Abstract
Animal trypanosomosis is a disease that is distributed worldwide which results in huge economic losses due to reduced animal productivity. Endemic regions are often located in the countryside where laboratory diagnosis is costly or inaccessible. The establishment of simple, effective, and accurate field tests is therefore of great interest to the farming and veterinary sectors. Our study aimed to develop a simple, rapid, and sensitive immunochromatographic test (ICT) for animal trypanosomosis utilizing the recombinant tandem repeat antigen TeGM6-4r, which is conserved amongst salivarian trypanosome species. In the specificity analysis, TeGM6-4r/ICT detected all of Trypanosoma evansi-positive controls from experimentally infected water buffaloes. As expected, uninfected controls tested negative. All sera samples collected from Tanzanian and Ugandan cattle that were Trypanosoma congolense- and/or Trypanosoma vivax-positive by microscopic examination of the buffy coat were found to be positive by the newly developed TeGM6-4r/ICT, which was comparable to results from TeGM6-4r/ELISA (kappa coefficient [κ] = 0.78). TeGM6/ICT also showed substantial agreement with ELISA using Trypanosoma brucei brucei (κ = 0.64) and T. congolense (κ = 0.72) crude antigen, suggesting the high potential of TeGM6-4r/ICT as a field diagnostic test, both for research purposes and on-site diagnosis of animal trypanosomosis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Animal trypanosomosis is a disease caused by various species of the hemoparasitic protozoan parasites of the genus Trypanosoma. Pathogenic trypanosomes are responsible for three disease syndromes in animals: nagana, surra, and dourine (Barrett et al. 2003). Nagana, or animal African trypanosomosis, is attributed to Trypanosoma congolense, Trypanosoma vivax, Trypanosoma simiae, Trypanosoma brucei brucei, and Trypanosoma suis and is usually severe in cattle, camels, and pigs. Surra is caused by Trypanosoma evansi and often leads to serious conditions in water buffalo, camels, and dogs. Dourine is caused by Trypanosoma equiperdum, which exclusively affects equid species. These diseases are considered one of the biggest constraints to worldwide livestock production. In Africa, animal trypanosomosis costs livestock producers and consumers an estimated $1340 million annually through direct losses such as reduced production of milk and meat (Kristjanson et al. 1999). Moreover, other factors, such as the choice between trypanotolerant and trypanosusceptible cattle and the avoidance of areas infested with tsetse flies for grazing and settling, have indirect effects on people’s livelihoods (Shaw 2009).
At present, animal trypanosomosis is widely distributed in Asia, South America, and Africa. Although several outbreaks of the disease have been reported in some European countries (Desquesnes et al. 2008; Tamarit et al. 2011), the highly endemic regions are still located in the developing world, specifically in rural areas. These areas are usually far from research centers or diagnostic laboratories, making disease diagnosis a difficult task. Many diagnostic methods for trypanosomosis have been developed and validated, including parasitological, serological, and molecular tests (OIE 2012). At present, however, the card agglutination test (CATT/T. evansi) is the only tool that is widely available in the field. Despite its efficiency and usefulness, CATT/T. evansi has some acknowledged limitations, which include varying degrees of specificity and sensitivity and its inability to detect T. evansi strain B, which does not express the variant surface antigen RoTat 1.2 (Njiru et al. 2010; Sullivan et al. 2013). A need therefore exists to develop a new field test to supplement the current CATT/T. evansi and to provide a greater choice of tests for the diagnosis of animal trypanosomosis.
An immunochromatographic test (ICT) utilizes the concept of chromatography and especially suited to simple and rapid diagnosis. The movement of a liquid sample past various zones along the test strip where molecules that exert specific interaction with the analyte have been attached is essential to current ICTs (Paek et al. 2000; Posthuma-Trumpie et al. 2009), in which results usually come within 10–20 min. The first commercially successful ICT was pregnancy test which rapidly detects human chorionic gonadotropin in urine when urine is simply added to the test strip. At present, with better methods and technology presented, a new generation of ICTs is widely used (for both medical and veterinary purposes) for qualitative and semi-quantitative detection. The present study aimed to develop an ICT for rapid serodiagnosis of animal trypanosomosis based on the typical ICT format, using the recombinant tandem repeat antigen GM6. The antigen is highly conserved among salivarian trypanosome species and exhibits strong antigenicity to T. evansi and T. congolense-infected water buffalo and cattle serum samples (Goto et al. 2011; Thuy et al. 2012). In contrast to previous studies which used an antigen consisting of two repeat units (TeGM6-2r) (Thuy et al. 2012), we expressed a new recombinant antigen GM6 named TeGM6-4r, which consisted of four repeat units and was derived from T. evansi. This study is also a follow-up of our previous assessment and preliminary evaluation of TeGM6-4r- and TeGM6-4r-based ICT (Nguyen et al. 2015; Nguyen et al. 2014). The ICT was produced with some modifications, utilizing non-tagged recombinant TeGM6-4r instead of 6× His-tagged TeGM6-4r and comprehensively evaluated using nine positive and six negative controls from three water buffaloes experimentally infected with T. evansi and 437 field samples from Tanzanian and Ugandan cattle. The performance of the test was also compared with the trypanosome crude antigen-based ELISA as recommended by the World Organisation for Animal Health (OIE) (OIE 2012).
Materials and methods
Production of TeGM6-4r
The gene encoding TeGM6-4r was amplified by PCR using genomic DNA from T. evansi Tansui strain and primer set 5′-GGA TCC ATG GAG CTT GCT AAA-3′ and 5′-GAA TTC CTA ATG TGA ATG CTC-3′ (underlined nucleotides are restriction sites of Bam HI and Eco RI). The PCR conditions have been previously described (Thuy et al. 2012). Briefly, the reactions were conducted for 30 cycles, at 94 °C for 30 s (denaturation), 54 °C for 30 s (annealing), and 72 °C for 1.5 min (extension). PCR products were separated by agarose gel electrophoresis and TeGM6 fragments, consisting of four repeat units, were extracted from the gel (Japan-QIAGEN K.K., Tokyo, Japan) for sequencing using the PCR primers, BigDye Terminator Ready Mix (Applied Biosystems, Life Technologies, Carlsbad, USA) and an ABI Prism 3100 Genetic Analyzer (Applied Biosystems, Life Technologies, Carlsbad, USA). Nucleotide and amino acid sequences were identified and analyzed using Genetyx version 8.0 (Genetyx Co., Tokyo, Japan) and BLAST (http://blast.ncbi.nlm.nih.gov/).
For protein expression, the gene fragment encoding the TeGM6-4r was inserted into the pGEX-6P-1 (GE Healthcare, Tokyo, Japan) and transformed into Escherichia coli BL 21. The transformed E. coli BL 21 was cultured in SOB medium (BD, Franklin Lakes, USA) to an OD600 of 0.4–0.6. The expression of the recombinant protein was induced by the addition of 0.5 mM isopropyl-thio-β-galactosidase. After cultivation for 3 h, the recombinant TeGM6-4r was purified in soluble form using glutathione Sepharose 4B and cleaved from glutathione S-transferase by PreScission Protease, which was performed in accordance with the manufacturer’s instructions (GE Healthcare, Tokyo, Japan). The integrity and purity of the protein were evaluated by 12 % sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Protein concentration was determined by bicinchoninic acid assay (Thermo Fisher Scientific, Waltham, USA). The purified recombinant protein was stored at −80 °C until use.
Production of trypanosome crude antigens
Bloodstream forms of T. b. brucei (GUTat3.1) and procyclic forms of T. congolense (IL3000) were maintained in the National Research Center for Protozoan Diseases, Obihiro University of Agriculture and Veterinary Medicine in in vitro culture medium HMI-9 and TVM-1 (Hirumi et al. 1980) and utilized as the sources of trypanosome crude antigens. Preparation of T. b. brucei and T. congolense crude antigens was described in the OIE Manual of Diagnostic Tests and Vaccines for Terrestrial Animals, 2012 (OIE 2012).
ELISA
ELISA was performed according to a previously described protocol (Thuy et al. 2012). Briefly, 1 μg of the trypanosome crude antigen or 200 ng of TeGM6-4r was coated on Maxisorp 96-well plates (Nalgene-Nunc, NY, USA) for 4 h and then blocked overnight with 1 % bovine serum albumin. After washing with phosphate-buffered saline containing 0.05 % Tween 20 (PBS-T), serum samples were added to each well (at 200 times dilution) and incubated at room temperature for 2 h. The plates were washed with PBS-T before the addition of horseradish peroxidase-conjugated protein G (Invitrogen, Tokyo, Japan) or anti-bovine IgM antibody (Bethyl Laboratories, Montgomery, USA). The washing step was then performed. Substrate tetramethylbenzidine (Kirkegaard & Perry Laboratories, Gaithersburg, USA) was applied and the reaction was stopped using 1 M phosphoric acid. The absorbance was read at 450 nm using an MTP-500 microplate reader (Corona Electric, Ibaraki, Japan).
Production of anti-TeGM6-4r polyclonal antibody
Each BALB/c mouse was immunized with 100 μl emulsion of purified recombinant TeGM6-4r (100 μg) and TiterMax® Gold adjuvant (Sigma-Aldrich, St. Louis, USA). After 2 weeks, antibody titer was determined using TeGM6-4r-based ELISA. Mice producing high antibody titer were sacrificed for blood collection and serum extraction. Sera were then subjected to polyclonal antibody purification using a MAbTrap kit (Sigma-Aldrich, St. Louis, USA) in accordance with the manufacturer’s instructions. The concentration of the polyclonal antibody was detected by bicinchoninic acid assay (Thermo Fisher Scientific, Waltham, USA). Anti-sera were stored at −80 °C until use.
Construction of TeGM6-4r-based ICT
TeGM6-4r-based ICT was designed for antibody detection. All of the membranes used for ICT (glass fiber, absorbance, and nitrocellulose membranes) were purchased from EMD Millipore Corporation (Billerica, USA). The recombinant TeGM6-4r was conjugated with 20 nm-sized gold colloid (British Biocell International, Cardiff, UK) by the following procedure: 100 μl of TeGM6-4r (at the concentration of 300 μg/ml) was gently mixed with 1 ml of gold colloid for initial binding, followed by blocking with 0.05 % polyethylene glycol (PEG) 20,000 and 1 % bovine serum albumin. The complex gold colloid-labeled TeGM6-4r was washed twice with blocking solution containing 0.05 % PEG 20,000 and 0.5 % bovine serum albumin, and the concentration was adjusted to OD520 1.5 with dilution buffer (10 mM Tris-HCl pH 8.2 and 5 % sucrose). Finally, the conjugate solution was absorbed into a glass fiber strip and dried in the dryer (VD-500R, TAITEC, Saitama, Japan) for 3 h. To create the test line and control line, TeGM6-4r (0.5 mg/ml) and anti-TeGM6-4r polyclonal antibody (1 mg/ml) were jetted on the nitrocellulose membrane using an XYZ Dispensing Platform (BioDot, Irvine, USA). The membrane was then blocked with 0.5 % casein and dried overnight at room temperature. Subsequently, all of the ICT components were assembled manually and cut into 3-mm strips by CM 4000 (BioDot, Irvine, USA). For running the ICT, 10 μl of serum sample was diluted five times with PBS and loaded on the sample pad. The result showed in 10–20 min. An ICT was determined to be positive if it displayed both test and control lines and negative if it only displayed the control line.
Serum samples
Negative and positive controls were obtained from three water buffaloes experimentally infected with T. evansi in China. Sera were collected at five time points during the infection: 22 days before infection (pre-infection) and 8, 15, 22, and 48 days post-infection (DPI). Pre-infection and 8 DPI sera were confirmed negative and the rest (15, 22, and 48 DPI) were confirmed to be positive by parasitological tests and ELISA. All of the 437 field samples were collected from cattle in Uganda and Tanzania from May to July in 2006. The handling of the experimental animals was in accordance with the guidelines on Animal Experimentation of the Ethics Committee of Obihiro University of Agriculture and Veterinary Medicine, Japan (Approval number: 26-170), and the Animal Welfare guidelines of Sun Yat-sen (Zhongshan) University, China.
Data analysis
Data were analyzed using GraphPad (http://graphpad.com/quickcalcs/). The kappa coefficient (κ) was used to determine the agreement between two diagnostic tests. Calculations and interpretations of κ values followed the methods of Viera and Garrett (2005). A kappa of 1 indicates perfect agreement, whereas a kappa of 0 indicates agreement equivalent to chance.
Results
Detection of anti-GM6 antibodies in experimentally infected water buffaloes by TeGM6-4r/ICT
Recombinant antigen, non-tagged TeGM6-4r, was successfully produced. Its molecular mass was estimated to be 28.9 kDa (Fig. 1). The recombinant TeGM6-4r contained four repeat units. Each repeat unit was constructed from 68 amino acids. A database search for the GM6 amino acid sequence revealed similarity to the GM6 from other salivarian trypanosomes. GM6 from T. evansi shares 100, 63.8, and 54.5 % identity with T. b. brucei-, T. congolense-, and T. vivax-derived GM6, respectively. Since GM6 is highly conserved among salivarian trypanosome species, T. evansi-derived GM6 was chosen as a diagnostic antigen for animal trypanosomosis. Antigenicity of the newly expressed TeGM6-4r was compared with the TeGM6-2r by ELISA using 15 positive and negative control sera from water buffaloes (WB) experimentally infected with T. evansi. The result showed that average OD values (mean ± SD) of WB sera under TeGM6-4r/ELISA and TeGM6-2r/ELISA were 1.08 ± 0.95 and 0.56 ± 0.57, respectively. The immunoreactivity of TeGM6-4r was significantly increased (P = 0.001; paired Student’s t test).
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) image of recombinant TeGM6-4r. Lane 1: protein size marker. Lane 2: glutathione S-transferase-tagged recombinant TeGM6-4r (estimated to be 54.8 kDa including a glutathione S-transferase of about 25.9 kDa in size). Lane 3: non-tagged recombinant TeGM6-4r which was estimated to be 28.9 kDa
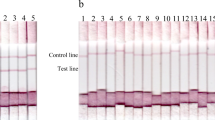
Next, the TeGM6-4r based ICT was assembled. In the preliminary evaluation of the test, TeGM6-4r/ICT was performed using the positive and negative control sera from WB experimentally infected with T. evansi. All of the negative WB sera (at pre-infection and 8 DPI) were negative and all of the positive sera (15, 22, and 48 DPI) were positive in the ICT (Fig. 2a). Although sera at 22 DPI were brown in color due to hemolysis, the test lines were still visible. TeGM6-4r/ICT was in strong agreement with TeGM6-4r/ELISA. In the TeGM6-4r/ELISA, the OD values of pre-infection and 8 DPI were lower than the cutoff value (0.3). They increased from 15 DPI and were considerably high at 48 DPI (Fig. 2b). To compare the performance of the present non-tagged TeGM6-4r/ICT with the previous 6× His-tagged/ICT and determine the sensitivity of the ICTs in comparison with ELISA, a positive sample was serially diluted and tested with the three tests (Fig. 3). The 6× His-tagged and non-tagged TeGM6-4r/ICT was able to detect the positive sample at 4 and 32 times dilution, respectively, whereas the ELISA capable of detecting the sample at 512 times dilution.
Recombinant TeGM6-4r-based immunochromatographic test (ICT) (a) and ELISA (b) using positive and negative controls from water buffalo experimentally infected with T. evansi. The two tests were in agreement in distinguishing negative controls (1 and 2) and positive controls (3, 4, and 5). Due to hemolysis in the sera 4, the background of ICT trips showed a brown color. BW1, BW2, and BW3 indicate water buffalo no. 1, 2, and 3, respectively. The numbers 1, 2, 3, 4, and 5 indicate serum samples collected 22 days before infection and 8, 15, 22, and 48 days post-infection, respectively. The ELISA cutoff value was 0.3, as indicated by the broken line
Sensitivity of the TeGM6-4r-based immunochromatographic test (ICT) and ELISA. The ELISA cutoff value was 0.3, as indicated by the broken line. The 6× His-tagged and non-tagged TeGM6-4r/ICTs could detect positive samples at a dilution factor of 4 and 32, respectively. The ELISA could detect positive samples at a dilution factor of 512
Detection of T. congolense and T. vivax infections in field-derived serum samples by TeGM6-4r/ICT
Field samples (n = 437) were collected from Tanzanian and Ugandan cattle from May to July in 2006. The samples were previously screened by microscopic examination of the buffy coat, which yielded the following positive detections: T. congolense (n = 15), T. vivax (n = 4), and mixed T. congolense and T. vivax (n = 5). Since T. vivax crude antigen was unavailable (and as recommended by the OIE), T. b. brucei and T. congolense crude antigen-based ELISAs were utilized as reference tests. In the first ELISA, we used HRP-conjugated protein G as the secondary antibody to detect IgG in the serum samples. The T. b. brucei and T. congolense crude antigen and TeGM6-4r-based tests detected 272, 262, and 235 positive samples, respectively. The TeGM6-4r/ICT detected 228 positive samples. However, there were 23 serum samples that were found to be positive by the ICT but negative by T. b. brucei crude antigen-based ELISA; while 24 and 32 of the samples found positive by ICT were found negative by T. congolense crude antigen and TeGM6-4r/ELISA, respectively. Thus, the second ELISA was performed using HRP-conjugated anti-bovine IgM antibody to detect IgM in the serum samples. The result showed that among the samples that were ICT positive but ELISA negative, 21/23, 14/24, and 32/32 sera became positive in T. b. brucei and T. congolense crude antigen and TeGM6-4r-based ELISA, respectively. Taken together, TeGM6-4r/ICT demonstrated 76.5 % sensitivity and 93.4 % specificity (κ = 0.64) with T. b. brucei crude antigen/ELISA and 79.6 % sensitivity and 98.7 specificity (κ = 0.72) with T. congolense crude antigen/ELISA (Table 1). The test showed substantial agreement (κ > 0.61) with crude antigen-based ELISA in the detection of trypanosomes in field samples. All of the samples that were found to be positive by microscopic examination of the buffy coat were also positive in the TeGM6-4r/ICT. In comparison with TeGM6-4r/ELISA, the TeGM6-4r/ICT had relatively lower sensitivity, but higher specificity. The κ value between the two tests was 0.78, indicating substantial agreement.
Discussion
Using sera experimentally infected with T. evansi, the TeGM6-4r/ICT could perfectly distinguish positive from negative samples, suggesting the test to be an accurate diagnostic tool for animal trypanosomosis. Our study demonstrated that the increase of the repeat units of GM6 by 4 significantly increased the immunoreactivity of the antigen. The result was consistent with a previous report (Valiente-Gabioud et al. 2011). The utilization of non-tagged TeGM6-4r instead of 6× His-tagged TeGM6-4r significantly increased the sensitivity of ICT. The ICT could detect positive serum at higher dilution factor (from 4 to 32 times dilution). The TeGM6-4r/ICT was in substantial agreement with TeGM6-4r/ELISA (κ = 0.78). Although the detection sensitivity of TeGM6-4r/ICT was lower than that of TeGM6-4r/ELISA (Fig. 3), this problem might be partially overcome by the use of higher concentrations of serum samples in the ICT (five times dilution in this study). Nevertheless, improving the sensitivity of the ICT is a matter of great concern. Several techniques have recently been proposed such as two-dimensional paper network format and gold nanoparticle enhancer; however, some of them were limited in that they were neither greatly effective nor convenient for use under field conditions (Fu et al. 2011; Tanaka et al. 2006). Further studies are still needed to improve the test sensitivity.
In comparison to CATT/T. evansi, the advantage of TeGM6-4r/ICT is that it can detect other species of salivarian trypanosomes, such as T. b. brucei, T. congolense, and T. vivax. When the ICT was utilized for screening of field-derived cattle serum samples, the TeGM6-4r/ICT showed positive in parasitologically positive samples, specifically 24 Ugandan and Tanzanian cattle samples that were found to be positive for T. congolense and/or T. vivax by the microhematocrit technique. In comparison to ELISA, TeGM6-4r/ICT showed substantial agreement with T. b. brucei (κ = 0.64) and T. congolense (κ = 0.72) crude antigen/ELISAs. Similarly, the TeGM6-4r/ICT and TeGM6-4r/ELISA also had substantial agreement (κ = 0.78). The results of our study indicated that there were a relatively high number of positive samples detected by both T. b. brucei and T. congolense crude antigen/ELISA (254/437 or 58.1 %). Moreover, microscopic examination of the buffy coat detected five samples that were positive for both T. congolense and T. vivax. This result is consistent with other reports which indicate that co-infection of animal trypanosomosis is present in African cattle (Gillingwater et al. 2010; Laohasinnarong et al. 2011). Since the therapeutic treatment for different animal trypanosomosis are quite similar, the TeGM6-4r/ICT, which can detect several pathogenic trypanosomes, represents a good choice of test.
Our study lacked a serodiagnostic test for T. vivax due to unavailability of T. vivax crude antigen. The parasitological test detected nine T. vivax positive sera which were also positive in TeGM6-4r/ICT; however, this number of positive samples was rather low. Therefore, performance of the ICT in detection of T. vivax has not been completely understood yet. Since GM6 antigen from T. vivax, TvGM6 was reported the excellent antigen for diagnosis of T. vivax, comparison of TeGM6-4r and TvGM6 also need to be taken into consideration (Pillay et al. 2013).
The present study attempted to improve and evaluate the previously produced ICT for the detection of animal trypanosomosis using both experimentally infected and field-derived water buffalo and cattle serum samples. The test utilized recombinant tandem repeat antigen TeGM6-4r, which was able to be produced in high quality and large quantity. Although the sensitivity of the TeGM6-4r/ICT was lower than that of TeGM6-4r/ELISA, the ICT could simultaneously detect IgG and IgM, which is advantageous. Moreover, the TeGM6-4r/ICT is able to diagnose T. evansi, T. brucei, T. congolense, and T. vivax infections. Since the TeGM6-4r antigen possesses high similarity to (63.8 %) with Trypanosoma brucei gambiense and Trypanosoma brucei rhodesiense (Thuy et al. 2012), it may also have diagnostic potential for human African trypanosomosis. Further studies and extensive evaluation of TeGM6-4r/ICT are required to improve the performance of the test. Together with CATT/T. evansi, TeGM6-4r/ICT would be a useful diagnostic tool, both for research purposes and for on-site diagnosis of animal trypanosome infections.
In conclusion, the results of our study indicated that TeGM6-4r/ICT is capable of diagnosing animal trypanosomosis. The test is relatively sensitive, reliable, and comparable to trypanosome crude antigen-based ELISAs, the reference tests which are recommended by OIE (2012). Moreover, it is suitable for field use and is simple to handle and interpret.
References
Barrett MP et al (2003) The trypanosomiases. Lancet 362(9394):1469–1480. doi:10.1016/s0140-6736(03)14694-6
Desquesnes M et al (2008) First outbreak of Trypanosoma evansi in camels in metropolitan France. Vet Rec 162(23):750–752
Fu E et al (2011) Enhanced sensitivity of lateral flow tests using a two-dimensional paper network format. Anal Chem 83(20):7941–7946. doi:10.1021/ac201950g
Gillingwater K, Mamabolo MV, Majiwa PAO (2010) Prevalence of mixed Trypanosoma congolense infections in livestock and tsetse in KwaZulu-Natal, South Africa. J S Afr Vet Assoc-Tydskr Suid-Afr Vet Ver 81(4):219–223
Goto Y et al (2011) Serological characterizations of tandem repeat proteins for detection of African trypanosome infection in cattle. Parasitol Int 60(4):538–540. doi:10.1016/j.parint.2011.09.003
Hirumi H, Hirumi K, Doyle JJ, Cross GA (1980) In vitro cloning of animal-infective bloodstream forms of Trypanosoma brucei. Parasitology 80(2):371–382
Kristjanson PM, Swallow BM, Rowlands GJ, Kruska RL, de Leeuw PN (1999) Measuring the costs of African animal trypanosomosis, the potential benefits of control and returns to research. Agric Syst 59(1):79–98. doi:10.1016/s0308-521x(98)00086-9
Laohasinnarong D et al (2011) Prevalence of Trypanosoma sp in cattle from Tanzania estimated by conventional PCR and loop-mediated isothermal amplification (LAMP). Parasitol Res 109(6):1735–1739. doi:10.1007/s00436-011-2513-2
Nguyen TT et al (2014) Diagnostic value of the recombinant tandem repeat antigen TeGM6-4r for surra in water buffaloes. Vet Parasitol 201(1–2):18–23. doi:10.1016/j.vetpar.2014.01.009
Nguyen TT et al (2015) Application of crude and recombinant ELISAs and immunochromatographic test for serodiagnosis of animal trypanosomosis in the Umkhanyakude District of KwaZulu-Natal Province, South Africa. J Vet Med Sci 77(2):217–220. doi:10.1292/jvms.14-0330
Njiru ZK, Ouma JO, Enyaru JC, Dargantes AP (2010) Loop-mediated isothermal amplification (LAMP) test for detection of Trypanosoma evansi strain B. Exp Parasitol 125(3):196–201. doi:10.1016/j.exppara.2010.01.017
OIE (2012) Trypanosoma evansi infection (surra). In: OIE (ed) Manual of diagnostic tests and vaccines for terrestrial animal, vol 1, 7th edn. OIE, Paris, pp 314–328
Paek SH, Lee SH, Cho JH, Kim YS (2000) Development of rapid one-step immunochromatographic assay. Methods 22(1):53–60. doi:10.1006/meth.2000.1036
Pillay D et al (2013) Trypanosoma vivax GM6 antigen: a candidate antigen for diagnosis of African animal trypanosomosis in cattle. PLoS One 8(10):10. doi:10.1371/journal.pone.0078565
Posthuma-Trumpie GA, Korf J, van Amerongen A (2009) Lateral flow (immuno) assay: its strengths, weaknesses, opportunities and threats. A literature survey. Anal Bioanal Chem 393(2):569–582. doi:10.1007/s00216-008-2287-2
Shaw APM (2009) Assessing the economics of animal trypanosomosis in Africa-history and current perspectives. Onderstepoort J Vet Res 76(1):27–32
Sullivan L, Wall SJ, Carrington M, Ferguson MAJ (2013) Proteomic selection of immunodiagnostic antigens for human African trypanosomiasis and generation of a prototype lateral flow immunodiagnostic device. Plos Neglect Trop Dis 7(2):14. doi:10.1371/journal.pntd.0002087
Tamarit A, Tejedor-Junco MT, Gonzalez M, Alberola J, Gutierrez C (2011) Morphological and biometrical features of Trypanosoma evansi isolates from an outbreak in mainland Spain. Vet Parasitol 177(1–2):152–156. doi:10.1016/j.vetpar.2010.11.044
Tanaka R et al (2006) A novel enhancement assay for immunochromatographic test strips using gold nanoparticles. Anal Bioanal Chem 385(8):1414–1420. doi:10.1007/s00216-006-0549-4
Thuy N, Goto Y, Lun ZR, Kawazu SI, Inoue N (2012) Tandem repeat protein as potential diagnostic antigen for Trypanosoma evansi infection. Parasitol Res 110(2):733–739. doi:10.1007/s00436-011-2632-9
Valiente-Gabioud AA, Veaute C, Perrig M, Galan-Romano FS, Sferco SJ, Marcipar IS (2011) Effect of repetitiveness on the immunogenicity and antigenicity of Trypanosoma cruzi FRA protein. Exp Parasitol 127(3):672–679. doi:10.1016/j.exppara.2010.11.011
Viera AJ, Garrett JM (2005) Understanding interobserver agreement: the kappa statistic. Fam Med 37(5):360–363
Acknowledgments
We thank Dr. Zhao-Rong Lun, Dr. Charles P. Otim, and Dr. Heriel A. Mbwambo for providing serum samples. This study was financially supported by the Ministry of Education, Cultural, Sports, Science, and Technology (MEXT), the Japan Society for the Promotion of Science (JSPS), and JST/JICA SATREPS.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nguyen, TT., Ruttayaporn, N., Goto, Y. et al. A TeGM6-4r antigen-based immunochromatographic test (ICT) for animal trypanosomosis. Parasitol Res 114, 4319–4325 (2015). https://doi.org/10.1007/s00436-015-4672-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-015-4672-z