.
Three cox1 gene sequences from three clones, each from an individual sarcocyst, had the same length of 1085 bp. The identities between the three clones were 99.8, 99.8, and 100%, respectively; therefore, only two sequences (KY711376 and KY711377) were submitted to GenBank. The differences included three nucleotide substitutions. For clone 1, the most similar sequence in GenBank was that of S. ovalis (KF241356) with 81.0% identity, followed by S. oviformis (KC209657; 80.5% identity) from roe deer (Capreolus capreolus) and S. hardangeri (KC209628; 80.4%). The same was true for clone 2, with same identities to S. ovalis (KF241356), S. oviformis (KC209657), and S. hardangeri (KC209628), respectively.
Three ITS-1 sequences (KY711378–KY711380) from three clones, each from a sarcocyst, were 614, 615, and 616 bp, respectively. The identities between them were 98.4, 98.5, and 99.8%, respectively. The differences included both insertions/deletions and nucleotide substitutions. BLAT searches only using the ITS-1 region (approximately 350 bp) of the three clones revealed that no sequences shared significant similarities with them.
In the phylogenetic tree inferred from 18S rDNA sequences (Fig. 2), or from cox1 gene sequences (Fig. 3), the species of Sarcocystis found in the present study was clustered together with S. hardangeri, S. ovalis, and S. oviformis in a clade basal to the large clade of Sarcocystis spp. using ruminants as intermediate hosts, and canids, felids, or humans as definitive hosts.
Phylogenetic tree based on 18S rDNA sequences. The tree was built using the maximum parsimony method with the Tree-Bisection-Regrafting algorithm. The tree was tested by selecting a bootstrap method that employed 1000 replicates. Sarcocystis dehongensis n. sp. (shown in bold) clustered consistently in a clade of Sarcocystis species that utilize transmission cycles between cervids intermediate hosts and corvids definitive hosts
Phylogenetic tree based on mitochondrial cox1 gene sequences. The tree was built using the maximum parsimony method with the Tree-Bisection-Regrafting algorithm. The tree was tested by selecting a bootstrap method that employed 1000 replicates. Sarcocystis dehongensis n. sp. (shown in bold) shared a close affinity with species of Sarcocystis using corvids as definitive hosts
On the basis of the host specificity, morphological, and molecular characteristics, a new species, S. dehongensis, is proposed for the unknown organism found in water buffalo from Dehong Prefecture, China.
Taxonomic summary of S. dehongensis n. sp. (Figs. 1, 2, and 3)
Diagnosis: Sarcocysts were macroscopic, up to 18.5 mm in length and 95 μm in diameter. The sarcocyst wall was thick with numerous 4.9–11.9 μm villar protrusions. TEM revealed that the sarcocyst had lancet- or leaf-like protrusions in longitudinal section, but the cross section showed that the protrusion appeared as mushroom-like shape with a core of tightly packed microtubules. This sarcocyst wall has been classified as “type 24” by Dubey et al. (2016).
Etymology: Species name is after the geographical locality where the species of Sarcocystis has been found in China.
Intermediate host: Water buffalo (B. bubalis)
Definitive host: Unknown
Locality: Dehong Autonomous Prefecture in southwestern China
Prevalence: Sarcocyst stages of S. dehongensis were found in 51 of 756 (6.7%) water buffaloes. The sarcocysts appeared in skeletal, abdominal, and neck muscles, but none in the hearts, tongues, or diaphragms.
Molecular characterization: Nucleotide sequences of the near full-length 18S rDNA (KY711373–KY711375), the partial mitochondrial cox1 gene (KY711376 and KY711377), and the full-length ITS-1 gene (KY711378–KY711380) have been deposited in GenBank. The species S. dehongensis may be unambiguously differentiated from Sarcocystis spp. from water buffalo or other ruminants on the basis of the three genetic markers.
Specimens deposited: Formalin-fixed tissues containing cysts of S. dehongensis, as well as photomicrographs from the LM and TEM examination of the sarcocysts, have been deposited at the Zoological Specimen Museum of Yunnan University, Kunming, China (collection number Prot 201608).
Discussion
The ultrastructure of the sarcocyst wall is a taxonomic criterion to differentiate Sarcocystis spp. within a given host. Dubey et al. (2016) grouped sarcocysts by the cyst wall ultrastructure into 42 wall types, with several subgroups. Following their proposals, the five Sarcocystis species of water buffalo have different TEM sarcocyst wall types, i.e., “type 21b” for S. fusiformis, “type 7a” for S. levinei, “type 28” for S. buffalonis, and “type 10c” for S. sinensis/S. dubeyi (Dubey et al. 2016). In the present study, the proposed S. dehongensis n. sp. was similar to “type 24,” that is, the cross section showed that the sarcocyst wall had mushroom-like villar protrusions with a core of tightly packed microtubules. The unique ultrastructural characteristics can easily distinguish S. dehongensis from other Sarcocystis species within water buffalo.
Up to now, five species of Sarcocystis, i.e., S. cornagliai from chamois (Rupicapra rupicapra) (Odening et al., 1996) and alpine ibex (Capra ibex) (Cornaglia et al., 1998), S. mihoensis from sheep (Ovis aries) (Saito et al., 1997), S. tragulusi from Williamson’s mouse deer (Tragulus williamsoni) (Hu et al., 2016), S. novaki from cattle (Bos taurus) (Novak et al., 1987; Odening, 1998), and S. atraii from common coot (Fulica atra) (El-Morsey et al., 2015), have the “type 24” cyst wall. The name S. tragulusi, misspelled as S. tuagulusi by us, should be amended to reflect the correct name of its intermediate host, mouse deer (T. williamsoni).
Using a detailed comparison of TEM from sarcocysts of these five species, they can be divided into three categories, that is, (1) the cyst wall protrusions of S. cornagliai, S. mihoensis, and S. tragulusi have a core formed from densely packed microtubules, that can penetrate into the ground substance layer; (2) the cyst wall protrusions of S. atraii have a core with loosely distributed microtubules; and (3) the cyst wall protrusions of S. novaki have a core with condensed microtubules, but not in ground substance layer. Ultrastructurally, sarcocysts of S. dehongensis n. sp. in the present study most closely resemble S. novaki sarcocysts from cattle in the USSR.
Sequence analysis has proved to be a useful tool to delineate or identify species of Sarcocystis from the same or different hosts, and different genetic markers have showed different levels of intra- or interspecific sequence diversities (Gjerde et al. 2015). In the present study, the sequence similarities of 18S rDNA, cox1, and ITS-1 genes among the different individual sarcocysts of S. dehongensis n. sp. were 98.7–99.6, 99.8–100, and 98.4–99.8%, respectively. While compared with other 18S rDNA and cox1 sequences in GenBank, the most similar were those of S. hardangeri and S. ovalis, respectively. However, the identities between them were not high, only 92.4 and 81.0%, respectively. Regarding the ITS-1 region, no any significant similarity sequences were found in GenBank using the BLST searches. The relationships between S. dehongensis and S. novaki were not clear, for the reasons that no any molecular data of S. novaki in GenBank could be used as reference materials up to now. Most Sarcocystis spp. infecting domestic animals are species-specific for their intermediate hosts and, recently, molecular evidences based on cox1 gene sequences suggested that the morphological indistinguishable sarcocysts in water buffalo and cattle should represent different Sarcocystis species (Gjerde et al. 2015).
Previous studies have suggested that phylogenetic analysis is a useful method in the search for unknown definitive hosts of Sarcocystis spp. (Hu et al. 2015). Based on its phylogenetic position using the sequences of the 18S rDNA and the mitochondrial cox1 gene, S. dehongensis n. sp. was in a clade with S. hardangeri, S. ovalis, and S. oviformis that originated from wild cervids, basal to the large clade of Sarcocystis spp. using ruminants as intermediate hosts, and canids, felids, or humans as definitive hosts. Corvids (Corvidae) are the definitive hosts of S. ovalis (Gjerde & Dahlgren, 2010); therefore, we surmise that S. dehongensis n. sp. probably uses corvids as its definitive hosts. The possibility of ruminant-bird life cycle for S. dehongensis n. sp. might be another reason that the 6.7% (51/756) prevalence for S. dehongensis n. sp. was dramatically lower than other species of Sarcocystis in water buffalo investigated previously in the same province, i.e., 74% (37/50) prevalence for S. fusiformis transmitted by cats, and 96% (48/50) prevalence for S. levinei transmitted by dogs (Zuo et al. 1988).
References
Chen X, Zuo Y, Rosenthal BM, He Y, Cui L, Yang Z (2011) Sarcocystis sinensis is an ultrastructurally distinct parasite of water buffalo that can cause foodborne illness but cannot complete its life-cycle in human beings. Vet Parasitol 178:35–39. doi:10.1016/j.vetpar.2010.12.026
Cornaglia E, Giaccherino AR, Peracino V (1998) Ultrastructural morphology of sarcosporidiosis in alpine ibex (Capra ibex). Vet Parasitol 75:21–32
Dubey JP, Calero-Bernal R, Rosenthal BM, Speer CA, Fayer R (2016) Sarcocystosis of animals and humans, 2nd edn. CRC Press, Boca Raton
El-Morsey A, El-Seify M, Desouky AR, Abdel-Aziz MM, El-Dakhly KM, Kasem S, Abdo W, Haridy M, Sakai H, Yanai T (2015) Morphologic and molecular characteristics of Sarcocystis atraii n. sp. (Apicomplexa: Sarcocystidae) infecting the common coot (Fulica atra) from Egypt. Acta Parasitol 60:691–699. doi:10.1515/ap-2015-0098
Fischer S, Odening K (1998) Characterization of bovine Sarcocystis species by analysis of their 18S ribosomal DNA sequences. J Parasitol 84:50–54
Gjerde B (2013) Phylogenetic relationships among Sarcocystis species in cervids, cattle and sheep inferred from the mitochondrial cytochrome c oxidase subunit I gene. Inter J Parasitol 43:579–591. doi:10.1016/j.ijpara.2013.02.004
Gjerde B (2014a) Sarcocystis species in red deer revisited: with a re-description of two known species as Sarcocystis elongata n. sp. and Sarcocystis truncata n. sp. based on mitochondrial cox1 sequences. Parasitology 141:441–452. doi:10.1017/S0031182013001819
Gjerde B (2014b) Molecular characterization of Sarcocystis rileyi from a common eider (Somateria mollissima) in Norway. Parasitol Res 113:3501–3509. doi:10.1007/s00436-014-4062-y
Gjerde B, Dahlgren SS (2010) Corvid birds (Corvidae) act as definitive hosts for Sarcocystis ovalis in moose (Alces alces). Parasitol Res 107:1445–1453. doi:10.1007/s00436-010-2017-5
Gjerde B, Hilali M, Mawgood SA (2015) Molecular characterisation of three regions of the nuclear ribosomal DNA unit and the mitochondrial cox1 gene of Sarcocystis fusiformis from water buffaloes (Bubalus bubalis) in Egypt. Parasitol Res 114:3401–3413. doi:10.1007/s00436-015-4566-0
Hu JJ, Liu TT, Liu Q, Esch GW, Chen JQ (2015) Sarcocystis clethrionomyelaphis Matuschka, 1986 (Apicomplexa: Sarcocystidae) infecting the large oriental vole Eothenomys miletus (Thomas) (Cricetidae: Microtinae) and its phylogenetic relationships with other species of Sarcocystis Lankester, 1882. Syst Parasitol 91:273–279. doi:10.1007/s11230-015-9572-1
Hu JJ, Huang S, Chen MY, Wen T, Esch GW, Liu Q, Liu TT (2016) Sarcocystis tuagulusi, n.sp. (Apicomplexa: Sarcocystidae) from Williamson’s mouse deer (Tuagulus williamsoni) (Artiodactyla:Tragulidae). Parasitol Res 115:1325–1330. doi:10.1007/s00436-015-4869-1
Medlin L, Elwood HJ, Stickel S, Sogin L (1988) The characterization of enzymatially eukaryotic 16S-like rRNA-coding regions. Gene 71:491–499
Novak MD, Fedoseenko VM, Orazalinova VA (1987) Cyst ultrastructure in Sarcocystis sp. in the cattle. Izvest Akad Nauk Kazakh SSR Ser Biol: 46–49
Odening K (1998) The present state of species-systematics in Sarcocystis Lankester, 1882 (Protista, Sporozoa, Coccidia). Syst Parasitol 41:209–233
Odening K, Stolte M, Bockhardt I (1996) On the diagnostics of Sarcocystis in chamois (Rupicapra rupicapra). Appl Parasitol 37:153–160
Saito M, Shibata Y, Kubo M, Itagaki H (1997) Sarcocystis mihoensis n. sp. from sheep in Japan. J Vet Med Sci 59:103–106
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Zuo YX, Chen FQ, Chen XW, Tang DH (1988) Studies on Sarcocystis species of water buffalo with description of Sarcocystis sp. J Yn Uni 10:91–92
Acknowledgements
This study was funded by the Natural Sciences Foundation of China (grant 31460557) and Southeast Asia Biodiversity Research Institute, Chinese Academy of Science (grant Y4zk111B01). We would like to thank Dr. J. P. Dubey for his positive comments regarding the content of the paper and his suggestions dealing with the presentation of the text.
Author information
Authors and Affiliations
Corresponding author
Additional information
Xinwen Chen and Tao Wen have the same contribution to this paper.
Rights and permissions
About this article
Cite this article
Chen, X., Wen, T., Hu, J. et al. Sarcocystis dehongensis n. sp. (Apicomplexa: Sarcocystidae) from water buffalo (Bubalus bubalis) in China. Parasitol Res 116, 2145–2150 (2017). https://doi.org/10.1007/s00436-017-5515-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-017-5515-x