Abstract
The parasite Trypanosoma cruzi (Kinetoplastida, Trypanosomatidae) can be classified based on biochemical and molecular markers, into six lineages or discrete typing units (DTUs), T. cruzi I–VI (TcI–VI), from which TcI and TcII are the parental genotypes. Trying to understand the dispersion of the subpopulations of T. cruzi in nature and its complex transmission cycles, the serine carboxypeptidase genes of T. cruzi were used as a molecular marker in the present study. DTUs of 25 T. cruzi isolates derived from different hosts and from different regions of Brazil were classified. Using specific primers, the complete serine carboxypeptidase open reading frame of 1401 bp was sequenced. The obtained data shows significant differences in the sequences of TcI and TcII. The analysis of the T. cruzi significantly different serine carboxypeptidase genes allowed distinguishing between the parental DTUs TcI to TcII and the hybrid DTU TcVI which grouped within the latter branch. The sequence diversity within the T. cruzi subpopulations was rather low. The analysis using the genes encoding proteases seems to be an interesting approach for the reconstruction of the origin and genotype evolution of T. cruzi.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The infection by the protozoan Trypanosoma cruzi (Kinetoplastida, Trypanosomatidae) is considered as an anthropozoonosis which occurs in nature as part of a complex transmission cycles that can be divided into different ecotopes such as sylvatic, peridomiciliar, and domiciliar (Garcia et al. 2007; Araújo et al. 2009; Coura 2013). T. cruzi is transmitted by various species from a group of blood-sucking insects which are known as kissing bugs or triatomines (Hemiptera, Reduviidae) (Jurberg and Galvão 2006).
The genetic variability of T. cruzi has been well demonstrated by many different aspects involving the parasite-vector interactions, the presence of clinical variations of the disease which are probably strongly associated with the intrinsic characteristic of the parasite and its host and the concomitant occurrence of different strains in the same host, as well as the large geographical dispersion of this parasite (Mello et al. 1996; Araújo et al. 2007, 2008, 2014; Garcia et al. 2007; Coura 2013; Coura et al. 2014). According to molecular biology data, the strains have been classified into six discrete typing units (DTUs), named TcI–TcVI (Zingales et al. 2012).
Proteases are a crucial part in many processes of differentiation of diverse microorganisms, parasite development and infection and mainly parasite-host interaction (McKerrow 1988; Teichert et al. 1989; Meirelles et al. 1992). So far, proteases from different classes have been identified in T. cruzi, mainly cruzipain, which is a major cysteine proteinase, characterized as endopeptidase with proteolytic activity in acidic conditions (Cazzulo et al. 1989; Murta et al. 1990; Parussini et al. 2003). Cathepsin B-like cysteine protease is another example of an endopeptidase of T. cruzi (Cazzulo 2002). The proteolytic activity is related to the differentiation of the parasite, mammalian host cells infection and the runaway of the parasite from the immune system of the host (Cicarelli and Lopes 1989; Bontempi and Cazzulo 1990; Bonaldo et al. 1991; Burleigh et al. 1997). Besides a strong attention to the endopeptidases in the literature, less is known about the exopeptidases in T. cruzi. The carboxypeptidases belong to the exopeptidase class and are a protease that cleaves the C-terminal amino acid residue from proteins and peptides, considered as major catalytic proteases, divided into metallo-caroxypeptidases (EC 3.4.17), cysteine-type carboxypeptidases (EC 3.4.18) and serine-type carboxypeptidases (SCPs, EC 3.4.16), where the latter have their major activity at acid pH (Breddam 1986; Remington and Breddam 1994). SCPs have been classified into subtypes C and D: C-type has affinity for hydrophobic C-terminal amino acid residues, while D-type hydrolyses basic residues at the same position (Mortensen et al. 1999).
Since only little is known about the carboxypeptidases composition in different T. cruzi genotypes, the main point of our study was to analyse differences in the gDNA level of the two parental subpopulation of T. cruzi (TcI and TcII) using the gene encoding SCP as a new molecular marker to differentiate these two groups. In the present study, a phylogenetic analysis was carried out for isolates from different hosts, regions and biomes of Brazil.
Material and methods
Parasites
Epimastigotes of 25 T. cruzi isolates derived from mammals, humans and vectors were grown in McNeal, Novy and Nicolle (NNN) medium with liver infusion tryptose (LIT) overlay supplemented with 10 % foetal calf serum (Chiari and Camargo 1984). Fifteen T. cruzi isolates were previously characterized by zymodeme, mini-exon polymerase chain reaction (PCR) and/or lectins, respectively (Pinho et al. 2000; Araújo et al. 2002, 2011; Herrera et al. 2004, 2005). The remaining isolates were characterized in the present work.
Genomic DNA extraction and characterization of Trypanosoma cruzi DTUs
Genomic DNA was extracted from 2 mL (approximately 1 × 106 parasites/mL) of axenic medium culture containing T. cruzi using the DNeasy Tissue Kit (Qiagen, Hilden, Germany) following the manufacture’s protocol and stored at −20 °C. So far, uncharacterized T. cruzi isolates used in the present study were determined as described previously (Brisse et al. 2001; Araújo et al. 2011). In brief, PCR amplification of the divergent domains of 24Sα ribosomal RNA (rRNA), 18S rRNA and the mini-exon gene was carried out on a Mastercycler (Eppendorf, Germany). Amplicons were separated by electrophoresis on 3 % ethidium bromide stained agarose gels. For determination of the PCR products size, the FragSize programme version 1.0.3 was used (www.bioinformatics.org). For standardization, the T. cruzi strains Y and F90 were used.
Polymerase chain reaction (PCR) of serine carboxypeptidase encoding genes
Specific forward and reverse primers were designed based on the highly conserved non-coding 5′- and 3′-regions of the sequences encoding T. cruzi SCP (GenBank accession numbers XM_797910 and AY310135, respectively): TCSCP-For (5′-GCGAAGGAAAAGACTCTGCAGG-3′) and TCSCP-Rev (5′-AGAAAACAACAGTCCACGTCA-3′). PCR reaction was carried out on a Mastercycler (Eppendorf). The cycling parameters were used according to Waniek et al. (2014).
Cloning and sequences
PCR products were excised from the gel, purified using a NucleoSpin® Gel and PCR Clean-up Kit (Macheray-Nagel, Germany) and cloned into the pGEM-T Easy Vector (Promega, USA) following the manufacture’s protocols. To exclude PCR and sequencing errors, at least three clones of each T. cruzi isolate were sequenced in both directions by Plataforma Genomica—Sequenciamento de DNA/PDTIS-FIOCRUZ/IOC, Rio de Janeiro, using the forward and reverse M13 primers.
Sequences and identity analyses
Because of possible nucleotide mismatches in the primer regions, these regions have been trimmed after sequencing and only the coding T. cruzi SCP regions were used for further analyses. Identity assessments were carried out by Blastx (Altschul et al. 1990) searches using the web servers of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/). The deduced amino acid sequences were aligned using Mafft version 6.937b. The respective amino acid sequences were deduced by using the translate tool on the Expasy homepage (http://web.expasy.org/translate/). All sequences generated for this study were submitted to the GenBank and received accession numbers: T. cruzi strain 832 (GU084438), JCA3 (GU084439), 659 (GU084440) and the other isolates have the respective numbers: KT327421-KT327454.
Phylogenetic analysis of Trypanosoma cruzi serine carboxypeptidases
For the SCP1 gene of different T. cruzi isolates, we created two data sets, one consisting of 41 ingroup taxa and 12 outgroup taxa and one consisting only of the ingroup. The final tree reconstruction showed that branches between the ingroup and outgroup are rather long, bearing the risk that the presence of the outgroup could alter the topology of the ingroup. In order to investigate and finally exclude this possibility, phylogenetic trees have been computed for both data sets and have finally been compared.
Both data sets were aligned with Mafft v6.937b using the “linsi” algorithm. For the full data set, a few obvious alignment problems have been edited by the eye. The full data set consists of 53 taxa and has a length of 1480 bp. The data set excluding the outgroup is gap free. Appling the mafft-linsi programme to it reproduced the input data set. This data set, which consists of 41 sequences, has a length of 1401 bp.
Bayesian phylogenetic analyses were conducted with MrBayes v. 3.1.2 (Ronquist and Huelsenbeck 2003) using 20 million MCMC generations for the data set with outgroup and 10 million generations for the data set without outgroup. Trees and model parameters where sampled every 100 generation. For both data sets, the Markov chains quickly converged. Convergence was tested as follows: split divergence <0.01 and likelihood values of MCMC runs were found to reach a plateau. A burning of 1000 (which corresponds to 100,000 generations) was chosen for both data sets.
Appropriate DNA substitution models were determined with jModeltest 2.1 (Darriba et al. 2012) using the Akaike’s information criterion (AIC) and Bayesian information criterion (BIC). Models were restricted to those available in MrBayes. Following the manual of MrBayes, substitution models were specified in MrBayes without fixing the parameter values, allowing them to vary during the annealing process. This should lead to more conservative but more realistic posterior probabilities.
The outgroup contains SCP sequences of the following species: Trypanosoma vivax (CCC51045), Trypanosoma brucei brucei (XM_817270), Trypanosoma brucei gambiense (FN554973-1, FN554973-2, FN554973-3, FN554973-4), Leishmania braziliensis (XM_001563925), Leishmania major (XM_001682399), Leishmania donovani (XM_003860011), Leishmania infantum (XM_001464824), Leishmania mexicana (XM_003874010) and Monosiga brevicollis (XM_001748835). Furthermore, we added following T. cruzi CL Brener (TcVI) strains in the ingroup (XM_797910, XM_812675, XM_812676, and AY184244).
Results
Sequencing data
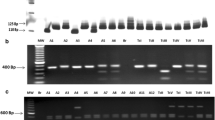
After PCR amplification with specific primers and separation on an agarose gel, a single distinct band became visible. All isolates showed the same PCR product size, lacking secondary bands (not shown). The sequencing procedure revealed that the size of all coding SCP regions is 1401 bp. In total, 426 sequences were obtained from 139 clones originating from 26 T. cruzi isolates and strains, respectively. From the isolates 593, 684, C45, F90, 645 and 328 two, from 657 and 661 three and from 518 four copies of SCP encoding nucleotide sequence were identified.
Trypanosoma cruzi DTUs
After amplification of 24Sα rRNA, 18S rRNA and mini-exon gene, electrophoresis of the amplicons and determination of band sizes, the T. cruzi isolate 518 found in humans was classified as TcII. The isolates 661, F90, 645, G05 and D7 found in Didelphis marsupialis and C45 found in Philander frenatus (both Didelphidae) were classified as TcI, respectively (Table 1). The rodent Proechimys sp. (Echimyidae) from the Amazon rain forest was infected with T. cruzi TcI (10268). The chiropterans Phyllostomus hastatus and Artibeus jamaicensis (both Phyllostomidae) were infected either with Trypanosoma cruzi marinkellei (323) only or were co-infected with two different trypanosomatid species, T. c. marinkellei and T. cruzi TcIII (328), respectively. From the isolate 8584 no PCR products could be amplified by any of the three methods mentioned above.
Serine carboxypeptidases gDNA and deduced amino acid sequences
The T. cruzi I group displayed clear differences in the genomic DNA (gDNA) of the SCP gene in comparison to the T. cruzi II group in the number of nucleotide substitutions. Throughout the T. cruzi SCP, nucleotide sequence 18 characteristic substitutions occurred (10 transitions, 8 transversions) which were present in more than 10 of the analysed clones and which differ in the two DTUs TcI and TcII. Most of these nucleotide substitutions are located in the region encoding the SCP signal peptide. From these point mutations, only four were missense mutations, leading to an amino acid replacement but without affecting the enzymatic function. Beside this nucleotide substitution pattern, there were single mutations distributed all over the T. cruzi SCP sequences. The open reading frames of all obtained T. cruzi SCP sequences are 1401-bp long, encoding a zymogen of 466 amino acid residues. All coding regions started with the initial ATG codon and terminated with the stop codon TGA. None of the nucleotide substitutions were located in regions encoding functional or structural important amino acid residues, e.g. disulphide bridges forming cysteine residues, catalytic triad or S1 and S1’ subsites. In all T. cruzi SCPs, the signal peptides were 21 amino acids long. Molecular weight and isoelectric point of the T. cruzi SCP amino acid sequences ranged from 51.5 to 51.8 kDa and 4.99 to 5.24, respectively.
On the nucleotide level, the pairwise similarities of T. cruzi SCP encoding sequences varied between 93.7 % (8584, XM_797910) and 100 %. Within the TcI group, we obtained the D7 sequence, whereas in the TcII group, five isolates were identical (593-2, 684-2, JCA6, Y, JCA3). On the amino acid level, T. cruzi SCP pairwise sequence similarities varied between 89.0 % (328-2, 8584) and 100 %. In addition, the TcII protein sequences were more homogenous (12 identical out of 16) than those from the TcI group (4 identical out of 16). The pairwise similarity of sequences obtained in the present study to the SCP of Trypanosoma brucei varied from 62.1 % (8584) to 63.4 % (661-1, 661-2, 291, C45-1, 645-1, 645-2, 657-2, 657-3, CL Brener AY184244 and XM_812675/6).
Phylogenetic analysis
For the full data set, the model with the best AIC value (which is also available in MrBayes) is the GTR+I+G model. The BIC was more conservative and chose the HKY+I+G model. For the data set without outgroup, the AIC suggested the GTR+I model and the BIC the K80+I model. In order to exclude a dependence of the phylogenetic tree on the model, both data sets were analysed with the GTR+I+G and the HKY+I+G models and the data set without outgroup additionally with the K80+I model. The resulting topologies were compared. For the full data set as well as the data set without outgroup, the resulting topologies were the same under all substitution models that have been tested. (Results are only shown for the GTR+I+G model).
Furthermore, the topologies of the ingroup were identical for the data sets with and without outgroup, (tree is not shown). Thus, the phylogenetic relations among the T. cruzi isolates does not depend on the choice of the model (for the models listed above) and the topology of the ingroup does not depend on the presence of the outgroup. Together with the high posterior probabilities of the resulting tree, this provided a high confidence into the phylogenetic relations among the T. cruzi isolates. Since the GTR+I+G model should provide most realistic posterior probabilities, this is the tree we show in Fig. 1 for the full data set with collapsed outgroup and in Supplemental Figure 1 with the full outgroup.
Bayesian phylogenetic tree of the full data set using the GTR+I+G model. In order to be able to see the branch lengths of the ingroup, the outgroup with its long branches has been collapsed. Branch labels depict Bayesian posterior probabilities. The full phylogenetic tree is shown in Supplemental Figure 1. The respective genotypes are shown after the name of the isolates. Parasites without genotype are T. c. marinkellei
The phylogenetic tree of the SCP encoding flagellate sequences shows a high posterior probability of 1 for the outgroup, which consists of different non-T. cruzi Kinetoplastida species. Similar high support values are found for the two most outer branches of the ingroup. The smaller of the two ingroup branches consists of taxa characterized as T. c. marinkellei (Fig. 1). The larger ingroup branch is the main T. cruzi branch within which TcI and TcII sequences are again separated by high Bayesian posterior probabilities. Inside the TcI branch the only differing genotype was 328-2, which is part of a mixed isolate (328-1, 328-2) characterized as a TcIII/T. c. marinkellei (Table 1). Genotype 328-2 can be clearly distinguished from the remaining TcI group. As expected, the other SCP encoding sequence of this isolate (328-1) was located in the T. c. marinkellei branch (Fig. 1). All analysed CL Brener (TcVI) SCP encoding sequences clustered in the TcII sub-branch (Fig. 1). Two of these sequences (XM_812676, XM_812675) clustered together with the isolate 10148, previously characterized as TcIII, a group that can clearly be distinguished from the remaining TcII sequences. Among the remaining TcII-like sequences, the CL Brener sequence AY184244 was again separated from the rest.
The SCP sequences inside the TcII sub-branch are more heterogeneous since the internal branch lengths are longer in comparison to the TcI sub-branch. As expected, the reference isolates F90 and Y clustered inside the TcI and TcII branches, respectively. In the T. c. marinkellei branch the isolates 328 and 323 grouped together whereas isolate 8584, which could not be characterize by standard methods was further separated.
Discussion
Though the association of different T. cruzi DTUs with the etiopathology of the Chagas disease is still controversial, some authors consider that the genetic constitution of different T. cruzi strains could be associated with the dissimilar clinical manifestations of the illness (Prata 2001; Marin-Neto et al. 2007). One important factor in the variation of clinical characteristics due to differing tissue tropism is related to molecular interactions between parasite and host at cell surface level (Macedo and Pena 1998; Andrade and Andrews 2005). Parasites that belong to the TcI genotype seem to be less pathogenic than TcII genotype and the hybrid DTUs that regularly cause more severe chagasic syndromes in Brazil (Yeo et al. 2005). This supports the hypothesis that the origin of the main T. cruzi subgroups might influence the course of the disease, although major genes determining parasite tropism and virulence remain to be identified (Macedo and Pena 1998; Briones et al. 1999; Andrade et al. 2010). Differing gene structure, gene expression, protein synthesis, and thus varying enzymatic specificity and activity might be one of the factors that influences the virulence of a T. cruzi isolate (Duschak et al. 2001; Lima et al. 2001; Uehara et al. 2012; Walker et al. 2014). Our sequence comparison of genes encoding SCP in T. cruzi shows a clear divergence between the parental T. cruzi subtypes TcI and TcII. How far these enzymes modulate the virulence of T. cruzi must be investigated. However, since the gene products have highly similar amino acid residues in the active regions, the modulation of virulence by these enzymes is probably low. SCPs are only a small part of the T. cruzi digestive enzyme portfolio. Therefore, the differences we see in SCP are likely to be only one of the many small differences, which only together accumulate to a noticeable synergetic effect on the parasites virulence.
Genome sequencing of the T. cruzi CL Brener (TcVI) clone has shown the presence of five SCP encoding genes (Parussini et al. 2003; El-Sayed et al. 2005). Unfortunately, the CL Brener SCP sequences XM_812678 and XM_797909 are incomplete and could belong to the same gene. However, by comparison of SCP sequences identified in the present study, it was always possible to determine the respective T. cruzi DTUs, even if more than one gene copy or allele were identified for one isolate. The CL Brener SCP genes, available in GenBank, show characteristics of different T. cruzi DTUs, but clustered all within the T. cruzi TcII-like branch. This finding is conclusive to the revisited model for T. cruzi evolution (Tomasini and Diosque 2015), in which TcVI is a product of hybridization between TcII and TcIII because this T. cruzi DTU does not possess SCP encoding genes, which cluster within both parental T. cruzi genotypes.
Infections by mixed T. cruzi genotypes are a common phenomenon in nature (Torres et al. 2004; Bosseno et al. 2006; Araújo et al. 2011). In different triatomine species, mixed infections may modulate the colonization pattern in the insect’s intestinal tract (Araújo et al. 2007, 2008, 2014). If the mixed infections are composed of highly divergent T. cruzi strains (e.g. TcI and TcII), its differentiation by PCR typing methods is simple. But if the T. cruzi DTUs are closely related (e.g. TcV and TcVI), its differentiation by simple characterization methods is much more difficult. Therefore, a mixed infection could be misinterpreted as a single infection, if the differentiation of the DTUs failed. Since mixed infections were identified in the present study, this scenario does not seem to be unlikely. Mixed infections with similar subgroups or different strains within the same DTU, might be called “cryptic mixed infections”.
Historically, two alternative hypotheses for the evolution of the TcIII, TcIV and TcV subtypes by hybridization events were proposed (Westenberger et al. 2005; Sturm and Campbell 2010). In the first hypothesis, the “two-hybridization” model, the homozygous DTUs TcIII and TcIV emerged by fusion from the parental genotypes TcI and TcII (Zingales et al. 2012). According to this vision, the heterozygous TcV and TcVI (former TcIId and TcIIe) are products of a second hybridization between TcII and TcIII (Westenberger et al. 2005; Freitas et al. 2006; Tomazi et al. 2009; Zingales et al. 2009). It has also been argued that any examined gene locus showed the separation into four sequence groups, identical with TcI to TcIV, whereas sequences obtained from TcV/VI fall either into the TcII or TcIII clades (Westenberger et al. 2005; Sturm and Campbell 2010). By Southern blot analyses, an intermediate profile has been shown for the hybrid T. cruzi DTUs TcV and TcVI, in which probes of certain genes produced either TcI- or TcII-like signals (Pedroso et al. 2003). A second hypothesis, the “three-ancestor” model, proposes three parental T. cruzi genotypes (TcI, TcII and TcIII) and two-hybridization events (Zingales et al. 2012). The “three-ancestor” model is supported by the grouping of all CL Brener SCP copies within the TcII-like branch. According to this model, TcVI would never have had contact to TcI, and therefore, it should strongly differ in the sequence of SCP. However, other factors like gene loss could have also led to the observed pattern. The “two-hybridization” model is supported by the grouping of the isolate 328-2 (TcIII). Based on the “three-ancestor” hypothesis, TcIII would never have been in contact to TcI and should therefore group in the TcII branch. In a more recent T. cruzi evolution model, the ancestral diversified into two different groups TcI-TcIII-TcIV and TcII. In two subsequent hybridization events between TcII and TcIII, the hybrids TcV and TcVI originated (Tomasini and Diosque 2015). Our results support this T. cruzi development model because TcI and TcII are well separated into two clades and all TcVI SCP sequences are grouping together with TcII. One TcIII isolate grouped in the TcI branch and another one in the TcII branch. This might be explained by the fact that TcIII is both one parental genotype of TcVI but also a sister group of TcI. A differing heredity of the analysed genes might cause an intermediate position of the TcIII genotype.
In the present study, we showed that T. cruzi DTUs could be separated and characterized by the comparison of SCP’s, which confirms previous findings. For heterogeneous T. cruzi hybrids, SCP’s of the parental T. cruzi DTUs TcI and TcII seem to be present in multiple copies/alleles, though always clustering within the respective DTU group (TcI or TcII). In this regard, the TcII branch (containing TcIII and TcVI) seems to be more heterogeneous than TcI. This is documented by the dendrograms showing a distinct heterogenous character of CL Brener and T. cruzi isolate 684, of which various SCP forms cluster within the same T. cruzi DTU branch. The acquired property of heterogeneous genes in hybrid TcVI stocks might also expand the tissue tropism of these T. cruzi subgroups and hence be responsible for its higher pathogenicity. Different T. cruzi stocks vary also in their total DNA content and in the size of homologous chromosomes (Dvorak et al. 1980; Pedroso et al. 2003). Genetic recombination due to an unusual mechanism of fusion and subsequent loss of alleles has been shown previously (Gaunt et al. 2003). Some coding sequences are maintained whereas other seems to be trimmed. In the case of SCP encoding genes, there is a clear difference in both the quantity and quality of genetic information between the parental T. cruzi genotypes and their hybrids. The present study is another contribution to the clarification that confirms both the laboratory model and the genetic T. cruzi structure in nature.
The SCP sequences of isolates 323, 328 and 8584 strongly differ from all other T. cruzi isolates. Since none of the CL Brener SCPs cluster with these three sequences, we assume that these isolates are not T. cruzi. In accordance with the characterization by Brisse et al. (2001), we classified these three isolates as T. c. marinkellei. In addition, our results are supported by previous studies of Phyllostomus and Artibeus species which are primarily infected by T. c. marinkellei (Barnabe et al. 2003; Maia da Silva et al. 2009). These genotypes have previously been characterized as TcII, TcII/TcIII or Trypanosoma rangeli, respectively (Lisboa et al. 2008; Fampa et al. 2010). According to previous investigations, TcIII is a DTU commonly associated with armadillos (Yeo et al. 2005; Llewellyn et al. 2009). In the present study, we showed that these isolates are in fact T. c. marinkellei. A possible reason for this discrepancy is the characterization method used in the previous studies. The authors have chosen the mini-exon gene technique, which only distinguishes between TcI and TcII. This less discriminative method might have revealed contradictory results when T. c. marinkellei, a trypanosomatid obviously closely related to T. cruzi, is used in the analysis. Altogether, we propose that the phylogenetic classification method used in this study should be the standard for further classifications of T. cruzi DTUs since this method is far less error prone than methods which rely only on partial sequences. In particular, the classification of bat trypanosmatids should be reconsidered in view of this study.
Looking for new treatment strategies of Chagas disease, T. cruzi cysteine proteases (cruzipain) recently became a possible new target for a medical treatment (Zingales et al. 2014). Specific inhibitors are able to obstruct crucial physiological functions of the parasite and thus disturb its establishment in the respective host. The enzymatic composition (protein primary structure, presence of enzyme isoforms) of the T. cruzi strain might be an important factor, which can modulate drug efficacy. The results of the present study emphasize the differences in proteases of different T. cruzi DTUs and confirm the importance of considering the respective T. cruzi strain.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Andrade LO, Andrews NW (2005) The Trypanosoma cruzi-host-cell interplay: location, invasion, retention. Nat Rev Microbiol 3:819–823
Andrade LO, Galvão LM, Meirelles Mde N, Chiari E, Pena SD, Macedo AM (2010) Differential tissue tropism of Trypanosoma cruzi strains: an in vitro study. Mem Inst Oswaldo Cruz 105:834–837
Araújo CAC, Mello CB, Jansen AM (2002) Trypanosoma cruzi I and Trypanosoma cruzi II: recognition of sugar structures by Arachis hypogaea (peanut agglutinin) lectin. J Parasitol 88:582–586
Araújo CAC, Cabello PH, Jansen AM (2007) Growth behaviour of two Trypanosoma cruzi strains in single and mixed infections: in vitro and in the intestinal tract of the blood-sucking bug, Triatoma brasiliensis. Acta Trop 101:225–231
Araújo CAC, Waniek PJ, Jansen AM (2008) Development of Trypanosoma cruzi (TcI) isolate in the digestive tract of an unfamiliar vector, Triatoma brasiliensis (Hemiptera, Reduviidae). Acta Trop 107:195–199
Araújo CAC, Waniek PJ, Jansen AM (2009) An overview of Chagas disease and the role of triatomines on its distribution in Brazil. Vector Borne Zoonotic Dis 9:227–234
Araújo CAC, Waniek PJ, Xavier SCC, Jansen AM (2011) Genotype variation of Trypanosoma cruzi isolates from different Brazilian biomes. Exp Parasitol 127:308–312
Araújo CAC, Waniek PJ, Jansen AM (2014) TcI/TcII co-infection can enhance Trypanosoma cruzi growth in Rhodnius prolixus. Parasit Vectors 7:94
Barnabe C, Brisse S, Tibayrenc M (2003) Phylogenetic diversity of bat trypanosomes of subgenus Schizotrypanum based on multilocus enzyme electrophoresis, random amplified polymorphic DNA, and cytochrome b nucleotide sequence analyses. Infect Genet Evol 2:201–208
Bonaldo MC, D’Escoffier NL, Sales JM, Goldenberg S (1991) Characterization and expression of proteases during Trypanosoma cruzi metacyclogenesis. Exp Parasitol 73:44–51
Bontempi E, Cazzulo JJ (1990) Digestion of human immunoglobulin G by the major cysteine proteinase cruzipain from Trypanosoma cruzi. FEMS Microbiol 70:337–342
Bosseno M-F, García LS, Baunaure F, Gastelúm EM, Gutierrez MS, Kasten FL, Dumonteil E, Brenière SF (2006) Identification in triatomine vectors of feeding sources and Trypanosoma cruzi variants by heteroduplex assay and a multiplex miniexon polymerase chain reaction. Am J Trop Med Hyg 74:303–305
Breddam K (1986) Serine carboxypeptidase. Rev Carlsberg Res Commun 51:83–128
Briones MRS, Souto RP, Stolf BS, Zingales B (1999) The evolution of two Trypanosoma cruzi subgroups inferred from rRNA genes can be correlated with the interchange of American mammalian to pathogenicity and host specificity. Mol Biochem Parasitol 104:219–232
Brisse S, Verhoef J, Tibayrenc M (2001) Characterization of large and small subunit rRNA and mini-exon genes further supports the distinction of six Trypanosoma cruzi lineages. Int J Parasitol 31:1218–1226
Burleigh BA, Caler EV, Webster P, Andrews NW (1997) A cytosolic serine endopeptidase from Trypanosoma cruzi is required for the generation of Ca2+ signaling in mammalian cells. J Cell Biol 136:609–620
Cazzulo JJ, Couso R, Raimundi A, Hellman U (1989) Further characterization and partial amino acid sequence of a cysteine proteinase from Trypanosoma cruzi. Mol Biochem Parasitol 33:33–42
Cazzulo JJ (2002) Proteinases of Trypanosoma cruzi: potential targets for the chemotherapy of Chagas disease. Curr Topics Med Chem 2:1257–1267
Chiari E, Camargo EP, Culture and cloning of Trypanosoma cruzi (1984) Genes and antigens of parasites. In: Morel CM (ed) A laboratory manual, Fundação Oswaldo Cruz. World Health Organization, Rio de Janeiro, pp 23–26
Cicarelli RMB, Lopes JD (1989) Characterisation of a protein from Trypanosoma cruzi trypomastigotes that cleaves non-immune IgG bound through its Fab fragment. J Immunol 142:1685–1690
Coura JR (2013) Chagas disease: control, elimination and eradication. Is it possible? Mem Inst Oswaldo Cruz 108:962–967
Coura JR, Viñas PA, Junqueira ACV (2014) Ecoepidemiology, short history and control of Chagas disease in the endemic countries and the new challenge for non-endemic countries. Mem Inst Oswaldo Cruz 109:856–862
Darriba D, Taboada GL, Doallo R, Posada DJ (2012) ModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772
Duschak VG, Ciaccio M, Nasser JR, Basombrío MA (2001) Enzymatic activity, protein expression, and gene sequence of cruzipain in virulent and attenuated Trypanosoma cruzi strains. J Parasitol 87:1016–1022
Dvorak JA, Hartman DL, Miles MA (1980) Trypanosoma cruzi: correlation of growth kinetics to zymodeme type in clones derived from various sources. J Protozool 27:472–474
El-Syaed NM, Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G, Tran A-N (2005) The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science 309:409–415
Fampa P, Lisboa CV, Zhaner V, Jansen AM, Ramirez MI (2010) Wide proteolytic activity survey reinforces heterogeneity among Trypanosoma cruzi TcI and TcII wild populations. Vector Borne Zoontic Dis 10:839–845
Freitas JM, Augusto-Pinto L, Pimenta JR, Bastos-Rodrigues L, Gonçalves VF, Teixeira SM, Chiari E, Junqueira AC, Fernandes O, Macedo AM, Machado CR, Pena SD (2006) Ancestral genomes, sex and the population structure of Trypanosoma cruzi. PLoS Pathog 2:e24
Garcia ES, Ratcliffe NA, Whitten MM, Gonzalez MS, Azambuja P (2007) Exploring the role of insect host factors in the dynamics of Trypanosoma cruzi–Rhodnius prolixus interactions. J Insect Physiol 53:11–21
Gaunt MW, Yeo M, Frame IA, Stothard JR (2003) Mechanism of genetic exchange in American trypanosomes. Nature 421:936–939
Herrera L, Xavier SCC, Viegas C, Martinez C, Cotias PM, Carrasco H, Urdaneta-Morales S, Jansen AM (2004) Trypanosoma cruzi in a caviomorph rodent: parasitological and pathological features of the experimental infection of Trichomys apereoides (Rodentia, Echimyidae). Exp Parasitol 107:78–88
Herrera L, D’Andrea PS, Xavier SCC, Mangia RH, Fernandes O, Jansen AM (2005) Trypanosoma cruzi infection in wild mammals of the National Park ‘Serra da Capivara’ and its surroundings (Piauí, Brazil), an area endemic for Chagas disease. Trans R Soc Trop Med Hyg 99:379–388
Jurberg J, Galvão C, Biology, ecology, and systematics of Triatominae (Heteroptera, Reduviidae), vectors of Chagas disease, and implications for human health (2006) Hug the bug. For love of true bugs. In: Rabitsch W (ed) Festschrift zum 70. Geburtstag von Ernst Heiss. Denisia 19, zugleich Kataloge der OÖ Landesmuseen 50, Linz, pp 1096–1116
Lima AP, dos Reis FC, Serveau C, Lalmanach G, Juliano L, Ménard R, Vernet T, Thomas DY, Storer AC, Scharfstein J (2001) Cysteine protease isoforms from Trypanosoma cruzi, cruzipain 2 and cruzain, present different substrate preference and susceptibility to inhibitors. Mol Biochem Parasitol 114:41–52
Lisboa CV, Pinho AP, Herrera HM, Gerhardt M, Cupolillo E, Jansen AM (2008) Trypanosoma cruzi (Kinetoplastida, Trypanosomatidae) genotypes in neotropical bats in Brazil. Vet Parasitol 156:314–318
Llewellyn MS, Lewis MD, Acosta N, Yeo M, Carrasco HJ, Segovia M, Vargas J, Torrico F, Miles MA, Gaunt MW (2009) Trypanosoma cruzi IIc: phylogenetic and phylogeographic insights from sequence and microsatellite analysis and potential impact on emergent Chagas disease. PLoS Negl Trop Dis 3:e510
Macedo AM, Pena SDJ (1998) Genetic variability of Trypanosoma cruzi: implications for the pathogenesis of Chagas disease. Parasitol Today 14:119–124
Maia da Silva F, Marcili A, Lima L, Cavazzana JM, Ortiz PA, Campaner M, Takeda GF, Paiva F, Nunes VLB, Camargo EP, Teixeira MMG (2009) Trypanosoma rangeli isolates of bats from Central Brazil: genotyping and phylogenetic analysis enable description of a new lineage using spliced-leader gene sequences. Acta Trop 109:199–207
Marin-Neto JA, Cunha-Neto EC, Maciel BC, Simões MV (2007) Pathogenesis of chronic Chagas heart disease. Circulation 115:1109–1123
McKerrow JH (1988) The role of proteinases in the pathogenesis of parasitic diseases. In: Sand MA, Leech JH, Root RK (eds) Parasitic Infections, vol 7, Contemporary issues in infectious disease. Churchill Livingstone, London, pp 51–59
Meirelles MNL, Juliano L, Carmona E, Silva SG, Costa EM, Murta ACM, Scharfstein J (1992) Inhibitors of the major cysteinyl proteinase (GP57/51) impair host cell invasion and arrest the intracellular development of Trypanosoma cruzi in vitro. Mol Biochem Parasitol 52:175–184
Mello CB, Azambuja P, Garcia ES, Ratcliffe NA (1996) Differential in vitro and in vivo behaviour of three strains of Trypanosoma cruzi in the gut and hemolymph of Rhodnius prolixus. Exp Parasitol 82:112–121
Mortensen UH, Olesen K, Breddam K (1999) Carboxypeptidase C including carboxypeptidase Y. In: Barrett AJ, Rawlings ND, Woessner JF (eds) Handbook of proteolytic enzymes. Academic Press Inc., London, pp 389–393
Murta ACM, Persechini PM, Souto-Padrón T, De Souza W, Guimarães JA, Scharfstein J (1990) Structural and functional identification of GP57/51 antigen of Trypanosoma cruzi as a cysteine proteinase. Mol Biochem Parasitol 43:27–38
Parussini F, García M, Mucci J, Agüero F, Sánchez D, Hellman U, Åslund L, Cazzulo JJ (2003) Characterization of a lysosomal serine carboxypeptidase from Trypanosoma cruzi. Mol Biochem Parasitol 131:11–23
Pedroso A, Cupolillo E, Zingales B (2003) Evaluation of Trypanosoma cruzi hybrid stocks based on chromosomal size variation. Mol Biochem Parasitol 129:79–90
Pinho AP, Cupolillo E, Mangia RH, Fernandes O, Jansen AM (2000) Trypanosoma cruzi in the sylvatic environment: distinct transmission cycles involving two sympatric marsupials. Trans R Soc Trop Med Hyg 94:1–6
Prata A (2001) Clinical and epidemiological aspects of Chagas disease. Lancet Infect Dis 1:92–100
Remington SJ, Breddam K (1994) Carboxypeptidase C and D. Meth Enzymol 244:231–248
Ronquist F, Huelsenbeck JP (2003) MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Sturm NR, Campbell DA (2010) Alternative lifestyles: the population structure of Trypanosoma cruzi. Acta Trop 115:35–43
Teichert U, Mechler B, Müller H, Wolf DH (1989) Lysosomal vacuolar proteinases of yeast are essential catalysts for protein degradation, differentiation and cell survival. J Biol Chem 264:16037–16045
Tomasini N, Diosque P (2015) Evolution of Trypanosoma cruzi: clarifying hybridisations, mitochondrial introgressions and phylogenetic relationships between major lineages. Mem Inst Oswaldo Cruz 110:403–413
Tomazi L, Kawashita SY, Pereira PM, Zingales B, Briones MRS (2009) Haplotype distribution of five nuclear genes based on network genealogies and Bayesian inference indicates that Trypanosoma cruzi hybrid strains are polyphyletic. Gen Mol Res 8:458–476
Torres JP, Ortiz S, Munoz S, Solari A (2004) Trypanosoma cruzi isolates from Chile are heterogeneous and composed of mixed populations when characterized by schizodeme and Southern analyses. Parasitology 128:161–168
Uehara LA, Moreira OC, Oliveira AC, Azambuja P, Lima AP, Britto C, dos Santos AL, Branquinha MH, d’Avila-Levy CM (2012) Cruzipain promotes Trypanosoma cruzi adhesion to Rhodnius prolixus midgut. PLoS Negl Trop Dis 6:e1958
Walker DM, Oghumu S, Gupta G, McGwire BS, Drew ME, Satoskar AR (2014) Mechanisms of cellular invasion by intracellular parasites. Cell Mol Life Sci 71:1245–1263
Waniek PJ, Araújo CAC, Momoli MM, Azambuja P, Jansen AM, Genta FA (2014) Serine carboxypeptidases of Triatoma brasiliensis (Hemiptera, Reduviidae): sequence characterization, expression pattern and activity localization. J Insect Physiol 63:9–20
Westenberger SJ, Barnabé C, Campbell DA, Sturm NR (2005) Two hybridization events define the population structure of Trypanosoma cruzi. Genetics 171:527–543
Yeo M, Acosta N, Llewellyn M, Sánchez H, Adamson S, Miles GAJ, López E, González N, Patterson JM, Gaunt MW, Arias RJ, Miles MA (2005) Origins of Chagas disease: Didelphis are natural hosts of Trypanosoma cruzi I and armadillos hosts of Trypanosoma cruzi II, including hybrids. Int J Parasitol 35:225–233
Zingales B, Andrade SG, Briones MRS, Campbell DA, Chiari E, Fernandes O, Guhl F, Lages-Silva E, Macedo AM, Machado CR, Miles MA, Romanha AJ, Sturm NR, Tibayrenc M, Schijman AG (2009) A new consensus for Trypanosoma cruzi intraspecific nomenclature: second revision meeting recommends TcI to TcVI. Mem Inst Oswaldo Cruz 104:1051–1054
Zingales B, Miles MA, Campbell DA, Tibayrenc M, Macedo AM, Teixeira MMG, Schijman AG, Llewellyn MS, Lages-Silva E, Machado CR, Andrade SG, Sturm NR (2012) The revised Trypanosoma cruzi subspecific nomenclature: rationale, epidemiological relevance and research applications. Infect Gen Evol 12:240–253
Zingales B, Miles MA, Moraes CB, Luquetti A, Guhl F, Schijman AG, Ribeiro I (2014) Drug discovery for Chagas disease should consider Trypanosoma cruzi strain diversity. Mem Inst Oswaldo Cruz 109:828–833
Acknowledgments
We are grateful to Dr. Octavio Fernandes and Dr. Adeílton Brandão (Laboratório Interdisciplinar de Pesquisas Médicas—IOC/FIOCRUZ) for the technical support. The present study received financial support from Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro—FAPERJ (Cientistas do Nosso Estado: E-26/100.456/2007 and Research Fellow: E-26/152.913/2005) and Conselho Nacional de Desenvolvimento Científico e Tecnológico—CNPq (Edital Universal: 472276/2006-9). All material has been collected before Oct. 12, 2014.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental figure 1
Bayesian phylogenetic tree of the full data set using the GTR+I+G model. The same tree but with collapsed outgroup is shown in Fig. 1. The respective genotypes are shown after the name of the isolates. Isolates without genotype are T. c. marinkellei. (PDF 148 kb)
Rights and permissions
About this article
Cite this article
de Araújo, C.A.C., Mayer, C., Waniek, P.J. et al. Differentiation of Trypanosoma cruzi I (TcI) and T. cruzi II (TcII) genotypes using genes encoding serine carboxypeptidases. Parasitol Res 115, 4211–4219 (2016). https://doi.org/10.1007/s00436-016-5198-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-016-5198-8