Abstract
Cathepsin L is a cysteine protease belonging to the papain family. In parasitic trematodes, cathepsin L plays essential roles in parasite survival and host–parasite interactions. In this study, cathepsin L of the lung fluke Paragonimus pseudoheterotremus (PpsCatL) was identified and its molecular biological and immunological features characterized. A sequence analysis of PpsCatL showed that the gene encodes a 325-amino-acid protein that is most similar to P. westermani cathepsin L. The in silico three-dimensional structure suggests that PpsCatL is a pro-enzyme that becomes active when the propeptide is cleaved. A recombinant pro-PpsCatL lacking the signal peptide (rPpsCatL), with a molecular weight of 35 kDa, was expressed in E. coli and reacted with P. pseudoheterotremus-infected rat sera. The native protein was detected in crude worm antigens and excretory–secretory products and was localized in the cecum and in the lamellae along the intestinal tract of the adult parasite. Enzymatic activity of rPpsCatL showed that the protein could cleave the fluorogenic substrate Z-Phe-Arg-AMC after autocatalysis but was inhibited with E64. The immunodiagnostic potential of the recombinant protein was evaluated with an enzyme-linked immunosorbent assay (ELISA) and suggested that rPpsCatL can detect paragonimiasis with high sensitivity and specificity (100 and 95.6 %, respectively). This supports the further development of an rPpsCatL-ELISA as an immunodiagnostic tool.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lung flukes in the genus Paragonimus are the causative agents of paragonimiasis and are endemic in Asia, Africa, and America (Waikagul 2007). Only 7 of more than 40 species have so far been reported to infect humans: Paragonimus africanus, P. kelikotti, P. mexicanus, P. westermani, P. uterobilateralis, P. heterotremus, and P. pseudoheterotremus (Intapan et al. 2012). P. pseudoheterotremus was first identified in the freshwater crab Larnaudia larnaudii from Kanchanaburi Province, Thailand (Waikagul 2007). The first human case was reported in a Thai male admitted to hospital with tuberculosis-like symptoms (Intapan et al. 2012). Fundamental information, including essential proteins, is still lacking for this novel human-infecting species, and intensive research is required to clarify the factors associated its biology and pathogenesis. In this context, the proteolytic enzyme cathepsin L of P. pseudoheterotremus (PpsCatL) was identified and its molecular biological and immunological features characterized here.
Cathepsin L is a cysteine protease that plays important roles in the entire life cycles of several trematodes (Collins et al. 2004; Dalton et al. 1996; Day et al. 1995; Park et al. 2002). In P. westermani, a variety of cathepsin L isoforms have been reported, which affect the parasite’s survival and virulence (Park et al. 2002). The cathepsin L proteins of Fasciola hepatica and F. gigantica are major secretory proteases that have contributed to the development of immunodiagnostic tools (Cornelissen et al. 2001; Varghese et al. 2012) and vaccines (Villa-Mancera et al. 2014; Sansri et al. 2015). An evolutionary study of Fasciola cathepsin L suggested a divergence time for F. hepatica and F. gigantica and the substrate specificities of the proteins (Irving et al. 2003). Using of F. hepatica cathepsins L1 and L3 to design a novel nonpeptidic inhibitors demonstrated a potential drug development against fascioliasis (Ferraro et al. 2016). Based on this information, identifying and characterizing cathepsin L of P. pseudoheterotremus should extend our understanding of the organism and the development of strategies to prevent and control this parasitic helminth.
Here, we amplified the cathepsin-L-encoding gene of P. pseudoheterotremus from its total RNA using random amplification of complementary DNA (cDNA) ends (RACE)–PCR and then analyzed the molecular biological features of the protein, including its sequence properties and in silico three-dimensional (3D) structure. The transcription level of PpsCatL in different developmental stages was determined with real-time RT–PCR. An expression of PpsCatL protein in the parasite was detected using western blot analysis. A recombinant pro-PpsCatL (rPpsCatL) was expressed in Escherichia coli and used to evaluate the immune response to the protein. The specific location of PpsCatL in the parasite tissues was analyzed with immunolocalization. The enzymatic activity of rPpsCatL was determined with fluorogenic peptide substrate, Z-Phe-Arg-AMC. To evaluate the utility of rPpsCatL in the development of immunodiagnostic tools, we reacted the protein with Paragonimus-infected human sera and sera infected with other parasites.
Materials and methods
Ethics statement
All animal works were conducted with the approval of the Faculty of Tropical Medicine Animal Care and Use Committee (no. FTM-ACUC 011/2012). Leftover helminth-infected human sera and healthy sera were used in this study with the permission of the Immunodiagnosis for Helminthiasis Unit, Department of Helminthology, Faculty of Tropical Medicine, Mahidol University, and the approval of the Ethics Committee of the Faculty of Tropical Medicine, Mahidol University (no. MUTM 2011-056-01).
Parasite
Natural waterfall crabs (Demanietta sp.) collected from Tak Province, Thailand were examined for P. pseudoheterotremus infection with the tissue compression technique (Sugiyama et al. 2004). To collect the metacercariae, the bodies of the P. pseudoheterotremus-positive crabs were separated into small pieces, homogenized in 0.85 % normal saline solution (NSS), and allowed to settle in a sediment jar with several changes of NSS (Sugiyama et al. 2004). The metacercariae of P. pseudoheterotremus in the sediment were collected under a stereoscopic microscope (Olympus, Japan) and used to infect the definitive host. Five 8-week-old female Wistar rats purchased from the National Laboratory Animal Center, Mahidol University were orally infected with five metacercariae each and then housed at the Animal Care Unit, Faculty of Tropical Medicine, Mahidol University. Eight weeks postinfection (wpi), a fecal examination was performed with a simple direct smear test and egg-positive rats were euthanized to isolate the adult worms from their respiratory tracts. The fresh recovered worms were washed thoroughly with NSS and stored at −80 °C until further analysis. To prepare newly excysted juvenile worms (NEJ), the metacercariae were washed thoroughly with 1× phosphate-buffered saline (PBS) and then digested with 1 % pepsin-HCl solution for 2 h at 37 °C. After the digested metacercariae were washed several times with 1× PBS, they were incubated at 37 °C for 6 h with complete medium comprising RPMI-1640 (Gibco™ Thermo Fisher Scientific, Watham, MA) supplemented with 10 % fetal bovine serum (FBS; Biowest SAS, Nuaillé, France) and 1× penicillin–streptomycin solution (Biowest SAS). The emerging NEJ were collected and incubated in complete medium at 37 °C for 3 h. The metacercariae, NEJ, and adult P. pseudoheterotremus were stored at −80 °C until further analysis.
Amplification of full-length PpsCatL cDNA
Degenerate primers were designed for PpsCatL by comparing the amino acid sequences of cathepsin L orthologs composed of PwCatL1, PwCatL2, PwCatL6, and PwCP7 (Table 1) using the Clustal Omega program (Goujon et al. 2010). Two highly conserved amino acid motifs with the lowest degree of degeneration, GSCWAF and WGTRWG, were selected and converted to potential nucleic acid codons. The degenerate primers designed were forward (Fwd) 5′-GGIWSITGYTGGGCITTY-3′ and reverse (Rev) 5′-ICCCCAICGIGTICCCCA-3′, where I is inosine, S is C or G, W is T or A, and Y is T or C.
The total RNA from adult P. pseudoheterotremus was isolated with TRIzol Reagent (Invitrogen, Carlsbad, CA), according to the manufacturer’s instructions. The total RNA (5 μg) was treated with 1 U of DNase I (Thermo Fisher Scientific) to eliminate any genomic DNA and then converted to first-strand cDNA using RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). Part of the PpsCatL cDNA was amplified with PCR in a total volume of 50 μl containing 1× Taq DNA polymerase buffer, 2 mM MgCl2, 0.2 mM each dNTP, 200 nM each degenerate primer, and 1 U of Taq DNA polymerase (Thermo Fisher Scientific). The cycling conditions were 95 °C for 5 min, 40 cycles of 95 °C for 30 s, 45 °C for 30 s, and 72 °C for 30 s, and a final extension step at 72 °C for 5 min. The partial cDNA sequence was subcloned into the pTZ57R/T plasmid (Thermo Fisher Scientific), and the DNA sequence was analyzed with DNA sequencing (AITbiotech Pte Ltd, Singapore). The sequence was used to design the primers with the Primer3 program (Untergasser et al. 2012) for 5′- and 3′-RACE–PCR, performed with the SMARTer™ RACE cDNA Amplification Kit (Clontech Laboratories, Inc., Mountain View, CA), according to manufacturer’s instructions. The 3′-RACE- and the 5′-RACE primers were 5′-GTTTGGAATCCGAAGCTGACTATCC-3′ and 5′-CGTGCTCCTCCTCATAAGCTCCTA-3′, respectively. The PCR products were subcloned into the pTZ57R/T plasmid and sequenced (AITbiotech Pte Ltd).
Bioinformatic analysis
The nucleotide and predicted amino acid sequences were submitted to the National Center for Biotechnology Information (NCBI) database with assigned accession numbers KX139301 and used for a bioinformatic analysis. The protein’s properties, including its molecular weight and isoelectric point (pI), signal peptide, and transmembrane helices, were predicted with Pepstats (Rice et al. 2000), SignalP 4.1 Server (Petersen et al. 2011), and TMHMM (Krogh et al. 2001), respectively. Any N- and O-glycosylation sites and disulfide bridges were identified with NetNGlyc 1.0 (http://www.cbs.dtu.dk/services/NetNGlyc/), NetOGlyc 4.0 (Steentoft et al. 2013), and the DiANNA 1.1 web server (Ferre and Clote 2006), respectively. The conserved protein motifs and catalytic triad of PpsCatL were analyzed with a multiple protein alignment constructed with Clustal Omega (Goujon et al. 2010). The evolutionary relationships of PpsCatL to other orthologs were determined with a phylogenetic tree based on a maximum likelihood analysis (ML) with 100 bootstrap replications in the MEGA version 5 program (Tamura et al. 2011). All the sequences used in this experiment are shown in Table 1. The 3D structure of PpsCatL was modeled in silico with Swiss-Model (Biasini et al. 2014), using the crystal structure of F. hepatica cathepsin L (PDB ID: 2o6x.1.A) as the template. The optimality of the predicted structure was tested with a Ramachandran plot analysis implemented at the RAMPAGE server (http://mordred.bioc.cam.ac.uk/~rapper/rampage.php).
Expression of rPpsCatL in E. coli
The first-strand cDNA of P. pseudoheterotremus was used as the template to amplify the PpsCatL sequence without the signal sequence in a reaction volume of 25 μl, containing 2 μl of cDNA, 1× Taq DNA polymerase buffer, 0.2 mM each dNTP, 2 mM MgCl2, 1 U of Taq DNA polymerase, and 100 nM PpsCatL Fwd and Rev primers. The Fwd and Rev primer sequences used to amplify pro-PpsCatL were 5′-GGGGATCCGTGAGCACTGTTCGGGTGC-3′ and 5′-AAAAAGCTTTTAACGGATGACTGCAGACG-3′, respectively. The BamHI restriction site in Fwd and the HindIII restriction site in Rev are underlined. The PCR was performed in a C1000™ thermal cycler (Bio-Rad Laboratories, Inc., Philadelphia, PA) with cycling parameters: 94 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min, followed by a final extension step at 72 °C for 5 min. The PCR product was cloned into pQE30 vector (Qiagen GmbH, Hilden, Germany) at BamHI and HindIII sites and then transformed into E. coli strain M15 (Froger and Hall 2007). The expression of rPpsCatL was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG; Thermo Fisher Scientific) for 4 h followed by purification it under denaturing conditions, as described previously (Pakchotanon et al. 2016). The purified rPpsCatL was used to produce a mouse polyclonal antibody and other downstream experiments.
Immunological analysis
The crude worm antigen (CWA) and the excretory–secretory product (ES) of adult P. pseudoheterotremus were prepared as described previously (Adisakwattana et al. 2007). CWA, ES, and rPpsCatL were size fractionated with 12 % SDS-PAGE and then transferred onto a polyvinylidene difluoride membrane (Pall Corporation, Washington, NY). The membranes were cut into small strips for an immunoblotting analysis, as described previously (Nuamtanong et al. 2012), with some modifications. Briefly, the membranes containing rPpsCatL, CWA, or ES were blocked with blocking solution (5 % skimmed milk in 1× PBS, 0.05 % Tween 20) and then incubated in blocking solution with P. pseudoheterotremus-infected rat serum (diluted 1:200), collected at 0 and 8 wpi, for rPpsCatL or with anti-rPpsCatL mouse serum (diluted 1:4000) for CWA and ES. After the membranes were washed, horseradish peroxidase (HRP)-conjugated goat anti-rat immunoglobulin G (IgG; diluted 1:1000; KPL Inc., Gaithersburg, MD) or HRP-conjugated goat anti mouse IgG (diluted 1:1000; Southern Biotech, Birmingham, AL) was added, and the membrane was incubated at room temperature for 1 h. The results were visualized by incubating the membranes with the colorimetric substrate, 2,6-dichloroindophenol (Sigma-Aldrich, St. Louis, MO).
PpsCatL transcripts detected in different developmental stages with SYBR Green real-time RT–PCR
Total RNAs from metacercariae, NEJ, and adult P. pseudoheterotremus were isolated with TRIzol Reagent, according to the manufacturer’s instructions (Invitrogen). The total RNA (1 μg) was treated with DNase I (Thermo Fisher Scientific) to eliminate any contaminating DNA. The DNA-free total RNA from each stage was used as the template to synthesize first-strand cDNA, as described above, and then to determine the level of PpsCatL transcription using SYBR® Green real-time PCR. The 20 μl reaction mixture contained 2 μl of first-strand cDNA, 1× iTaq Universal SYBR® Green (Bio-Rad Laboratories, Inc.), and 300 nM each of the Fwd and Rev primers. The specific primers for the PpsCatL qPCR were Fwd 5′-CTGTTCGGGTGCCAGATAAT-3′ and Rev 5′-CTCGTTCGTGCATCTGGTAG-3′. 18S ribosomal RNA (rRNA) was used as the internal control against which to normalize gene expression. The specific primers for P. pseudoheterotremus 18S rRNA were Fwd 5′-GATAACGGGTAACGGGGAAT-3′ and Rev 5′-AGCCTCTGTTGAGTCCCGTA-3′. Amplification was performed with the LightCycler® 480 II Real-Time PCR System (Roche Applied Science, Mannheim, Germany), with preincubation at 95 °C for 5 min, followed by 40 cycles of 95 °C for 20 s and 60 °C for 1 min. A melting curve analysis was performed at 65–95 °C. The level of gene expression was calculated with the 2–∆∆Ct formula (Livak and Schmittgen 2001). The experiments were performed with three replicates.
Immunolocalization of PpsCatL in parasite tissues
Paraffin-embedded sections of adult P. pseudoheterotremus were prepared and immunolocalization was performed as described previously (Adisakwattana et al. 2007), with some modifications. In brief, 5-μm-thick paraffin-embedded sections were dewaxed, the antigenic epitopes were retrieved, and the endogenous peroxidase was neutralized. Nonspecific binding sites were blocked with blocking solution (10 % [w/v] bovine serum albumin in PBS, pH 7.4) at room temperature for 20 min, and the sections were then incubated with anti-rPpsCatL mouse serum (1:200) or preimmune serum (1:200) in blocking solution. The sections were then incubated with HRP-conjugated goat anti-mouse IgG (1:1000; SouthernBiotech, Birmingham, AL), and the color signal was developed with an aminoethyl carbazole (AEC) staining kit (Sigma-Aldrich), according to the manufacturer’s instructions. The results were observed under a light microscope.
Activity assay of rPpsCatL
rPpsCatL was refolded and autocatalyzed as described previously with some modification (Hwang and Chung 2002; Kodera et al. 2005). Briefly, rPpsCatL was diluted in refolding buffer containing 5 mM EDTA, 10 mM GSH, 1 mM GSSG, 0.7 M l-arginine at 4 °C for 16 h. After dialysis and concentration, refolded rPpsCatL was mixed with 50 mM sodium acetate buffer (pH 4.0) containing 200 mM NaCl, 10 mM 2-mercaptonethanol, and then incubated at 37 °C for 16 h. The activated rPpsCatL was concentrated and subsequently used for activity assay. Enzymatic activity of rPpsCatL was performed by hydrolysis of fluorogenic peptide substrate, Z-Phe-Arg-AMC (Calbiochem® EMD Millipore Corporation, Temecula, CA) as mentioned previously (Kodera et al. 2005). The enzymatic activity was detected by a spectrofluorometer and expressed as increased fluorescence units per minute during incubation. Cysteine protease inhibitor, E64, was used as a control experiment. The assay was performed in triplicate.
Evaluation of the immunodiagnostic potential of rPpsCatL
A total of 255 serum samples from 29 diseased individuals were classified into three groups: group 1 contained sera from 28 patients with paragonimiasis, group 2 contained 197 serum samples from helminth- and protozoa-infected patients, and group 3 contained sera from 30 healthy individuals. The details are given in Table 2.
A 100-μl sample of rPpsCatL (2.5 μg/ml) or CWA (5 μg/ml) in 0.05 M carbonate buffer (pH 9.6) was coated onto each well of a 96-well microtiter plate (Nunc; Thermo Scientific, Denmark) and incubated at 37 °C for 1 h and then at 4 °C for 16–18 h. An enzyme-linked immunosorbent assay (ELISA) was performed as previously described, with some modifications. In brief, nonspecific binding was blocked with blocking solution (0.5 % skim milk in 1× PBS) at 37 °C for 1 h, and the sections were then washed with PBS–Tween. Human sera (1:400) were added to the wells and incubated at 37 °C for 1 h. After the wells were washed, HRP-conjugated goat anti-human IgG antibody (1:2000) was added. The samples were incubated at 37 °C for 1 h and then visualized with the addition of ABTS substrate (Sigma-Aldrich). The reaction was developed in the dark at room temperature for 30 min and then stopped with 1 % SDS. The optical density at a wavelength at 405 nm was measured with the Sunrise™ absorbance reader (Tecan Group Ltd, Männedorf, Switzerland).
Results
Sequence analysis and 3D structure prediction
The full-length PpsCatL cDNA was amplified with RACE–PCR, and the nucleotide composition was analyzed with DNA sequencing. It contained 978 bp and encoded an open reading frame encoding 325 amino acid residues (Fig. 1). A homology comparison of the deduced amino acid sequence with the available database using the protein BLAST program demonstrated its highest homology to P. westermani cathepsin L6 (accession no. AAY81944.1), with 79.69 % identity. The homology shared by PpsCatL and other cysteine protease orthologues in the genus Paragonimus was determined and is shown with a heatmap (Fig. 2). The full-length PpsCatL cDNA and deduced amino acid sequence were submitted to the NCBI database under accession number KX139301. The protein properties were predicted with the Pepstat program, which showed that the molecular mass of the full-length PpsCatL protein is approximately 36.5 kDa, with a pI of 5.4. The potential signal peptide was predicted at amino acid positions 1–18 at the N-terminus (MTLHTIRCLAFLLACACA), but no transmembrane helix was detected. No posttranslational modification sites, including for N- or O-glycosylation, were observed in PpsCatL. Ten cysteine residues occur in the PpsCatL amino acid sequence at positions 8, 15, 17, 133, 136, 167, 174, 207, 265, and 313, with two potential disulfide bridges at Cys133–Cys174 and Cys167–Cys207.
Nucleotide sequence of full-length PpsCatL (accession no. KX139301) consists of 978 nucleotides that encode 325 amino acid residues. The signal peptide identified at amino acid residues 1–18 is underlined. The cleavage site between the propeptide and mature protein is indicated with an arrowhead. Potential disulfide bonds are boxed
A multiple sequence alignment of PpsCatL and its orthologues suggested a putative propeptide cleavage site between Ala111 and Ala112. The mature protease is composed of 214 amino acid residues, with a calculated molecular mass of approximately 23.7 kDa and a theoretical pI of 5.3. The conserved ERFNIN and GNFD motifs, signatures of cathepsin L, were observed in the propeptide region of PpsCatL, at amino acid positions of 48–63 and 76–82, respectively. A conserved structural motif, GCNGG, is presented in PpsCatL at positions 173–177. The catalytic triad is located at Cys136, His272, and Asn292, and an S1 subsite of the protease active site was identified at Gln130, Cys136, His272, Asn292, and Trp294. The S2 subsite of the active site contains Leu192, Glu193, Ala244, and Leu270 (Fig. 3).
Multiple alignment of deduced amino acid sequences identified the conserved motifs within orthologous cathepsin L proteins. Identical and similar residues are indicated with black and gray shadings, respectively. Gaps are indicated with a dash. Putative amino acid residues of the catalytic triad (Cys [C], His [H], Asn [N]) are indicated with asterisks above the residues. ERFNIN, GNFD, and GCNGG motifs are labeled above the alignment with number signs, pilcrow signs, and plus signs, respectively. S1 subsites are indicated with a black arrow below the alignment, and S2 subsites are indicated with a white arrow below the alignment
The evolutionary relationships between full-length enzymes of PpsCatL and its orthologues were analyzed with the ML method and 100 bootstrap replications. PpsCatL clustered on the same branch as P. westermani cathepsin L and was most closely related to PwCatL6. The Paragonimus cathepsins L were more closely related to Schistosoma cathepsins L than to Fasciola cathepsins L. The trematode cathepsins B were distinctly separated from the cathepsins L on the phylogenetic tree (Fig. 4).
The 3D structures of the pro- and mature PpsCatL were predicted using the crystal structure of F. hepatica cathepsin L (PDB ID: 2o6x.1.A) as the template. The pro-PpsCatL 3D structure showed that the propeptide region folds onto the cathepsin L surface to protect the active site from substrates (Fig. 5a). After the propeptide (amino acid residues 19–111) is removed, PpsCatL becomes a mature protease (amino acid residues 112–325), with its active site exposed to the substrate (Fig. 5b). The structure of mature PpsCatL consists of two domains (left and right). The left domain contains three α-helices and the right domain contains a β-sheet, with a front helix that forms a coiled structure. An interaction at the top of the two domains creates the active-site cleft (Fig. 5b). Ramachandran plots suggested that this structural model is optimal (data not shown).
Expression and purification of rPpsCatL
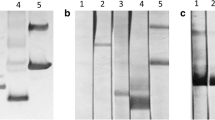
After protein expression was induced with IPTG, rPpsCatL was predominantly expressed with an approximately molecular mass of 35 kDa (Fig. 6a). Its water solubility was tested and showed that rPpsCatL was expressed in an insoluble form in E. coli, which was detected with an anti-His-tag antibody (Fig. 6a). rPpsCatL was purified under denaturing condition, and a major band of 35 kDa was eluted (Fig. 6b). The purified rPpsCatL reacted with the anti-His-tag antibody (data not shown). Eluted fractions 1 to 10 (E1–E10) were pooled and then stepwise dialyzed to remove the excess urea and then used for production of mouse polyclonal antibody and further studies.
rPpsCatL was heterologously expressed in E. coli (a) and then purified with a Co2+ affinity column (b). M broad-range prestained protein marker, NI noninducing, ID inducing, WB western blot analysis with anti-His-tag antibody, FT flow-through, E elution. c Western blotting analysis of native PpsCatL in CWA and ES using anti-rPpsCatL mouse serum. CWA crude worm antigen, ES excretory–secretory product, rPpsCatL recombinant P. pseudoheterotremus cathepsin L. d Detection of rPpsCatL with P. pseudoheterotremus-infected rat sera. M broad-range prestained protein marker, NI noninfected sera, IF infected sera
Detection of PpsCatL in native parasite antigens and the immune response to rPpsCatL
The presence of PpsCatL in the CWA and ES of P. pseudoheterotremus was detected on western blots probed with anti-rPpsCatL mouse serum. In the CWA, the anti-rPpsCatL serum detected the native protein at molecular weights of approximately 35, 28, and 25 kDa. In the ES, a protein band with a molecular weight of 25 kDa predominantly reacted with the anti-rPpsCatL serum, and also in the range of 10–18 kDa, but most strongly at 14 kDa (Fig. 6c).
The immune response of P. pseudoheterotremus-infected rats to rPpsCatL was observed with a western blotting analysis. All five infected rat sera, collected at 8 wpi, reacted with 35 kDa rPpsCatL and other truncated proteins (Fig. 6d). Normal rat and mouse sera were used as the negative controls.
PpsCatL transcripts in different developmental stages of P. pseudoheterotremus
The transcription of PpsCatL in different developmental stages of P. pseudoheterotremus (metacercaria, NEJ, and adult) was determined to define its stage-specific expression. PpsCatL was transcribed most strongly in the adult parasite, approximately sixfold more strongly than in the metacercariae or NEJ. The expression of PpsCatL mRNA in the metacercariae and NEJ did not differ significantly (Fig. 7).
Stage-specific transcription of PpsCatL was monitored in different developmental stages of P. pseudoheterotremus using SYBR Green real-time RT–PCR. MET metacercaria, NEJ newly excysted juvenile, Adult adult worm. Relative expression levels were calculated by comparison with the transcription levels in the metacercariae
Tissue-specific localization of PpsCatL in adult P. pseudoheterotremus
PpsCatL expression was immunolocalized in the adult stage based on the transcription of the gene. PpsCatL was specifically expressed in the digestive system, including the esophagus and intestine, but not in other tissues. PpsCatL accumulated strongly at the lamellae lining the esophagus and along the intestine throughout the whole parasite body (Fig. 8a, b). Immunolocalization with preimmunized mouse serum gave negative results (Fig. 8c).
Enzymatic activity of rPpsCatL
rPpsCatL was refolded and activated by autocatalytic processes prior to determination of proteolytic activity. Hydrolysis of fluorogenic peptide substrate Z-Phe-Arg-AMC was measured, and the result demonstrated that activated rPpsCatL could cleave Z-Phe-Arg-AMC, while inactive pro-enzyme did not show proteolytic activity. The enzymatic activity of activated rPpsCatL was eliminated when incubated with cysteine protease inhibitor, E64 (Fig. 9).
Enzymatic activity of rPpsCatL was determined by hydrolysis of Z-Phe-Arg-AMC substrate. Inactivated (iPpsCatL) or activated rPpsCatL (aPpsCatL) was incubated with the substrate, and the enzymatic activity was measured as increased fluorescence units/min. Incubating activated rPpsCatL with 5 μM E64 (aPpsCatL + E64) was performed as a control. Neg negative control
ELISA
The dilutions of the human sera and the secondary antibody were optimized with checkerboard titration (data not shown). The optimal dilution of human serum and the secondary antibody were 1:400 and 1:2000, respectively. In this study, ELISAs using rPpsCatL or the CWA as the antigen were performed to compare their sensitivities and specificities. The cutoff points for CWA- and rPpsCatL-ELISAs were 0.232 (mean + 5SD) and 0.412 (mean + 6SD), respectively. The frequency distributions of the negative, positive, and samples were plotted to indicate the number of true positive results, true negative results, false positive results, and false negative results (Fig. 10a left, b left). The sensitivity and specificity of CWA were 100 and 84.1 %, respectively, and those of rPpsCatL were 100 and 95.6 %, respectively (Fig. 10a right, b right).
Evaluation of CWA-ELISA (a) and rPpsCatL-ELISA (b). Scatter plot indicates the absorbance values for parasite-infected sera (a left and b left). Dashed lines indicate cutoff values for the CWA-ELISA and rPpsCatL-ELISA at 0.232 and 0.412, respectively. Sensitivity and specificity were calculated with a 2 × 2 table, which showed that the sensitivity and specificity of the CWA-ELISA were 100 and 84.1 %, respectively, and those of the rPpsCatL-ELISA were 100 and 95.6 %, respectively (a right and b right). Abbreviations on the x-axis are described in the Table 2
Discussion
In parasitic trematodes, cathepsin L is an essential cysteine protease that is involved in a variety of activities throughout their life cycles, including encystation, excystation, migration, digestion, maturation, fertilization, and immune evasion (Grams et al. 2001; Shin et al. 2001; Collins et al. 2004; Na et al. 2006; Chung et al. 2008). Several cathepsin L isoforms were previously identified in a medically important lung fluke, P. westermani, but not in other Paragonimus species (Park et al. 2002). Here, cathepsin L of the agent of zoonotic paragonimiasis, P. pseudoheterotremus, was identified and characterized with bioinformatic and molecular biology approaches. The amplification of PpsCatL using degenerate-primer-based RT–PCR combined with RACE–PCR isolated the full-length gene, which showed high homology to P. westermani cathepsins L, especially PwCatL6 and PwCatL5. PwCatL5 is a well-characterized cysteine protease found in the adult P. westermani worm and is classified in the cathepsin L family (Park et al. 2002). An amino acid analysis of PpsCatL demonstrated that the protein has a molecular weight of 36 kDa and contains a signal peptide but no transmembrane helix, indicating its excretory–secretory potential. These data suggest an extracellular role for this protein, involving digestion, migration, invasion, and host–parasite interactions (Day et al. 1995; Dalton et al. 1996; Collins et al. 2004). The presence of ten cysteine residues, with two potential disulfide bonds, in PpsCatL possibly facilitates its tertiary structure, forming intermolecular interactions that create a dimeric structure, as seen in other cathepsin L orthologues. The cathepsin L found in the embryos and larvae of the brine shrimp (Artemia franciscana) forms a heterodimer with cathepsin L-associated protein (CLAP), which is more active and more stable than the cathepsin L monomer (Warner et al. 2004). A multiple alignment of PpsCatL with other orthologues identified the motifs ERFNIN and GNFD in the propeptide of PpsCatL, which have been described as signatures of cathepsin L-like proteases but not cathepsin B-like proteases (Kollien et al. 2004; Pandey et al. 2009). The ERFNIN and GNFD motifs are considered to facilitate the inhibition of the falcipain 2 propeptide, which was confirmed when the deletion of both motifs impaired the inhibition of falcipain 2 (Pandey et al. 2009). GCNGG is another conserved motif, found at amino acid residues 173–177 of mature PpsCatL. This motif was previously reported to be associated with the formation of the globular protein, which requires disulfide bridges (Karrer et al. 1993). The catalytic triad of mature PpsCatL is composed of Cys136, His272, and Asn292, which are conserved within the papain family (clan CA) (Pandey and Dixit 2012). S1 and S2 subsites were predicted at the active site of PpsCatL, which are conserved in other cathepsin L orthologues. The S1 and S2 subsites of cathepsin L, especially S2, contribute to its substrate preference (Alves et al. 2001; Lecaille et al. 2007). Mutation of the S2 pocket (L67Y/A205L) in human cathepsin L caused a change in its enzymatic specificity, affecting its amino acid preferences in both peptidyl substrates and small selective inhibitors (Lecaille et al. 2007).
A phylogenetic tree demonstrated that PpsCatL is closely related to the cathepsins L of P. westermani, PwCatL6, PwCatL5, and PwCatL8, which may suggest biological and biochemical associations of this protein in the genus Paragonimus. The in silico tertiary structure of mature PpsCatL has been reported (reviewed by Turk et al. 2012). However, a crystallographic analysis of PpsCatL and other Paragonimus cathepsins L is required to clearly define the structure of this protein in the genus Paragonimus.
In a prokaryotic expression system, pro-PpsCatL without the signal peptide was expressed as an insoluble product with an approximate molecular weight of 35 kDa, which is similar to the calculated molecular weight of pro-PpsCatL (34.5 kDa). The detection of PpsCatL in CWA and ES with anti-rPpsCatL mouse serum indicates that the pro-PpsCatL produced by E. coli was the same size as that found in the CWA. These data are evidence of an unglycosylated PpsCatL, which is consistent with the lack of potential glycosylation site predicted in this protein. A cathepsin L lacking potential glycosylation sites was previously observed in F. hepatica, procathepsin L1, and was detectable in the ES of the adult parasite (Collins et al. 2004). The lack of glycosylation in human and mouse procathepsins L does not disturb the structural folding, stability, or secretory properties of these proteases (Kane 1993; Smith et al. 1989). PpsCatL was present in the ES of P. pseudoheterotremus, with a predominant molecular weight of 25 kDa, which is the predicted molecular weight of mature PpsCatL. In F. hepatica, the mature cathepsins L1 (FhCatL1) and L2 (FhCatL2), but not the full-length proteins, were detected in the ES of the adult parasite (Collins et al. 2004). The ladder-like pattern of the protein observed in the CW and ES of adult P. pseudoheterotremus is attributed to catalytic of pro-enzyme or degradation. The immune response to PpsCatL was studied by treating P. pseudoheterotremus-infected rats with the recombinant protein. A strong signal was observed in all the infected animals, suggesting the potential utility of this protein in the development of immunodiagnostic tools. When F. hepatica cathepsin L was used for immunodiagnosis, fascioliasis was successfully detected in both humans and ruminants (Cornelissen et al. 2001; Gonzales Santana et al. 2013).
The transcriptional upregulation of PpsCatL was mainly detected in the adult stage of P. pseudoheterotremus and less in the other developmental stages. The differential expression of the cathepsin L isoforms in different developmental stages is frequently reported in Fasciola species (Dowd et al. 1994). FgCatL1G is mainly expressed in the metacercariae and NEJ of F. gigantica, but not in the adult (Norbury et al. 2011; Sansri et al. 2013). The expression of PpsCatL in the adult P. pseudoheterotremus was localized to the gastrodermal tissues along the parasite gut, especially in the gut lamellae. Secretory PpsCatL was detected in the gut lumen and ES of the parasite. These data suggest important roles for PpsCatL in the parasite itself and in the environment of the host. Cathepsin L1 secreted from F. hepatica is considered to be a major virulence factor, with essential roles in helminth pathogenicity, such as in nutrient acquisition by the digestion of host proteins, the cleavage of the host extracellular matrix, allowing parasite migration, and the impairment of the host immunity by destroying immunoglobulin and suppressing the Th1 immune response (Collins et al. 2004).
Enzymatic activity of rPpsCatL was determined by measuring an increment of fluorescent units after substrate cleavage. The proteolytic activity of rPpsCatL was observed after autoactivation in acidic pH but not inactive enzyme. The autocatalysis is required for activation of pro-cathepsin L to mature enzyme by removing a pro-peptide region (reviewed by Turk et al. 2012). In Fasciola, activation of pro-cathepsin Ls at acidic pH (approximately pH 4–5) exhibited mature enzyme that actively digested substrates (Collins et al. 2004; Sansri et al. 2013; Yamasaki et al. 2002). Inhibition of rPpsCatL activity with cysteine protease inhibitor, E64, but not other inhibitor families (data not shown) confirmed a property of PpsCatL as cysteine protease.
The rPpsCatL-ELISA was highly sensitive and specific in the diagnosis of human paragonimiasis. However, the results presented here could not confirm the species specificity of the ELISA because no species-confirmed sera were available. The cross-reaction of cathepsin L within the same genus has been reported for F. hepatica- and F. gigantica-infected ruminants. The detection of F. gigantica with an FhCatL-ELISA elicited similar outcome for F. hepatica (Cornelissen et al. 2001). Therefore, the positive results shown in this experiment may be attributable to diverse Paragonimus species, such as P. westermani, P. heterotremus, and P. pseudoheterotremus. Further evidence of cross-reactivity is the high percentage identity (>45 %) between the cathepsin L amino acid sequences within the genus Paragonimus. However, validation of the rPpsCatL-ELISA using sera with confirmed species-specific paragonimiasis, especially from animals infected with P. heterotremus, is required to clarify this issue. P. heterotremus and P. pseudoheterotremus are sister species with high genetic similarity (Intapan et al. 2012). Therefore, the high homology of the cathepsin L proteins in both species might cause cross-reactivity of the rPpsCatL-ELISA. However, cathepsin L has not yet been identified in P. heterotremus, and its discovery will allow the basic functions and properties of the cathepsin L protein in these sibling species to be compared, including their sequence homology, protein structures, biochemistry, and immune response. Only 10 from 197 samples of heterologous sera produced false positive results with the rPpsCatL-ELISA, including sera from individuals infected with O. viverrini (three cases), Ascaris lumbricoides (two cases), B. malayi (two cases), S. stercoralis (one case), Toxocara sp. (one case), and Fasciola sp. (one case). Nine heterologous-infected sera showed weak positive with rPpsCatL, perhaps because the high antibody titers raised against these diseases affect the specificity of antibodies. As demonstrated previously, false positive HIV tests (Murex HIV Ag/Ab combination EIA) were significantly associated with increased levels of S. haematobium IgG1 (Everett et al. 2010). One serum from an individual infected with A. lumbricoides reacted strongly with rPpsCatL in our study, which may be attributable to a previous history of paragonimiasis or another closely related helminthic infection.
The high sensitivity (100 %) and specificity (95.6 %) of the rPpsCatL-ELISA suggest its potential utility in screening for paragonimiasis. The higher specificity of the rPpsCatL-ELISA (95.6 %) compared with that of the CWA-ELISA (84.1 %) suggests that CWA be replaced with rPpsCatL for clinical immunodiagnosis. However, the rPpsCatL-ELISA must be evaluated with nonparasitic infectious diseases (e.g., viral, bacterial, and fungal) and noninfectious diseases (e.g., lung cancer, lung diseases) in future studies.
In summary, a novel cathepsin L-like protease was identified in P. pseudoheterotremus, which is predominantly expressed in the adult stage. This cathepsin L was found in ES products and specifically localizes to the intestinal brush border of the parasite. The recombinant protein expressed in E. coli reacted with rat-infected sera and differentially diagnosed human paragonimiasis with high sensitivity and specificity. Further studies that evaluate the utility of the rPpsCatL-ELISA in species-specific paragonimiasis, nonparasitic infections, and some noninfectious lung diseases will further the development of an rPpsCatL-based immunodiagnostic tool in the near future.
References
Adisakwattana P, Viyanant V, Chaicumpa W, Vichasri-Grams S, Hofmann A, Korge G, Sobhon P, Grams R (2007) Comparative molecular analysis of two asparaginyl endopeptidases and encoding genes from Fasciola gigantica. Mol Biochem Parasitol 156(2):102–116. doi:10.1016/j.molbiopara.2007.07.006
Alves LC, Melo RL, Cezari MH, Sanderson SJ, Mottram JC, Coombs GH, Juliano L, Juliano MA (2001) Analysis of the S(2) subsite specificities of the recombinant cysteine proteinases CPB of Leishmania mexicana, and cruzain of Trypanosoma cruzi, using fluorescent substrates containing non-natural basic amino acids. Mol Biochem Parasitol 117(2):137–143
Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, Schwede T (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 42(Web Server issue):W252–W258. doi:10.1093/nar/gku340
Chung YB, Kita H, Shin MH (2008) A 27 kDa cysteine protease secreted by newly excysted Paragonimus westermani metacercariae induces superoxide anion production and degranulation of human eosinophils. Korean J Parasitol 46(2):95–99. doi:10.3347/kjp.2008.46.2.95
Collins PR, Stack CM, O’Neill SM, Doyle S, Ryan T, Brennan GP, Mousley A, Stewart M, Maule AG, Dalton JP, Donnelly S (2004) Cathepsin L1, the major protease involved in liver fluke (Fasciola hepatica) virulence: propetide cleavage sites and autoactivation of the zymogen secreted from gastrodermal cells. J Biol Chem 279(17):17038–17046. doi:10.1074/jbc.M308831200
Cornelissen JB, Gaasenbeek CP, Borgsteede FH, Holland WG, Harmsen MM, Boersma WJ (2001) Early immunodiagnosis of fasciolosis in ruminants using recombinant Fasciola hepatica cathepsin L-like protease. Int J Parasitol 31(7):728–737
Dalton JP, Clough KA, Jones MK, Brindley PJ (1996) Characterization of the cathepsin-like cysteine proteinases of Schistosoma mansoni. Infect Immun 64(4):1328–1334
Day SR, Dalton JP, Clough KA, Leonardo L, Tiu WU, Brindley PJ (1995) Characterization and cloning of the cathepsin L proteinases of Schistosoma japonicum. Biochem Biophys Res Commun 217(1):1–9
Dowd AJ, Smith AM, McGonigle S, Dalton JP (1994) Purification and characterisation of a second cathepsin L proteinase secreted by the parasitic trematode Fasciola hepatica. Eur J Biochem 223(1):91–98
Everett DB, Baisely KJ, McNerney R, Hambleton I, Chirwa T, Ross DA, Changalucha J, Watson-Jones D, Helmby H, Dunne DW, Mabey D, Hayes RJ (2010) Association of schistosomiasis with false-positive HIV test results in an African adolescent population. J Clin Microbiol 48(5):1570–1577. doi:10.1128/JCM.02264-09
Ferraro F, Merlino A, Dell Oca N, Gil J, Tort JF, Gonzalez M, Cerecetto H, Cabrera M, Corvo I (2016) Identification of chalcones as Fasciola hepatica cathepsin L inhibitors using a comprehensive experimental and computational approach. PLoS Negl Trop Dis 10(7):e0004834. doi:10.1371/journal.pntd.0004834
Ferre F, Clote P (2006) DiANNA 1.1: an extension of the DiANNA web server for ternary cysteine classification. Nucleic Acids Res 34(Web Server issue):W182–W185. doi:10.1093/nar/gkl189
Froger A, Hall JE (2007) Transformation of plasmid DNA into E. coli using the heat shock method. J Vis Exp 6:253. doi:10.3791/253
Gonzales Santana B, Dalton JP, Vasquez Camargo F, Parkinson M, Ndao M (2013) The diagnosis of human fascioliasis by enzyme-linked immunosorbent assay (ELISA) using recombinant cathepsin L protease. PLoS Negl Trop Dis 7(9):e2414. doi:10.1371/journal.pntd.0002414
Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R (2010) A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res 38(Web Server issue):W695–W699. doi:10.1093/nar/gkq313
Grams R, Vichasri-Grams S, Sobhon P, Upatham ES, Viyanant V (2001) Molecular cloning and characterization of cathepsin L encoding genes from Fasciola gigantica. Parasitol Int 50(2):105–114
Hwang HS, Chung HS (2002) Preparation of active recombinant cathepsin K expressed in bacteria as inclusion body. Protein Expr Purif 25(3):541–546
Intapan PM, Sanpool O, Thanchomnang T, Imtawil K, Pongchaiyakul C, Nawa Y, Maleewong W (2012) Molecular identification of a case of Paragonimus pseudoheterotremus infection in Thailand. Am J Trop Med Hyg 87(4):706–709. doi:10.4269/ajtmh.2012.12-0235
Irving JA, Spithill TW, Pike RN, Whisstock JC, Smooker PM (2003) The evolution of enzyme specificity in Fasciola spp. J Mol Evol 57(1):1–15. doi:10.1007/s00239-002-2434-x
Kane SE (1993) Mouse procathepsin L lacking a functional glycosylation site is properly folded, stable, and secreted by NIH 3T3 cells. J Biol Chem 268(15):11456–11462
Karrer KM, Peiffer SL, DiTomas ME (1993) Two distinct gene subfamilies within the family of cysteine protease genes. Proc Natl Acad Sci U S A 90(7):3063–3067
Kodera T, Asano M, Kawai M, Miwa T, Nio N (2005) The effective methods in refolding and activation of cathepsin L-like soybean protease D3. J Food Sci 70(8):c495–c502
Kollien AH, Waniek PJ, Nisbet AJ, Billingsley PF, Schaub GA (2004) Activity and sequence characterization of two cysteine proteases in the digestive tract of the reduviid bug Triatoma infestans. Insect Mol Biol 13(6):569–579. doi:10.1111/j.0962-1075.2004.00504.x
Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305(3):567–580. doi:10.1006/jmbi.2000.4315
Lecaille F, Chowdhury S, Purisima E, Bromme D, Lalmanach G (2007) The S2 subsites of cathepsins K and L and their contribution to collagen degradation. Protein Sci 16(4):662–670. doi:10.1110/ps.062666607
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25(4):402–408. doi:10.1006/meth.2001.1262
Na BK, Kim SH, Lee EG, Kim TS, Bae YA, Kang I, Yu JR, Sohn WM, Cho SY, Kong Y (2006) Critical roles for excretory-secretory cysteine proteases during tissue invasion of Paragonimus westermani newly excysted metacercariae. Cell Microbiol 8(6):1034–1046. doi:10.1111/j.1462-5822.2006.00685.x
Norbury LJ, Beckham S, Pike RN, Grams R, Spithill TW, Fecondo JV, Smooker PM (2011) Adult and juvenile Fasciola cathepsin L proteases: different enzymes for different roles. Biochimie 93(3):604–611. doi:10.1016/j.biochi.2010.12.004
Nuamtanong S, Dekumyoy P, Adisakwattana P (2012) Evaluation of recombinant serine protease inhibitor from Trichinella spiralis for immunodiagnosis of swine trichinosis. Southeast Asian J Trop Med Public Health 43(5):1094–1104
Pakchotanon P, Molee P, Nuamtanong S, Limpanont Y, Chusongsang P, Limsomboon J, Chusongsang Y, Maneewatchararangsri S, Chaisri U, Adisakwattana P (2016) Molecular characterization of serine protease inhibitor isoform 3, SmSPI, from Schistosoma mansoni. Parasit Res. doi:10.1007/s00436-016-5053-y
Pandey KC, Dixit R (2012) Structure-function of falcipains: malarial cysteine proteases. J Trop Med 2012:345195. doi:10.1155/2012/345195
Pandey KC, Barkan DT, Sali A, Rosenthal PJ (2009) Regulatory elements within the prodomain of Falcipain-2, a cysteine protease of the malaria parasite Plasmodium falciparum. PLoS One 4(5):e5694. doi:10.1371/journal.pone.0005694
Park H, Kim SI, Hong KM, Kim MJ, Shin CH, Ryu JS, Min DY, Lee JB, Hwang UW (2002) Characterization and classification of five cysteine proteinases expressed by Paragonimus westermani adult worm. Exp Parasitol 102(3-4):143–149
Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8(10):785–786. doi:10.1038/nmeth.1701
Rice P, Longden I, Bleasby A (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16(6):276–277
Sansri V, Changklungmoa N, Chaichanasak P, Sobhon P, Meemon K (2013) Molecular cloning, characterization and functional analysis of a novel juvenile-specific cathepsin L of Fasciola gigantica. Acta Trop 128(1):76–84. doi:10.1016/j.actatropica.2013.06.013
Sansri V, Meemon K, Changklungmoa N, Kueakhai P, Chantree P, Chaichanasak P, Lorsuwannarat N, Itagaki T, Sobhon P (2015) Protection against Fasciola gigantica infection in mice by vaccination with recombinant juvenile-specific cathepsin L. Vaccine 33(13):1596–1601. doi:10.1016/j.vaccine.2015.02.010
Shin MH, Kita H, Park HY, Seoh JY (2001) Cysteine protease secreted by Paragonimus westermani attenuates effector functions of human eosinophils stimulated with immunoglobulin G. Infect Immun 69(3):1599–1604. doi:10.1128/IAI.69.3.1599-1604.2001
Smith SM, Kane SE, Gal S, Mason RW, Gottesman MM (1989) Glycosylation of procathepsin L does not account for species molecular-mass differences and is not required for proteolytic activity. Biochem J 262(3):931–938
Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT, Lavrsen K, Dabelsteen S, Pedersen NB, Marcos-Silva L, Gupta R, Bennett EP, Mandel U, Brunak S, Wandall HH, Levery SB, Clausen H (2013) Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J 32(10):1478–1488. doi:10.1038/emboj.2013.79
Sugiyama H, Morishima Y, Kameoka Y, Arakawa K, Kawanaka M (2004) Paragonimus ohirai metacercariae in crabs collected along the Arakawa River in Tokyo, Japan. J Vet Med Sci 66(8):927–931
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739. doi:10.1093/molbev/msr121
Turk V, Stoka V, Vasiljeva O, Renko M, Sun T, Turk B, Turk D (2012) Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim Biophys Acta 1824(1):68–88. doi:10.1016/j.bbapap.2011.10.002
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3—new capabilities and interfaces. Nucleic Acids Res 40(15):e115. doi:10.1093/nar/gks596
Varghese A, Raina OK, Nagar G, Garg R, Banerjee PS, Maharana BR, Kollannur JD (2012) Development of cathepsin-L cysteine proteinase based Dot-enzyme-linked immunosorbent assay for the diagnosis of Fasciola gigantica infection in buffaloes. Vet Parasitol 183(3-4):382–385. doi:10.1016/j.vetpar.2011.07.032
Villa-Mancera A, Reynoso-Palomar A, Utrera-Quintana F, Carreon-Luna L (2014) Cathepsin L1 mimotopes with adjuvant Quil A induces a Th1/Th2 immune response and confers significant protection against Fasciola hepatica infection in goats. Parasitol Res 113(1):243–250. doi:10.1007/s00436-013-3650-6
Waikagul J (2007) A new species of Paragonimus (Trematoda: Troglotrematidae) from a cat infected with metacercariae from mountain crabs Larnaudia larnaudii. J Parasitol 93(6):1496–1500. doi:10.1645/GE-1054.1
Warner AH, Pullumbi E, Amons R, Liu L (2004) Characterization of a cathepsin L-associated protein in Artemia and its relationship to the FAS-I family of cell adhesion proteins. Eur J Biochem 271(20):4014–4025. doi:10.1111/j.1432-1033.2004.04338.x
Yamasaki H, Mineki R, Murayama K, Ito A, Aoki T (2002) Characterisation and expression of the Fasciola gigantica cathepsin L gene. Int J Parasitol 32(8):1031–1042
Acknowledgments
This study was supported by grant from Thailand Research Fund (TRF) in the program of initiative career development through Dr. Tippayarat Yoonuan (TRG5480011) and also partially supported by ICTM grant of the Faculty of Tropical Medicine. We thank Jaruchat Boonnachot from Department of Tropical Pathology, Faculty of Tropical Medicine, Mahidol University for tissue processing in immunolocalization. Our gratitude also goes to the Faculty of Tropical Medicine for funding the proofreading, editing, and page charging of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
All animal works were conducted with the approval of the Faculty of Tropical Medicine Animal Care and Use Committee (no. FTM-ACUC 011/2012). Leftover helminth-infected human sera and healthy sera were used in this study with the permission of the Immunodiagnosis for Helminthiasis Unit, Department of Helminthology, Faculty of Tropical Medicine, Mahidol University, and the approval of the Ethics Committee of the Faculty of Tropical Medicine, Mahidol University (no. MUTM 2011-056-01).
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Yoonuan, T., Nuamtanong, S., Dekumyoy, P. et al. Molecular and immunological characterization of cathepsin L-like cysteine protease of Paragonimus pseudoheterotremus . Parasitol Res 115, 4457–4470 (2016). https://doi.org/10.1007/s00436-016-5232-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-016-5232-x