Abstract
Background
Hepatocellular carcinoma (LIHC) has severe consequences due to late diagnosis and the lack of effective therapies. Currently, potential biomarkers for the diagnosis and prognosis of LIHC have not been systematically evaluated. Previous studies have reported that RAC1 is associated with the B cell receptor signaling pathway in various tumor microenvironments, but its relationship with LIHC remains unclear. We investigated the relationship between RAC1 and the prognosis and immune infiltration microenvironment of LIHC, exploring its potential as a prognostic biomarker for this type of cancer.
Methods
In this study, we analyzed data from The Cancer Genome Atlas (TCGA) using the Wilcoxon signed-rank test and logistic regression to assess the association between RAC1 expression and clinical characteristics in LIHC patients. Additionally, Kaplan-Meier and Cox regression methods were employed to confirm the impact of RAC1 expression levels on overall survival. Immunohistochemistry was used to validate RAC1 protein expression in LIHC. We constructed RAC1 knockdown LIHC cells and studied the effects of RAC1 protein on cell proliferation and migration at both cellular and animal levels.
Results
RAC1 expression levels were significantly elevated in LIHC tissues compared to normal tissues. High RAC1 expression was strongly associated with advanced pathological stages and was identified as an independent factor negatively affecting overall survival. At both cellular and animal levels, RAC1 knockdown significantly inhibited the proliferation and migration of LIHC cells. Furthermore, RAC1 expression was positively correlated with the infiltration of Th2 cells and macrophages in the tumor microenvironment, suggesting that RAC1 may contribute to the deterioration of the tumor immunosuppressive microenvironment and potentially lead to reduced patient survival.
Conclusion
These findings indicate that RAC1 expression promotes LIHC proliferation and migration and influences the landscape of immune cell infiltration in the tumor microenvironment. Based on these results, RAC1 is proposed as a potential prognostic biomarker for LIHC, associated with both cancer progression and tumor immune cell infiltration.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Rho family of GTPases, a subfamily of the Ras superfamily, consists of small (~ 21 kD) signaling G proteins. These GTPases typically switch between active and inactive states by exchanging guanosine diphosphate (GDP) and guanosine triphosphate (GTP) molecules(Jin et al. 2024), reliably binding to specific proteins and activating signaling cascades. Among them, RAC1 plays a crucial role in regulating cell growth, angiogenesis, tissue remodeling, and cell migration(Tátrai et al. 2024). In the unstimulated state, RAC1 exists in the cytosol as a complex with guanine nucleotide dissociation inhibitors (GDIs). Upon activation by soluble chemoattractant, chemokines, phagocytic particles, or any upstream regulators, RAC1 initiates signaling. This activation involves the dissociation of RAC1 from GDIs and the exchange of GDP for GTP facilitated by membrane-localized guanine nucleotide exchange factors. Consequently, RAC1, in its GTP-bound form, becomes anchored to the membrane(Yamauchi et al. 2005; Alvarez and Agaisse 2014).
Furthermore, RAC1 is associated with tumor progression and tumor immune microenvironment(Shi et al. 2024). Immune cell infiltration is a major component of the tumor microenvironment, and the activation or inactivation of RAC1 within this environment not only affects the plasticity of tumor cell movement but may also influence immune cell infiltration by regulating various signaling pathways, thereby impacting the overall landscape of the tumor microenvironment(Sanz-Moreno et al. 2008; De et al. 2019). Therefore, a better understanding of the relationship between RAC1 gene expression and immune cell infiltration within the tumor microenvironment is essential for deciphering immunotherapeutic mechanisms, defining predictive biomarkers, and identifying new therapeutic targets(Zhou et al. 2021; Sauzeau et al. 2022b).
Previous studies have shown that RAC1 gene expression is higher in melanoma, glioblastoma, colon cancer and ovarian cancer compared to adjacent normal tissues(Hudson et al. 2018; Al-Koussa et al. 2020; Kotelevets and Chastre 2020; Cannon et al. 2020). Unfortunately, the crucial role of RAC1 in the development and progression of liver hepatocellular carcinoma (LIHC) remains largely uncharacterized. Our study aims to discover whether RAC1 genes expression level is a potential diagnostic and prognostic marker and its association with immune infiltration in LIHC.
Materials and methods
Dataset source and preprocessing
The expression levels of RAC1 in 424 LIHC samples with 50 adjacent normal tissue samples were analyzed based on the dataset of The Cancer Genome Atlas (TCGA-LIHC) mRNA sequencing (RNA-seq) database. Clinical characteristics such as tumor statue, histologic grades, pathologic stages and vascular invasion were assessed with respect to their respective clinical patterns. Differential transcriptional expression was compared to Student`s t-test.
Immune infiltration algorithm
Based on the ssGSEA algorithm provided in R-packet-GSVA [1.46.0], the provided markers of 24 immune cells were used to calculate the immune infiltration of the corresponding data.
Diagnostic analysis
The Receiver Operating Characteristic (ROC) curve was applied to assess the specificity and sensitivity of gene prediction accuracy, using the area under the ROC curve (AUC) as a diagnostic value based on the “pROC” package in the statistical software (version 1.18.0) was used.
Survival analysis
The Kaplan-Meier curve was performed to compare overall survival (OS) between the differential expression groups of RAC1 including 424 LIHC samples in the TCGA database and KM Plotter, respectively. The correlation between RAC1 expression and survival was analyzed to discover the significant prognostic factors. The hazard ratio (HR) with 95% confidence interval (CI) and log-rank P value were also calculated.
Correlation analysis
Gene expression correlation analysis was performed for given sets of mRNA expression data in TCGA-LIHC were used for analysis. The correlation coefficient was determined by the Spearman method.
Immunohistochemical staining (IHC)
We have obtained the IHC result of hepatocellular carcinoma sample (ID: 2766) from The Human Protein Atlas (https://www.proteinatlas.org/). Meanwhile, we also explored the RAC1 protein level in LIHC by IHC using a commercial tissue microarray (Bioaitech Co., Xi’an, China, cat #D202Lv01) with 10 LIHC and 10 normal liver tissues. IHC procedure was conducted as follows: tissues were first dewaxed and hydrated, followed by antigen retrieval and blocking of endogenous peroxidase activity. The slides were then incubated with the primary antibody overnight at 4 °C. After incubation with the secondary antibody, the slides were stained with diaminobenzidine (DAB). For the evaluation and scoring, two pathologists independently assessed the staining results. RAC1 protein expression was scored based on both the percentage of positively stained tumor cells and the staining intensity. Specifically, the percentage of immunoreactive tumor cells was scored as 1 (< 10%), 2 (10–25%), 3 (26–49%), and 4 (≥ 50%). Staining intensity was visually assessed and scored as 1 (negative), 2 (light yellow), 3 (light brown), and 4 (dark brown). The final immunoreactivity score for each case was obtained by multiplying the percentage score by the intensity score.
Cell culture
The LIHC cell lines utilized in this study, HepG2 and Huh7, were procured from the China Center for Type Culture Collection. The complete culture medium formulation used for cell culture consisted of Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. The cells were cultured in an incubator maintained at 37 °C with 5% CO2. Cells in the logarithmic growth phase were used for subsequent experiments.
Transfection of small interfering RNA
The siRNAs used in this study were sourced from Genscript Co., Ltd., located in Nanjing, China. For the transfection of siRNAs into liver hepatocellular carcinoma (LIHC) cell lines, we utilized Lipofectamine 2000 (Invitrogen, Carlsbad, USA), following the manufacturer’s instructions.
The transfection procedure began with seeding LIHC cells onto cell culture plates. The cells were cultured in appropriate growth medium until they reached approximately 50% confluence. At this stage, the siRNAs were prepared for transfection. Each siRNA was diluted in an opti-MEM medium and mixed with Lipofectamine 2000 reagent in a separate tube. The siRNA-Lipofectamine 2000 mixture was then incubated at room temperature for 20 min to allow for the formation of siRNA-lipid complexes.
Following incubation, the siRNA-lipid complex was added to the cells in their culture plates. The final concentration of siRNA in the transfection mixture was set to 100 nM. The cells were then incubated with this transfection mixture for 48 h to ensure sufficient uptake and effective gene silencing.
The siRNAs used in this study had the following sequences:
siRAC1-1: 5’-AAGACAAGCCGATTGCCGACG-3’.
siRAC1-2: 5’-AAGCCGATTGCCGACGTGTTC-3’.
Quantitative real-time polymerase chain reaction
The cDNA was amplified using SYBR-Green PCR Master Mix (TAKARA, Tokyo, Japan) on a QuantStudio 5 system (ABI, Carlsbad, USA) under the following conditions: an initial denaturation at 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 15 s and annealing/extension at 60 °C for 1 min. The level of RAC1 mRNA was calculated by 2−ΔΔCt method using GAPDH as internal control.
The sequences of the specific primers used for amplification were as follows:
GAPDH forward, 5’-AGATCCCTCCAAAATCAAGTGG-3’;
GAPDH reverse, 5’- GGCAGAGATGATGACCCTTTT-3’;
RAC1 forward, 5’-ATGTCCGTGCAAAGTGGTATC − 3’;
RAC1 reverse, 5’-CTCGGATCGCTTCGTCAAACA-3’.
Western blotting
After transfecting cells with siRNA for 72 h, cells were collected and lysed using RIPA buffer. Equal amounts of protein from each group were separated by 12% SDS-PAGE and transferred onto PVDF membranes. The membranes were blocked with 5% BSA at room temperature for 1 h, followed by incubation with anti-RAC1 antibody (Abcam, ab155938) and then with HRP-conjugated secondary antibody (Abcam, ab288151). Signal detection was performed using ECL chemiluminescence reagents, and results were recorded with a chemiluminescent imaging system. The membranes were then stripped of antibodies and re-blocked, followed by sequential incubation with anti-GAPDH antibody (Abcam, ab8245) and HRP-conjugated secondary antibody (Abcam, ab205719). ECL detection and imaging were subsequently performed.
Wound healing assay
The migratory ability of different groups of LIHC cells was evaluated using a wound-healing assay. When the cells covered 95% of the bottom of a six-well plate, a 10 µL pipette tip was used to create a straight scratch in the monolayer. The cells were then washed twice with PBS and replenished with complete medium. After imaging, the cells were cultured for an additional 24 h before being imaged again. The migration index was calculated using Image J software with the following formula:
Proliferation assay
The proliferation capability of different LIHC cell groups was assessed using the CCK-8 assay. When the cells reached 50% confluence, culture medium containing 10% CCK-8 solution was added to each well and incubated at 37 °C for 30 min. Absorbance at 450 nm was then measured for each well. After the measurement, the medium was replaced with fresh culture medium, and cells were cultured further. Proliferation was assessed at various time points, including 1, 2, 4, 8, 12, 24, 36, 48, and 96 h. The proliferation rate of each cell line was calculated relative to the absorbance value at the 0-hour time point.
Subcutaneous tumor xenograft model of LIHC cells
After 72 h of transfection, the cells were collected for tumor xenograft implantation. The LIHC xenograft model was established by subcutaneously injecting different groups of LIHC cells into the right armpits of six-week-old BALB/c nude mice (1 × 10^7 cells per mouse; three animals per group). Tumor size was recorded every other day, and tumor volume was calculated according to the formula: width^2 × length/2. Twelve days after tumor cell implantation, all mice were euthanized, and tumor tissues were collected.
Statistical analysis
The Wilcoxon rank sum test was used to assess the significant differential expression levels of the RAC1 in LIHC with the threshold of gene expression being selected as the median method. Correlation of gene expression was evaluated by Spearman’s correlation coefficient. Univariate Cox analysis was used to screen for potential risk factors, and multivariate Cox analysis was used to verify the independent variate of RAC1 expression on overall survival. All statistical analyzes were performed with the R statistical software (version 4.2.1). A P-value of less than 0.05 is considered statistically significant.
Results
Expression of the RAC1 in LIHC
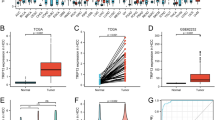
We analyzed RAC1 mRNA levels in LIHC and adjacent normal tissue samples using TCGA RNA-seq datasets. RAC1 was significantly upregulated in LIHC compared to normal tissues (Fig. 1A and B, Table S1). Immunohistochemical staining further confirmed the elevated RAC1 protein level in LIHC tissues compared to normal liver tissues (Fig. 1C and D).
Expression of the RAC1 in LIHC (A-B) Evaluation of RAC1 expression in patients with LIHC compared to adjacent normal tissue samples: (A) unpaired samples, (B) paired samples; (C-D) RAC1 protein level in patients with LIHC compared to normal tissue: (C) normal liver tissue, (D) hepatocellular carcinoma tissue. ***p < 0.001
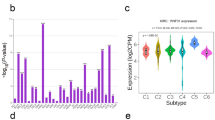
Correlation between RAC1 expression and LIHC clinicopathological parameters
To further assess the role of RAC1 in clinical traits, we examined RAC1 expression by analyzing the TCGA RNA-seq data in subgroups. The results indicate that RAC1 expression is significantly upregulated in higher histological grades, advanced pathological stages, higher AFP levels, and in samples with vascular invasion. These findings suggest that elevated RAC1 expression is closely associated with more aggressive clinical features. (Fig. 2).
Diagnostic value and prognostic potential of RAC1 in LIHC
We examined RAC1 protein levels in a tissue microarray consisting of 10 normal liver tissues and 10 LIHC tissues using immunohistochemistry (IHC). The results showed that RAC1 expression was significantly higher in LIHC tissues compared to normal tissues at the protein level (Fig. 3A and B). Consequently, we considered RAC1 as a potential diagnostic and prognostic marker for LIHC. We used the ROC curves to assess the performance of RAC1 in predicting result of LIHC. When predicting outcomes from normal or tumor tissue, the predictability of RAC1 (AUC = 0.915, 95% CI: 0.881–0.949) showed higher accuracy compared to AFP (AUC = 0.724, 95% CI: 0.671–0.776) (Fig. 3C). To determine the prognostic value of mRNA expression levels in LIHC, Kaplan-Meier curves were adopted to assess expression level in relation to patient survival and clinical follow-up period. Log-rank tests were performed to assess statistical significance. Our study revealed that high mRNA expression of RAC1 (HR = 1.973, 95% CI: 1.398–2.786, p < 0.001) was significantly associated with poor prognosis. (Fig. 3D)
Diagnostic value and prognostic potential of RAC1 in LIHC (A) Expression and (B) scoring of RAC1 protein, as detected by IHC in 10 LIHC and 10 normal liver tissues (scale bar = 50 μm). (C) ROC curves and the predictability of RAC1; (D) Kaplan-Meier curve of overall survival and expression levels of RAC1; ROC, receiver operating characteristics; ***p < 0.001
High-level expression of RAC1 is an independent risk factor for OS of LIHC
Univariate Cox analysis showed that high RAC1 expression was significantly correlated with poor OS (HR = 1.977, 95% CI = 1.384–2.824, p < 0.001). Multivariate Cox analysis confirmed RAC1 gene expression was an independent risk factor for OS in patients with LIHC (HR = 1.847, 95% CI = 1.244–2.740, p = 0.002) (Table 1). Besides, tumor status (HR = 1.769, 95% CI = 1.182–2.647, p = 0.006) and pathological stages (HR = 2.139, 95% CI = 1.442–3.173, p < 0.001) were another independent risk factors in LIHC.
Correlation between RAC1 expression and immune infiltration in LIHC
We have established a RAC1-associated immune-infiltrating landscape of tumor micro environment (TME) immune cells in LICH. The results showed that the expression of RAC1 was positively correlated with Th2, NK CD56 bright, macrophages and Tem cells, but negatively with Th17 cells (Fig. 4A) (Table S1). Kaplan-Meier plot curves were used to estimate the prognostic value of RAC1 expression levels in LIHC associated with infiltrating immune cells. Our analysis revealed that high mRNA expressions of RAC1 are significantly associated with poor prognosis of Th2-enriched patients (HR = 2.44, 95% CI: 1.42–4.19, p = 0.0008) (Fig. 4B and D) and macrophage-enriched patients (HR = 2.15, 95% CI: 1.33–3.49, p = 0.001) (Fig. 4C and E), but not the infiltrating immune cells decreased subgroups.
Correlation between RAC1 expression and immune infiltration in LIHC. (A) Landscape of RAC1 correlation to infiltrating immune cells. *p < 0.05; **p < 0.01; ***p < 0.001. (B-C) Scatter diagram demonstrated the correlation between RAC1 genes expression and infiltrating immune cells. (B) Enrichment of Th2 cells; (C) Enrichment of macrophages; (D-E) Kaplan-Meier curve of overall survival for RAC1 gene expression associated with immune cell infiltration in LIHC. (D) subset enriched in Th2 cells; (E) subset enriched in macrophage cells
Knockdown of RAC1 inhibits the proliferation and migration of LIHC cells
To further investigate the biological role of RAC1 in LIHC development, we used siRNAs to knock down RAC1 in HepG2 and Huh7 cells. The efficiency of the knockdown was verified by qRT-PCR (Fig. 5A and B) and western blotting (Fig. 5C and D). Upon successful knockdown of RAC1, the migratory abilities of both LIHC cell lines were significantly inhibited (Fig. 5E and F). We also monitored the proliferation of LIHC cell lines after RAC1 knockdown using the CCK-8 assay. The results demonstrated that successful knockdown of RAC1 led to a significant reduction in the proliferation capacity of both HepG2 and Huh7 cells (Fig. 5H and I). Additionally, we investigated the effect of RAC1 knockdown on the proliferative capacity of LIHC cells in an animal model. The knockdown of RAC1 significantly slowed LIHC proliferation (Fig. 5I-K). These findings demonstrate that RAC1 is involved in the migration and proliferation of LIHC cells in vivo.
Knockdown of RAC1 inhibits the proliferation and migration of LIHC cells. Interference efficiency of siRAC1 in HepG2 and Huh7 was assessed by qRT-PCR (A, B) and western blot (C, D) (n = 5). (E, F) Wound healing assays were performed in HepG2 and Huh7 cells with RAC1 knocking down (n = 5). (G, H) Proliferation rate of HepG2 and Huh7 cells with RAC1 knocking down (n = 3). (I, J) Tumor growth curves of HepG2 and Huh7 cells with RAC1 knocking down (n = 3). (K) Images of the tumor tissues collected after 12 days. ***p < 0.001
Relationships between RAC1 expression and immune markers
To further investigate the relationship between RAC1 and the infiltrating immune cells, we analyzed the correlations between several immune cell markers and Rac1 by coexpression heatmap. These markers have been used to characterize immune cells including Th2 cells and tumor-associated macrophages. The results showed that RAC1 was associated with Th2 cell markers GATA3 (r = 0.312, p = 8.14e-10), STAT6 (r = 0.227, p = 1.07e-05) and macrophage markers CCL2 (r = 0.250, p = 1.14e-06), IL10 (r = 0.243, p = 2.13e-06), VSIG4 (r = 0.292, p = 9.91e-09), MS4A4A (r = 0.266, p = 2.05e-07). In general, RAC1 was positively correlated with the tumor-infiltrating immune markers. We also analyzed the markers of exhausted T cells including PDCD1 (r = 0.314, p = 6.36e-10), CTLA4 (r = 0.307, p = 1.49e-09), TIGIT (r = 0.245, p = 1.75e-06), HAVCR2 (r = 0.401, p = 8.84e-16). (Fig. 6) (Table S2).
Discussion
Rac1 is a key member of the Rho GTPase family. As a cytoskeletal regulatory protein, Rac1 primarily regulates cell adhesion and movement by promoting actin cytoskeleton remodeling(Machacek et al. 2009; Martin et al. 2016). It plays a crucial role in transducing migration signals to the cytoskeleton(Zegers and Friedl 2014; Navarro-Lérida et al. 2015; Swaminathan et al. 2021). Disruption of RAC1 interferes with cell adhesion and lamellipodia formation, affecting cytoskeletal dynamics and disrupting the dynamic interactions between cells and the extracellular matrix (ECM), thereby driving tumor cell invasion and migration(Arthur et al. 2002; Du et al. 2019; Liu et al. 2021). In head and neck squamous cell carcinoma (HNSCC), Rac1 participates in cell-matrix adhesion and promotes cancer cell migration through fibronectin, mediated by Ras-related protein 1 (Rap1)(Liu et al. 2020). Additionally, upregulation of Rac1 expression accelerates hepatocellular carcinoma metastasis by promoting lamellipodia formation in liver cancer cells via activation of downstream targets such as the Scar/Wave complex(Guo et al. 2019). Moreover, external mechanical stimuli regulate cytoskeletal remodeling and contraction in pancreatic cancer cells through Rac1 and other cytoskeletal remodeling factors, maintaining their motility(Kalli et al. 2022).
The role of RAC1 in the tumor immune microenvironment is complex and diverse, particularly in its regulation of immune cell functions and influence on tumor immune evasion(Carrizzo et al. 2014; Haga and Ridley 2016). RAC1 affects the activity and infiltration of tumor-associated immune cells such as plasma cells, lymphocytes, and macrophages, directly shaping the nature and strength of tumor immune responses(Porter et al. 2016; Lawson and Ridley 2017; Fionda et al. 2021). Research indicates that RAC1 can regulate the migration capacity of these immune cells within tumor tissues, impacting their distribution and functional roles(Maldonado and Dharmawardhane 2018; Zhu et al. 2021). This not only alters the composition and behavior of immune cells but also their ability to detect and target tumor cells(Wang et al. 2013).Furthermore, RAC1 is pivotal in mediating interactions between immune cells and tumor cells(Bosco et al. 2008). For instance, it modulates the phagocytic activity of macrophages, influencing their efficacy in killing tumor cells(Song et al. 2024; Bailly et al. 2024). Additionally, RAC1 plays a critical role in regulating the expression and function of immune checkpoint molecules, thereby affecting T cell activation and the ability of tumor cells to evade immune detection(Mishra et al. 2023; Tao et al. 2024). Of paramount importance is the finding that aberrant activation of the RAC1 signaling pathway is closely linked to the development and progression of various types of cancers, including breast cancer, melanoma, and prostate cancer(Sauzeau et al. 2022a, b; Bailly et al. 2022; Ma et al. 2023). Therefore, understanding and targeting RAC1 in the tumor immune microenvironment could provide a solid foundation for developing new immune-based therapies(Arnst et al. 2017).
In our study, we observed that RAC1 gene expression in LIHC was significantly higher compared to normal tissues. This upregulated RAC1 gene expression was closely associated with various pathological parameters, including histological grade, clinical stage, AFP level, and vascular invasion status. Importantly, multivariate Cox analysis demonstrated that RAC1 expression serves as an independent factor influencing overall survival in LIHC patients. ROC analysis confirmed the potential prognostic value of RAC1 in LIHC, and Kaplan-Meier survival analysis revealed that patients with high RAC1 gene expression had significantly shorter overall survival (p < 0.001). Concurrently, we constructed RAC1 knockdown LIHC cell lines to investigate its impact on cell proliferation and migration. Knockdown of RAC1 significantly inhibited LIHC migration and also suppressed the proliferation of LIHC in subcutaneous tumor models. Therefore, high RAC1 gene expression severely worsens patient prognosis by promoting LIHC migration and proliferation.
To further investigate the relationship between RAC1 and tumor immune infiltration, we established a RAC1-associated tumor immune microenvironment profile. We found that RAC1 was significantly differentially expressed in various tumor-infiltrating immune cells, particularly in Th2 cells and tumor-associated macrophages. Incorporating RAC1 expression in immune cells into our prognostic assessment, we found that high RAC1 expression in Th2 cells and macrophages was associated with reduced survival in LIHC patients. This indicates that RAC1 influences the prognosis of LIHC patients by modulating the tumor immune microenvironment.
In summary, this study demonstrates that RAC1 gene expression can serve not only as a diagnostic factor for the development of LIHC but also as an independent prognostic marker. The correlation analysis of RAC1 with various clinicopathological parameters further substantiates this hypothesis. High RAC1 gene expression promotes LIHC proliferation and migration and is significantly associated with poor prognosis. Furthermore, our research indicates that RAC1 gene expression exhibits diverse associations with tumor-infiltrating immune cells, such as Th2 cells, tumor-associated macrophages, and even exhausted T cells, suggesting its potential as a target for immune therapy in LIHC. However, the underlying signaling pathways require further investigation.
Data availability
Data is provided within the manuscript or supplementary information files.
References
Al-Koussa H, Atat OE, Jaafar L, Tashjian H, El-Sibai M (2020) The role of rho GTPases in Motility and Invasion of Glioblastoma Cells. Anal Cell Pathol 2020:1–9. https://doi.org/10.1155/2020/9274016
Alvarez DE, Agaisse H (2014) A role for the small GTPase Rac1 in vaccinia actin-based motility. Small GTPases. https://doi.org/10.4161/sgtp.29038
Arnst JL, Hein AL, Taylor MA, Palermo NY, Contreras JI, Sonawane YA, Wahl AO, Ouellette MM, Natarajan A, Yan Y (2017) Discovery and characterization of small molecule Rac1 inhibitors. Oncotarget 8(21):34586–34600. https://doi.org/10.18632/oncotarget.16656
Arthur WT, Noren NK, Burridge K (2002) Regulation of rho family GTPases by cell-cell and cell-matrix adhesion. Biol Res 35(2):239–246. https://doi.org/10.4067/s0716-97602002000200016
Bailly C, Beignet J, Loirand G, Sauzeau V (2022) Rac1 as a therapeutic anticancer target: promises and limitations. Biochem Pharmacol 203:115180. https://doi.org/10.1016/j.bcp.2022.115180
Bailly C, Degand C, Laine W, Sauzeau V, Kluza J (2024) Implication of Rac1 GTPase in molecular and cellular mitochondrial functions. Life Sci 342:122510. https://doi.org/10.1016/j.lfs.2024.122510
Bosco EE, Mulloy JC, Zheng Y (2008) Rac1 GTPase: a rac of all trades. Cell Mol Life Sci 66(3):370. https://doi.org/10.1007/s00018-008-8552-x
Cannon AC, Uribe-Alvarez C, Chernoff J (2020) RAC1 as a therapeutic target in malignant melanoma. Trends Cancer 6(6):478–488. https://doi.org/10.1016/j.trecan.2020.02.021
Carrizzo A, Forte M, Lembo M, Formisano L, Puca AA, Vecchione C (2014) Rac-1 as a new therapeutic target in cerebro- and cardio-vascular diseases. Curr Drug Targets 15(13):1231–1246. https://doi.org/10.2174/1389450115666141027110156
De P, Aske JC, Dey N (2019) RAC1 takes the lead in solid tumors. Cells 8(5):382. https://doi.org/10.3390/cells8050382
Du G-S, Qiu Y, Wang W-S, Peng K, Zhang Z-C, Li X-S, Xiao W-D, Yang H (2019) Knockdown on aPKC-ι inhibits epithelial-mesenchymal transition, migration and invasion of colorectal cancer cells through Rac1-JNK pathway. Exp Mol Pathol 107:57–67. https://doi.org/10.1016/j.yexmp.2018.11.007
Fionda C, Stabile H, Molfetta R, Kosta A, Peruzzi G, Ruggeri S, Zingoni A, Capuano C, Soriani A, Paolini R, Gismondi A, Cippitelli M, Santoni A (2021) Cereblon regulates NK cell cytotoxicity and migration via Rac1 activation. Eur J Immunol 51(11):2607–2617. https://doi.org/10.1002/eji.202149269
Guo Y, Zhu J, Wang X, Li R, Jiang K, Chen S, Fan J, Xue L, Hao D (2019) Orai1 promotes Osteosarcoma Metastasis by activating the Ras-Rac1-WAVE2 signaling pathway. Med Sci Monit Int Med J Exp Clin Res 25:9227–9236. https://doi.org/10.12659/MSM.919594
Haga RB, Ridley AJ (2016) Rho GTPases: regulation and roles in cancer cell biology. Small GTPases 7(4):207–221. https://doi.org/10.1080/21541248.2016.1232583
Hudson LG, Gillette JM, Kang H, Rivera MR, Wandinger-Ness A (2018) Ovarian Tumor Microenvironment Signaling: convergence on the Rac1 GTPase. Cancers 10(10):358. https://doi.org/10.3390/cancers10100358
Jin C, Chen H, Xie L, Zhou Y, Liu L, Wu J (2024) GPCRs involved in metabolic diseases: pharmacotherapeutic development updates. Acta Pharmacol Sin 1–16. https://doi.org/10.1038/s41401-023-01215-2
Kalli M, Li R, Mills GB, Stylianopoulos T, Zervantonakis IK (2022) Mechanical stress signaling in pancreatic Cancer cells triggers p38 MAPK- and JNK-Dependent cytoskeleton remodeling and promotes Cell Migration via Rac1/cdc42/Myosin II. Mol Cancer Res 20(3):485–497. https://doi.org/10.1158/1541-7786.MCR-21-0266
Kotelevets L, Chastre E (2020) Rac1 signaling: from intestinal homeostasis to Colorectal Cancer Metastasis. Cancers 12(3):665. https://doi.org/10.3390/cancers12030665
Lawson CD, Ridley AJ (2017) Rho GTPase signaling complexes in cell migration and invasion. J Cell Biol 217(2):447–457. https://doi.org/10.1083/jcb.201612069
Liu M, Banerjee R, Rossa C, D’Silva NJ (2020) RAP1-RAC1 signaling has an important role in Adhesion and Migration in HNSCC. J Dent Res 99(8):959–968. https://doi.org/10.1177/0022034520917058
Liu S, Gu L, Wu N, Song J, Yan J, Yang S, Feng Y, Wang Z, Wang L, Zhang Y, Jin Y (2021) Overexpression of DTL enhances cell motility and promotes tumor metastasis in cervical adenocarcinoma by inducing RAC1-JNK-FOXO1 axis. Cell Death Dis 12(10):929. https://doi.org/10.1038/s41419-021-04179-5
Ma N, Xu E, Luo Q, Song G (2023) Rac1: a Regulator of Cell Migration and a potential target for Cancer Therapy. Mol Basel Switz 28(7):2976. https://doi.org/10.3390/molecules28072976
Machacek M, Hodgson L, Welch C, Elliott H, Pertz O, Nalbant P, Abell A, Johnson GL, Hahn KM, Danuser G (2009) Coordination of rho GTPase activities during cell protrusion. Nature 461(7260):99–103. https://doi.org/10.1038/nature08242
Maldonado M, del Dharmawardhane M S (2018) Targeting Rac and Cdc42 GTPases in Cancer. Cancer Res 78(12):3101–3111. https://doi.org/10.1158/0008-5472.CAN-18-0619
Martin K, Reimann A, Fritz RD, Ryu H, Jeon NL, Pertz O (2016) Spatio-temporal co-ordination of RhoA, Rac1 and Cdc42 activation during prototypical edge protrusion and retraction dynamics. Sci Rep 6:21901. https://doi.org/10.1038/srep21901
Mishra AK, Rodriguez M, Torres AY, Smith M, Rodriguez A, Bond A, Morrissey MA, Montell DJ (2023) Hyperactive rac stimulates cannibalism of living target cells and enhances CAR-M-mediated cancer cell killing. Proc Natl Acad Sci U S A 120(52):e2310221120. https://doi.org/10.1073/pnas.2310221120
Navarro-Lérida I, Pellinen T, Sanchez SA, Guadamillas MC, Wang Y, Mirtti T, Calvo E, Del Pozo MA (2015) Rac1 nucleocytoplasmic shuttling drives nuclear shape changes and tumor invasion. Dev Cell 32(3):318–334. https://doi.org/10.1016/j.devcel.2014.12.019
Porter AP, Papaioannou A, Malliri A (2016) Deregulation of rho GTPases in cancer. Small GTPases 7(3):123–138. https://doi.org/10.1080/21541248.2016.1173767
Sanz-Moreno V, Gadea G, Ahn J, Paterson H, Marra P, Pinner S, Sahai E, Marshall CJ (2008) Rac Activation and Inactivation Control Plasticity of Tumor Cell Movement. Cell 135(3):510–523. https://doi.org/10.1016/j.cell.2008.09.043
Sauzeau V, Beignet J, Bailly C (2022a) Rac1 as a target to treat dysfunctions and Cancer of the bladder. Biomedicines 10(6):1357. https://doi.org/10.3390/biomedicines10061357
Sauzeau V, Beignet J, Vergoten G, Bailly C (2022b) Overexpressed or hyperactivated Rac1 as a target to treat hepatocellular carcinoma. Pharmacol Res 179:106220. https://doi.org/10.1016/j.phrs.2022.106220
Shi X, Chen W, Yin Y, Cao H, Wang X, Jiang W, Li Q, Li X, Yu Y, Wang X (2024) RAC1high NK cell-based immunotherapy in hepatocellular carcinoma via STAT3-NKG2D axis. Cancer Lett 592:216909. https://doi.org/10.1016/j.canlet.2024.216909
Song Y, Liu Y, Guo F, Zhao L, Qin G (2024) Single-cell RNA sequencing reveals RAC1 involvement in Macrophages Efferocytosis in Diabetic kidney disease. Inflammation 47(2):753–770. https://doi.org/10.1007/s10753-023-01942-y
Swaminathan K, Campbell A, Papalazarou V, Jaber-Hijazi F, Nixon C, McGhee E, Strathdee D, Sansom OJ, Machesky LM (2021) The RAC1 target NCKAP1 plays a crucial role in the progression of Braf;Pten-Driven Melanoma in mice. J Invest Dermatol 141(3):628–637e15. https://doi.org/10.1016/j.jid.2020.06.029
Tao L, Xu X, Fang Z, Christopoulos P, Cortinovis D, Lu Y (2024) RAC1 as a potential pan-cancer diagnostic, prognostic, and immunological biomarker. Transl Cancer Res 13(3):1533–1553. https://doi.org/10.21037/tcr-23-2016
Tátrai E, Ranđelović I, Surguta SE, Tóvári J (2024) Role of Hypoxia and Rac1 inhibition in the Metastatic Cascade. Cancers 16(10):1872. https://doi.org/10.3390/cancers16101872
Wang J-Y, Yu P, Chen S, Xing H, Chen Y, Wang M, Tang K, Tian Z, Rao Q, Wang J (2013) Activation of Rac1 GTPase promotes leukemia cell chemotherapy resistance, quiescence and niche interaction. Mol Oncol 7(5):907–916. https://doi.org/10.1016/j.molonc.2013.05.001
Yamauchi A, Marchal CC, Molitoris J, Pech N, Knaus U, Towe J, Atkinson SJ, Dinauer MC (2005) Rac GTPase isoform-specific regulation of NADPH oxidase and Chemotaxis in murine neutrophils in vivo: ROLE OF THE C-TERMINAL POLYBASIC DOMAIN *. J Biol Chem 280(2):953–964. https://doi.org/10.1074/jbc.M408820200
Zegers MM, Friedl P (2014) Rho GTPases in collective cell migration. Small GTPases 5:e28997. https://doi.org/10.4161/sgtp.28997
Zhou H-C, Liu C-X, Pan W-D, Shang L-R, Zheng J-L, Huang B-Y, Chen J-Y, Zheng L, Fang J-H, Zhuang S-M (2021) Dual and opposing roles of the androgen receptor in VETC-dependent and invasion-dependent metastasis of hepatocellular carcinoma. J Hepatol 75(4):900–911. https://doi.org/10.1016/j.jhep.2021.04.053
Zhu Y, Xie J, Shi J (2021) Rac1/ROCK-driven membrane dynamics promote natural killer cell cytotoxicity via granzyme-induced necroptosis. BMC Biol 19(1):140. https://doi.org/10.1186/s12915-021-01068-3
Author information
Authors and Affiliations
Contributions
Yuan Li and Aidong Gu are equal for the contribution for this work. Conceptualization, Yuan Li and Aidong Gu; methodology, Yuan Li; software, Aidong Gu; validation, Aidong Gu and Yuan Li; resources, Lili Yang; data curation, Yuan Li; writing—original draft preparation, Aidong Gu and Yuan Li; writing—review and editing, Qingbo Wang; supervision, Qingbo Wang; project administration, Qingbo Wang. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethical approval
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of Nanjing University of Chinese Medicine.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, Y., Gu, A., Yang, L. et al. RAC1 serves as a prognostic factor and correlated with immune infiltration in liver hepatocellular carcinoma. J Cancer Res Clin Oncol 150, 418 (2024). https://doi.org/10.1007/s00432-024-05933-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00432-024-05933-w