Abstract
During development, Schistosoma japonicum undergoes many morphological and physiological transformations as a result of profound changes in gene expression. Proteins containing zinc finger motifs usually play an important role in DNA recognition, RNA packaging, and transcriptional activation. In our current study, we cloned the open reading frame (ORF) of SjZFP1 of S. japonicum, which encodes a zinc finger protein. We analyzed the complementary DNA (cDNA) sequence of SjZFP1 and examined the expression of SjZFP1 messenger RNA (mRNA) at various developmental stages. We also tested the effects of RNA interference (RNAi) silencing on worm burden, spawning, and egg hatching. The ORF in the SjZFP1 cDNA was 1017 bp in length and was predicted to encode a 338-aa protein with a molecular mass of approximately 38.5 kDa and theoretical isoelectric point (pI) of 7.08. Several conserved regions, including a B-box-type zinc-binding domain, two bipartite nuclear localization signal domains, a paired amphipathic helix repeat, and overlapping RING and PHD finger domains, were identified in the predicted amino acid sequence of SjZFP1. Using real-time PCR, we showed that the SjZFP1 mRNA was expressed across all of the developmental stages of the parasite and that the level of transcription was highest in the cercariae, eggs, schistosomula, and mature adult worms. The level of SjZFP1 mRNA expression in cultured schistosomula treated with one of two SjZFP1-specific small interfering RNAs (siRNAs; AY770 and AY546) was reduced by over 80 %, compared with that in the controls. In RNAi experiments in BALB/c mice, the level of SjZFP1 mRNA increased significantly when the mice were treated with the same SjZFP1-specific siRNAs during the early stages of infection. By contrast, the level of SjZFP1 mRNA decreased significantly when the mice were treated with the SjZFP1-specific siRNAs during the middle to late stages of infection. In four independent experiments, fewer worms were recovered from mice treated with the SjZFP1-specific siRNAs, compared with the number of worms recovered from the control mice. Both the average number and hatching rates of liver eggs recovered from mice treated with the SjZFP1-specific siRNAs during the middle to late stages of infection were significantly lower than those of the liver eggs recovered from the control mice. Our results suggest that the SjZFP1 gene might be important for parasite development, spawning in the vertebrate host, and egg hatching.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Human schistosomiasis is a chronic debilitating disease that is prevalent in tropical and subtropical countries. Schistosomiasis is caused by blood-dwelling flukes of the genus Schistosoma (Gryseels et al. 2006). Schistosoma japonicum is one of five schistosome species that infect humans and is prevalent in China and other Asian countries. The control of S. japonicum is considered to be a major public health priority in China (Wang et al. 2008). Because of the zoonotic nature of schistosomiasis, integrated control strategies have been employed in most endemic areas for several decades, and considerable efforts have been made in the development of vaccines and new drugs for the treatment of the schistosomiasis (Loukas and Bethony 2008). However, the addition of infrastructure and educational components would augment current efforts to control schistosomiasis.
Schistosome worms have a complex life cycle, involving molluscous and vertebrate hosts as well as short periods of larvae free-swimming in water. The transition from one developmental stage to another involves modifications in the morphology, physiology, and biochemistry of the parasite, which are associated with the activation or inactivation of developmental-stage-specific gene expression. Previous studies have focused on the identification and characterization of tissue-, sex-, and developmental-stage-specific differential gene expression using microarray and proteomic approaches (El-Ansary and Al-Daihan 2005; Hong et al. 2013; Gobert et al. 2009). The regulation of gene expression plays a key role in development, especially with regard to the adaptation of the parasite larvae to different environments, larval maturation to the adult, sexual differentiation, and oogenesis. However, studies of transcriptional control in S. japonicum are scanty (Fantappie et al. 2007).
Zinc finger proteins are among the most abundant proteins in eukaryotic genomes. Their functions are extremely diverse and include DNA recognition, RNA packaging, transcriptional activation, the regulation of apoptosis, protein folding and assembly, and lipid binding (Laity et al. 2001). The SmZF1 gene of Schistosoma mansoni encodes a zinc finger protein that is expressed at different developmental stages of the parasite (Eleutério de Souza et al. 2001; Drummond et al. 2009). The SmZF1 protein binds to both double- and single-stranded DNA and also binds RNA oligonucleotides with lower affinity (Calzavara-Silva et al. 2004). Previous studies have suggested that SmZF1 acts as a transcription factor in S. mansoni (Drummond et al. 2009).
In our previous study, we constructed a phage display library using the complementary DNA (cDNA) of S. japonicum. We screened the library using fresh sera and liver and lung extracts from Microtus fortis, a nonpermissive host of S. japonicum (Sun et al. 2008a, b, c), and obtained 10, 19, and 13 phages, respectively, that displayed unique antigens. We used some of these phage clones to vaccinate mice. We found that vaccination with the phage displaying a zinc finger protein of S. japonicum, which we have named SjZFP1, induced a high level protection against S. japonicum infection (Song et al. 2009).
In our current study, we investigated the biological function of the SjZFP1 gene further. We cloned the open reading frame (ORF) of SjZFP1 from S. japonicum and characterized the structure of the SjZFP1 protein based on its predicted amino acid sequence. We also analyzed the messenger RNA (mRNA) expression profile of SjZFP1 at different stages of development and examined the effects of SjZFP1-specific RNA interference (RNAi) on worm burden, spawning in the vertebrate liver, and the egg hatching rate. Our results show that SjZFP1 plays important roles in parasite development, spawning, and egg hatching.
Materials and methods
Ethical statement
All procedures performed on animals within this study were conducted following guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC). The animal study protocol was approved by the Animal Care and Use Committee of the Shanghai Veterinary Research Institute (permit number 2010-12), Chinese Academy of Agricultural Sciences, People’s Republic of China.
Parasites and hosts
A strain of S. japonicum originally collected in Anhui Province in China was maintained in our laboratory using Oncomelania hupensis and New Zealand rabbits as the mollusk and vertebrate host, respectively. Cercariae were prepared routinely by exposing infected snails to light to induce shedding. Cercarial numbers and viability were determined using a light microscope prior to infection. Inbred 6- to 8-week-old male BALB/c mice and adult male New Zealand rabbits were purchased from Shanghai Experimental Animal Center (Chinese Academy of Sciences). Eggs and worms at different developmental stages were recovered from experimentally infected rabbits.
Molecular cloning and characterization of SjZFP1
The complete sequence of the ORF of SjZFP1 was predicted based on the SjZFP1 mRNA sequence from S. japonicum in GenBank (accession no. AY222909.1). The cDNA of the ORF was amplified using reverse transcription polymerase chain reaction (RT-PCR) and mRNA template prepared from adult S. japonicum. The forward (5′-GTGGATCCATGGGTTTATGTAAGTGTGAA-3′) and reverse (5′-CCACTCGAGTCAAGCATCGTTTAACCCATC-3′) primers used for RT-PCR annealed to sequences flanking the start and stop cordons in the SjZFP1 mRNA and contained a BamHI or XhoI restriction site (underlined), respectively. The PCR products were cloned into the pMD19-T easy vector (Takara-Bio, Shiga, Japan), and competent DH5α Escherichia coli were transformed using the ligation reactions. The SjZFP1 and flanking sequences of recombinant clones were confirmed by DNA sequencing.
The amino acid sequence of the SjZFP1 protein was predicted from the cloned cDNA sequence of SjZFP1 using DNAsis program. The molecular weight and isoelectric point (pI) were calculated using the Compute pI/Mw computational tool (http://www.expasy.ch/tools/pitool.html). The conserved domains were identified using the Motif scan program (http://myhits.isb-sib.ch/cgi-bin/motif_scan).
Analysis of SjZFP1 transcription at various developmental stages
Total RNA was extracted from S. japonicum at various stages of its life cycle using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions. After removing residual DNA using RNase-free DNase (Takara-Bio), the SjZFP1 cDNA was synthesized using the PrimeScript RT Reagent Kit with gDNA Eraser (TaKaRa Bio, Inc.) and 1 μg of total RNA according to standard protocols. The forward (5′-CAGGATGCCAGATAGAT-3′) and reverse (5′-AGAAGTGCGATGAATGT-3′) primers used for the amplification of a 201-bp sequence of SjZFP1 during real-time PCR were designed using the Oligo 6 software. Forward (5′-CTGATTTTCCATTCGTTTG-3′) and reverse (5′-GTTGTCTACCATGAAGGCA-3′) primers were used to amplify a 213-bp sequence of α-tubulin as the internal control. Real-time PCR was performed using 1.0 μL cDNA produced from 1 ng of total RNA, 5 pmol of each gene-specific primer, and 10 μL of Power SYBR Premix Ex Taq (Takara-Bio) in a total volume of 20 μL. Thermal cycling was performed at 95 °C for 10 min, followed by 40 cycles of 95 °C for 10 s, 65 °C for 15 s, and 72 °C for 20 s. A melting curve analysis of the specific PCR product was performed. The quantitative RT-PCR experiments were technically repeated three times. The relative quantification of the real-time PCR product was performed using the Realplex software (Eppendorf, Hamburg, Germany).
RNAi analysis of SjZFP1 expression in schistosomula
Three SjZFP1-specific small interfering RNAs (siRNAs), AY359, AY546, and AY770 (Table 1), and a nonspecific siRNA negative control (NC) were obtained from Shanghai GenePharma (Shanghai, China). Schistosomula were produced by mechanically removing the tails from cercariae, as described previously (Brink et al. 1977). The schistosomula were cultured in 24-well plates (200 parasites/well) in complete RPMI 1640 medium (Invitrogen) containing 10 % rabbit serum (Gibco, Invitrogen), 1 % penicillin/streptomycin (Sigma-Aldrich, St. Louis, MO, USA), 10 μM hydrocortisone (Sigma-Aldrich), 10 μM 5-hydroxytryptamine (Sigma-Aldrich), 0.1 % lactalbumin hydrolysate (Sigma-Aldrich), and 0.2 U/mL insulin (Sigma-Aldrich) at 37 °C in an atmosphere of 5 % CO2. The schistosomula were soaked in a solution containing 100 nM SjZFP1-specific siRNA, 100 nM NC siRNA, or no siRNA (blank) for 72 h. The gene-silencing effect of each siRNA was determined at the end of the cultivation period using real-time PCR, as described above.
RNAi analysis of SjZFP1 expression in vivo
Four independent experiments were performed using four groups of 6-week-old male BALB/c mice, with five to eight mice per group. Infection was induced via abdominal skin penetration using 40 ± 5 freshly shed cercariae per mouse. In the first three experiments, the mice received a 0.1-mL injection of diethylpyrocarbonate (DEPC)-treated water (blank control), the AY770 siRNA (OD = 1), the AY546 siRNA (OD = 1), or the NC siRNA (OD = 1) via the caudal vein on day 3, day 5, day 7, day 18, day 31, and day 35 post-infection. In the fourth experiment, the mice received an intravenous injection of one of the siRNAs or DEPC-treated water, as described above, on day 19 post-infection and every 3 days thereafter. On day 42 post-infection, the mice were exanguinated, and the number of worms recovered from each mouse (worm burden) was recorded.
To determine the liver egg counts, the liver was removed from each mouse after exsanguination, and the weight was recorded. The liver was homogenized at 10,000 rpm for 1 min and diluted to 20 mL with chlorine-free water. A 1-mL aliquot of the homogenate was combined with 1 mL of 10 % NaOH and incubated at 37 °C for 30 min. The suspensions were agitated, and 100-μL aliquots were collected from the center of each tube. The 100-μL aliquots were diluted to 1 mL with PBS, and the numbers of eggs in the diluted suspensions were microscopically counted three times.
To hatch the eggs, 4 mL of liver homogenate was transferred to a flat-bottomed flask. The flask was filled with chlorine-free water, and a layer of absorbent cotton was used to plug the neck of the bottle, avoiding air bubbles. The bottle was exposed to light and incubated at 27 °C for 4 h to induce egg hatching. The supernatant above the cotton plug, which contained the miracidia, was collected and combined with one to two drops of iodine to fix the miracidia. The fixed miracidia were collected by centrifugation at 4000 rpm for 5 min and microscopically counted at ×40 magnification. The hatching rate was calculated as the ratio of the number of miracidia obtained to the total number of eggs.
Reductions in worm burden, liver egg count, and hatching rate in the siRNA-treated mice, relative to those of the control mice, were calculated as RR = (NOCG – NOEG) ÷ NOEG, where RR was the reduction rate, NOCG was the value obtained for the control group (blank control or NC siRNA), and NOEG was the value obtained for the experimental group.
Statistical analysis
The data are expressed as the mean ± standard deviation. The results for each group were compared using Student’s t tests to examine the intergroup differences. The result of comparisons with P < 0.05 was considered to be statistically significant.
Results
Cloning, sequencing, and molecular characterization of SjZFP1
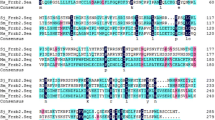
A single 1053-bp product was obtained by RT-PCR. The sequence analysis revealed that this fragment contained the sequence of the SjZFP1 gene of S. japonicum. A 1017-nt ORF was identified in the SjZFP1 cDNA (Fig. 1), which was predicted to encode a 338-aa protein with an estimated molecular mass of 38.5 kDa and theoretical pI of 7.08. The comparison of the SjZFP1 mRNA sequence with that of other zinc finger proteins from S. japonicum in GenBank (accession nos. AY222909 and EZ000159) showed that it shared 99.7 and 99.4 % nucleotide sequence identity and 99.4 and 98.5 % amino acid sequence identity, respectively. The protein blast analysis also revealed that the predicted amino acid sequence of SjZFP1 shared 37 and 36 % similarity with zinc finger protein-like 1 from Homo sapiens (accession no. NP_006773.2) and Bos taurus (accession no. NP_001069740), respectively.
Nucleotide sequence and predicted amino acid sequence of the SjZFP1 gene from Schistosoma japonicum. The sequences corresponding to zinc finger motifs (B-box-type, residues 1–33; PHD-type, residues 51–107; RING-type, residues 54–104) are shaded. The cDNA sequence containing the full open reading frame has been submitted to GenBank (accession no. GQ901167)
The predicted amino acid sequence of SjZFP1 was rich in leucine (10.9 %), asparagine (7.4 %), aspartic acid (7.1 %), and serine (7.4 %), but was poor in methionine (1.8 %) and tryptophan (1.2 %). Several conserved regions were identified in the SjZFP1 protein, which included a B-box-type zinc-binding domain (residues 1–33), two bipartite nuclear localization signal domains (residues 245–259 and 258–275), a paired amphipathic helix repeat (residues 250–258), and overlapping RING finger (RING-type, residues 54–104) and PHD finger (PHD-type with residues 51–107) domains.
Transcription of SjZFP1 at different developmental stages
Eight developmental stages were examined using real-time PCR, including eggs, cercariae, and 7-, 13-, 18-, 23-, 31-, and 42-day-old worms. Male and female 42-day-old worms were separated to examine sex-specific expression in the adult. The SjZFP1 mRNA was expressed in all the developmental stages, and the highest levels of transcription (P < 0.01) were observed in the cercariae, eggs, 7-day-old schistosomula, and mature adult worms (Fig. 2a). At 42 days of age, the level of transcription in females was higher (P <0.01) than that in males (Fig. 2b).
Levels of SjZFP1 mRNA at the various developmental stages of Schistosoma japonicum. a SjZFP1 mRNA levels in samples collected throughout the eight developmental stages of S. japonicum (E egg; C cercariae; 7d, 13d, 18d, 23d, 31d, and 42d represent 7-, 13-, 18-, 23-, 31- and 42-day-old worms, respectively). b Adult male and female worms (42 days old) were quantified using real-time PCR. The data were normalized based on an internal control (α-tubulin). The data are expressed as the mean ± standard deviation
RNAi-mediated SjZFP1 gene knockdown in cultured schistosomula
RNAi-mediated SjZFP1 gene knockdown was performed using three SjZFP1-specific siRNAs and the NC siRNA. The effects of the SjZFP1-specific RNAi on SjZFP1 mRNA expression were analyzed using real-time PCR (Fig. 3), and the inhibition of SjZFP1 transcription in the siRNA-treated groups was calculated. Treatment with the AY770 and AY546 siRNAs reduced the level of SjZFP1 transcription by 82.8 and 81.6 %, compared with that of the blank control (P < 0.01), and by 73.7 and 71.8 % (P < 0.01), compared with that of the NC siRNA. Only the AY770 and AY546 siRNAs were selected for the mouse RNAi experiments.
Effects of siRNA-mediated gene silencing on SjZFP1 mRNA expression. Schistosomula were transformed from cercariae and soaked for 72 h in medium containing control siRNA (NC), one of the three SjZFP1-specific siRNAs, or no siRNA (BC). The level of SjZFP1 mRNA after the gene knockdown experiments was quantified using real-time PCR and normalized based on an internal control (α-tubulin). The data are expressed as the mean ± standard deviation of three experiments
RNAi-mediated SjZFP1 gene knockdown in mice
The level of transcription in the 42-day-old parasites recovered from mice treated with the siRNAs in experiments 2, 3, and 4 was examined using real-time PCR (Fig. 4). In experiments 2 and 3, the administration of the SjZFP1-specific siRNAs in the early phases of infection increased the level of SjZFP1 transcription (P < 0.05) in the adult parasites by 94.0 and 22.2 % in the AY546 group and by 27.6 and 94.7 % in the AY770 group, respectively, compared with blank control group. Compared with the NC group, the level of SjZFP1 mRNA increased by 52.9 and 34.9 % in the AY546 group and by 0.6 and 114.9 % in the AY770 group in experiments 2 and 3, respectively. By contrast, in experiment 4, the administration of the SjZFP1-specific siRNAs in the middle to late stages of infection reduced the level of SjZFP1 mRNA by 56 and 38.3 % in the AY546 and AY770 groups, respectively (P <0.05).
Effect of SjZFP1 gene silencing on life cycle parameters
The worm burden, number of liver eggs, and average egg hatching rate were determined from the mice in the four in vivo experiments (Table 2). The worm burden was lower in the mice treated with SjZFP1-specific siRNAs than that in the mice treated with the blank control or NC siRNA. Compared with the blank control, the worm burden was reduced by 24, 35.6, 14.4, and 11.9 % in the AY546 group (P < 0.05) and by 21.3, 14.9, −2.5, and 8.5 % in the AY770 group (P > 0.05).
In experiments 1, 2, and 3, the average number of liver eggs in the AY546 and AY770 groups was not significantly different than that of the blank control and NC siRNA groups. However, the overall average number of liver eggs recovered from the AY770 group (experiments 1–3) was 23.8 % higher (P < 0.01) than that of the blank group. In experiment 4, the average number of liver eggs recovered from the AY546 and AY770 groups was 22.9 and 24.5 % lower than that of the blank control group (P < 0.05), but was not significantly different than that of the NC siRNA group.
The hatching rates of the liver eggs recovered in experiments 2, 3, and 4 are shown in Table 2. In experiments 2 and 3, the hatching rates of the eggs recovered from the AY546, AY770, and NC groups did not differ significantly from that of the blank control group. In experiment 4, the hatching rates of the eggs recovered from the AY546 and AY770 groups were 29.6 and 37.5 % lower than that of the NC siRNA and blank control group (P < 0.05).
Discussion
Zinc finger proteins are widespread in eukaryotes, representing an important class of DNA-binding proteins that are frequently involved in transcriptional regulation. In S. mansoni, these regulatory proteins are known to modulate morphological and physiological changes, playing crucial roles in parasite development (Bitar et al. 2013). In our current study, multiple conserved regions, including a B-box-type zinc-binding domain, two bipartite nuclear localization signal domains, a paired amphipathic helix repeat, and overlapping RING and PHD finger domains, were identified in the amino acid sequence of SjZFP1.
The RING finger motif is present in many chromatin-associated proteins, and RING finger domains are also involved in protein-protein interactions (Aasland et al. 1995). Because PHD fingers have a structure similar to that of the RING finger, PHD fingers may have similar functions (Aasland et al. 1995). Increasing evidence indicates that PHD fingers bind to specific nuclear proteins and nucleosomes, suggesting that they function in the regulation of chromatin structure or transcription (Bienz 2006). The B-box zinc finger domain consists of approximately 40 amino acids. One or two copies of this motif are often associated with a RING finger domain and a coiled-coil domain, which form the so-called tripartite motif. The tripartite motif is found in transcription factors, ribonucleoproteins, and proto-oncoproteins. However, its function remains unclear.
Ko et al. (2007) showed that nuclear localization signals regulate the expression and subcellular localization of the transcriptional repressor Snail through GSK3-β-dependent phosphorylation. Using real-time PCR, we demonstrated that the SjZFP1 mRNA is expressed in all the developmental stages of S. japonicum and that the level of SjZFP1 transcription was highest in the cercariae, eggs, 7-day-old schistosomula, and mature adult worms. The presence of the conserved motifs in the SjZFP1 protein suggests that it might play important roles in the regulation of chromatin structure or transcription throughout schistosome development.
RNAi is a process in which gene-specific double-stranded RNA (dsRNA) or siRNA triggers the degradation of homologous mRNAs (Corrêa et al. 2010). RNAi-mediated gene silencing has become a valuable tool for determining the biological role of genes, especially for organisms in which knockouts cannot be made, such as parasitic trematodes. In our current study, we performed RNAi gene silencing of SjZFP1 transcription and observed the effects of RNAi-mediated SjZFP1 knockdown on worm burden, the number of liver eggs, and the egg-hatching rate in infected mice. The level of SjZFP1 mRNA in the cultured parasites in the AY770 and AY546 groups was reduced by 82.8 and 81.6 %, compared with that of the blank control, and by 73.7 and 71.8 %, compared with that of the NC group. In the RNAi experiments in mice, the level of SjZFP1 mRNA on day 42 post-infection was significantly higher than that of the controls when the siRNA treatment was administered mainly in the early stages of infection, and it was significantly lower than that of the controls when the siRNA treatment was administered in the middle to late stages of infection. These results showed that the RNAi-mediated reduction in SjZFP1 mRNA expression may have been transient and that the level of SjZFP1 mRNA increased as the SjZFP1-specific siRNAs were eliminated. Mourão et al. (2009) demonstrated that the efficacy of dsRNA-mediated gene silencing for producing consistent phenotypic changes and/or altering levels of gene expression in sporocysts of S. mansoni is highly dependent on the target gene, the specific dsRNA sequence used, and the timing of the post-treatment evaluation of target gene expression.
In the four mouse experiments, the worm burden in AY546 group was significantly lower than that in blank group (P < 0.05). Both the average number of liver eggs and the hatching rate of the eggs recovered from the mice treated with the SjZFP1-specific siRNAs in the middle to late stages of infection were significantly lower than those of the controls. In the experiments in which the mice were treated with the SjZFP1-specific dsRNAs mainly in the early stages of infection, the overall average number of liver eggs in the AY770 group, based on all three experiments, was 23.8 % higher than that of the blank control group. These results may have been caused by a higher level of SjZFP1 expression on day 42, compared with that immediately following the dsRNA treatments. These data suggest that SjZFP1 might play an important role in parasite development, spawning in the vertebrate host, and egg hatching.
References
Aasland R, Gibson TJ, Stewart AF (1995) The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci 20(2):56–59
Bienz M (2006) The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci 31(1):35–40
Bitar M, Drummond MG, Costa MG, Lobo FP, Calzavara-Silva CE, Bisch PM, Machado CR, Macedo AM, Pierce RJ, Franco GR (2013) Modeling the zing finger protein SmZF1 from Schistosoma mansoni: insights into DNA binding and gene regulation. J Mol Graph Model 39:29–38
Brink LH, McLaren DJ, Smithers SR (1977) Schistosoma mansoni: a comparative study of artificially transformed schistosomula and schistosomula recovered after cercarial penetration of isolated skin. Parasitology 74(1):73–86
Calzavara-Silva CE, Prosdocimi F, Abath FG, Pena SD, Franco GR (2004) Nucleic acid binding properties of SmZF1, a zinc finger protein of Schistosoma mansoni. Int J Parasitol 34(11):1211–1219
Corrêa RL, Steine FA, Berezikov E, Ketting RF (2010) MicroRNA-directed siRNA biogenesis in Caenorhabditis elegans. PLoS Genet 6(4):e1000903
Drummond MG, Calzavara-Silva CE, D’Astolfo DS, Cardoso FC, Rajão MA, Mourão MM, Gava E, Oliveira SC, Macedo AM, Machado CR, Pena SD, Kitten GT, Franco GR (2009) Molecular characterization of the Schistosoma mansoni zinc finger protein SmZF1 as a transcription factor. PLoS Negl Trop Dis 3(11):e547
El-Ansary A, Al-Daihan S (2005) Stage-specifically expressed schistosome proteins as potential chemotherapeutic targets. Med Sci Monit 11:RA94–RA103
Eleutério de Souza PR, Valadão AF, Calzavara-Silva CE, Franco GR, de Morais MA, Jr AFG (2001) Cloning and characterization of SmZF1, a gene encoding a Schistosoma mansoni zinc finger protein. Mem Inst Oswaldo Cruz 96:123–130
Fantappie MR, de Oliveira FM, Santos RD, Mansure JJ, Furtado DR, de Abreu da Silva IC, Rumjanek FD (2007) Control of transcription in Schistosoma mansoni: chromatin remodeling and other regulatory elements. Acta Trop 108:186–193
Gobert GN, Moertel L, Brindley PJ, McManus DP (2009) Developmental gene expression profiles of the human pathogen Schistosoma japonicum. BMC Genomics 10:128. doi:10.1186/1471-2164-10-128
Gryseels B, Polman K, Clerinx J, Kestens L (2006) Human schistosomiasis. Lancet 368:1106–1118
Hong Y, Sun A, Zhang M, Gao F, Han Y, Fu Z, Shi Y, Lin J (2013) Proteomics analysis of differentially expressed proteins in schistosomula and adult worms of Schistosoma japonicum. Acta Trop 126(1):1–10
Ko H, Kim HS, Kim NH, Lee SH, Kim KH, Hong SH, Yook JI (2007) Nuclear localization signals of the E-cadherin transcriptional repressor Snail. Cells Tissues Organs 185(1–3):66–72
Laity JH, Lee BM, Wright PE (2001) Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol 11(1):39–46
Loukas A, Bethony JM (2008) New drugs for an ancient parasite. Nat Med 14:365–367
Mourão MM, Dinguirard N, Franco GR, Yoshino TP (2009) Phenotypic screen of early-developing larvae of the blood fluke, Schistosoma mansoni, using RNA interference. PLoS Negl Trop Dis 3(8):e502
Sun H, Liu J, Sun S, Song Z, Shi Y, Lu K, Li H, Jin Y, Lin J (2008a) Screening a phage display cDNA library of Schistosoma japonicum with extract from liver of Microtus fortis. Chin J Prev Vet Med 30(8):592–598 (In Chinese)
Sun H, Liu J, Sun S, Song Z, Shi Y, Lu K, Li H, Jin Y, Lin J (2008b) Screening a phage display cDNA library of Schistosoma japonicum with extract from lung of Microtus fortis. Chin J Vet Parasitol 16(2):9–13 (In Chinese)
Sun Y, Sun H, Jia R, Liu J, Yun C, Shi Y, Lu K, Li H, Jin Y, Lin J (2008c) Screening the target genes of Schistosoma japonicum related to the naturely resistance of Microtus fortis. Chin J Schisto Control 20(1):26–31 (In Chinese)
Song Z, Liu J, He J, Sun S, Shi Y, Li H, Lu K, Lin J, Yue C (2009) Vaccination of mice with phage-displayed schistosomiasis japonica antigens identified by the sera of Microtus fortis. Chin J Prev Vet Med 31(12):963–968 (In Chinese)
Wang L, Utzinger J, Zhou XN (2008) Schistosomiasis control: experiences and lessons from China. Lancet 372:1793–1795
Acknowledgments
This research was supported by a grant from the National Natural Science Foundation of China (grant no. 31072129).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liu, X., Liu, JM., Song, Zy. et al. The molecular characterization and RNAi silencing of SjZFP1 in Schistosoma japonicum . Parasitol Res 114, 903–911 (2015). https://doi.org/10.1007/s00436-014-4255-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-014-4255-4