Abstract
Purpose
The advent of immune checkpoint blockade (ICB) therapies this year has changed the way glioblastoma (GBM) is treated. Meanwhile, some patients with strong PD-L1 expression remain immune checkpoint resistant. To better understand the molecular processes that influence the immune environment, there is an urgent need to characterize the immunosuppressive tumor microenvironment and identify biomarkers to predict patient survival outcomes.
Patients and methods
Our study analyzed RNA-sequencing data from 178 GBM samples. Their unique gene expression patterns in the tumor microenvironment were analyzed by an unsupervised clustering algorithm. Through these expression patterns, a panel of T-cell exhaustion signatures, immunosuppressive cells, and clinical features correlates with immunotherapy response. The presence or absence of immune status and prognostic signatures was then validated with the test dataset.
Results
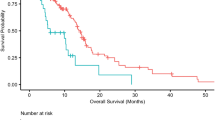
38.2% of GBM patients showed increased expression of anti-inflammatory cytokines, significant enrichment of T cell exhaustion signals, higher proportion of immunosuppressive cells (macrophages and CD4 regulatory T cells) and nine inhibitory checkpoints (CTLA4, PDCD1, LAG3, BTLA, TIGIT, HAVCR2, IDO1, SIGLEC7, and VISTA). The immunodepleted class (IDC) was used to classify these immunocompromised individuals. Despite the high density of tumor-infiltrating lymphocytes shown by IDC, such patients have a poor prognosis. Although PD-L1 was highly expressed in IDC, it suggested that there might be ICB resistance. There are many IDC predictive signatures to discover.
Conclusion
PD-1 is strongly expressed in a novel immunosuppressive class of GBM, but this cluster may be resistant to ICB therapy. A comprehensive description of this drug-resistant tumor microenvironment could provide new insights into drug resistance mechanisms and improved immunotherapy techniques.









Similar content being viewed by others
Data availability
Datasets analyzed during the current study are available in The Cancer Genome Atlas (TCGA) repository: https://portal.gdc.cancer.gov/, http://www.cgga.org.cn/.
Abbreviations
- ICB:
-
Immune checkpoint blockade
- GBM:
-
Glioblastoma
- IDC:
-
Immune depletion class
- CNS:
-
Central nervous system
- FDA:
-
Food and drug administration
- PD-1:
-
Programmed cell death protein 1
- PD-L1:
-
Programmed death-ligand 1
- GM-CSF:
-
Colony-stimulating factor
- UCC:
-
Unsupervised consistent clustering
- TCGA:
-
The cancer genome atlas
- CGGA:
-
Chinese glioma genome atlas
- ssGSEA:
-
Gene set enrichment analysis
- TILs:
-
Tumor-infiltrating lymphocytes
- GO:
-
Gene ontology
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- Tex:
-
T cell exhaustion
- OS:
-
Overall survival
References
Aran D, Hu Z, Butte AJ (2017) xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol 18:220
Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF et al (2009) Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462(7269):108-U22
Bota DA, Chung J, Dandekar M, Carrillo JA, Kong X-T, Fu BD et al (2018) Phase II study of ERC1671 plus bevacizumab versus bevacizumab plus placebo in recurrent glioblastoma: interim results and correlations with CD4+ T-lymphocyte counts. CNS Oncol. https://doi.org/10.2217/cns-2018-0009
Cameron F, Whiteside G, Perry C (2011) Ipilimumab: frst global approval. Drugs 71:867–104
Chen YP, Wang YQ, Lv JW, Li YQ, Chua MLK, Le QT et al (2019a) Identifcation and validation of novel microenvironment-based immune molecular subgroups of head and neck squamous cell carcinoma: implications for immunotherapy. Ann Oncol 30(1):68–75
Chen YP, Wang YQ, Lv JW, Li YQ, Chua MLK, Le QT et al (2019b) Identification and validation of novel microenvironment-based immune molecular subgroups of head and neck squamous cell carcinoma: implications for immunotherapy. Ann Oncol 30(1):68–75
Choi BD, Maus MV, June CH, Sampson JH (2019) Immunotherapy for Glioblastoma: adoptive T-cell strategies. Clin Cancer Res 25(7):2042–2048
Clancy T, Dannenfelser R, Troyanskaya O, Malmberg KJ, Hovig E, Kristensen V (2017) Bioinformatics approaches to profile the tumor microenvironment for immunotherapeutic discovery. Curr Pharm Des 23:4716–4725
Cloughesy TF, Mochizuki AY, Orpilla JR, Hugo W, Lee AH, Davidson TB et al (2019) Neoadjuvant anti-PD-1 immunotherapy promotes a survival beneft with intratumoral and systemic immune responses in recurrent glioblastoma. Nat Med 25:477–486
Cote DJ, Ostrom QT, Gittleman H, Duncan KR, Crevecoeur TS, Kruchko C et al (2019) Glioma incidence and survival variations by county-level socioeconomic measures. Cancer 125(19):3390–3400
DeMaria PJ, Bilusic M (2019) Cancer vaccines. Hematol Oncol Clin North Am 33:199–214
Du L, Lee JH, Jiang H, Wang C, Wang S, Zheng Z, Shao F, Xu D, Xia Y, Li J, Zheng Y, Qian X, Li X, Kim HR, Xing D, Liu P, Lu Z, Lyu J (2020) β-Catenin induces transcriptional expression of PD-L1 to promote glioblastoma immune evasion. J Exp Med 217(11):e20191115
Duan Z, Luo Y (2021) Targeting macrophages in cancer immunotherapy. Signal Transduct Target Ther 6(1):127
Ferris RL, Blumenschein G, Fayette J, Guigay J, Colevas AD, Licitra L et al (2016) Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med 375:1856–1867
Galon J, Angell HK, Bedognetti D, Marincola FM (2013) The continuum of cancer immunosurveillance: prognostic, predictive, and mechanistic signatures. Immunity 39:11–26
Hanzelmann S, Castelo R, Guinney J (2013) GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinformatics 14(1):7
Haslam A, Prasad V (2019) Estimation of the percentage of US patients with cancer who are eligible for and respond to checkpoint inhibitor immunotherapy drugs. JAMA Netw Open 2:e192535
Heynckes S, Daka K, Franco P, Gaebelein A, Frenking JH, Doria-Medina R et al (2019) Crosslink between temozolomide and PD-L1 immune-checkpoint inhibition in glioblastoma multiforme. BMC Cancer 19:117
Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB et al (2010) Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 363:711–723
Horvath L, Thienpont B, Zhao L, Wolf D, Pircher A (2020) Overcoming immunotherapy resistance in non-small cell lung cancer (NSCLC) - novel approaches and future outlook. Mol Cancer 19(1):141
Kahan SM, Wherry EJ, Zajac AJ (2015) T cell exhaustion during persistent viral infections. Virology 479–480:180–93
Kazandjian D, Suzman DL, Blumenthal G, Mushti S, He K, Libeg M et al (2016) FDA approval summary: nivolumab for the treatment of metastatic non-small cell lung cancer with progression on or after platinum-based chemotherapy. Oncologist 21:634–642
Kim H, Park H (2007) Sparse non-negative matrix factorizations via alternatingnon-negativity-constrained least squares for microarray data analysis. Bioinformatics 23(12):1495–1502
Le Rhun E, Preusser M, Roth P, Reardon DA, van den Bent M, Wen P, Reifenberger G, Weller M (2019) Molecular targeted therapy of glioblastoma. Cancer Treat Rev 80:101896
Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q et al (2020) TIMER2.0 for analysis of tumor-infltrating immune cells. NuclIDC Acids Res. 48(W1):W509–W14
Mahmoud AB, Ajina R, Aref S, Darwish M, Alsayb M, Taher M, AlSharif SA, Hashem AM, Alkayyal AA (2022) Advances in immunotherapy for glioblastoma multiforme. Front Immunol 12(13):944452
Mao X, Xu J, Wang W, Liang C, Hua J, Liu J, Zhang B, Meng Q, Yu X, Shi S (2021) Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: new findings and future perspectives. Mol Cancer 20(1):131
Marenco-Hillembrand L, Wijesekera O, Suarez-Meade P, Mampre D, Jackson C, Peterson J et al (2020) Trends in glioblastoma: outcomes over time and type of intervention: a systematic evidence based analysis. J Neurooncol 147(2):297–307
Mariathasan S, Turley S, Nickles D et al (2018) TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 554:544–548
Mathewson ND, Ashenberg O, Tirosh I, Gritsch S, Perez EM, Marx S, Jerby-Arnon L, Chanoch-Myers R, Hara T, Richman AR, Ito Y, Pyrdol J, Friedrich M, Schumann K, Poitras MJ, Gokhale PC, Gonzalez Castro LN, Shore ME, Hebert CM, Shaw B, Cahill HL, Drummond M, Zhang W, Olawoyin O, Wakimoto H, Rozenblatt-Rosen O, Brastianos PK, Liu XS, Jones PS, Cahill DP, Frosch MP, Louis DN, Freeman GJ, Ligon KL, Marson A, Chiocca EA, Reardon DA, Regev A, Suvà ML, Wucherpfennig KW (2021) Inhibitory CD161 receptor identified in glioma-infiltrating T cells by single-cell analysis. Cell 184(5):1281-1298.e26
McGranahan T, Therkelsen KE, Ahmad S, Nagpal S (2019) Current state of immunotherapy for treatment of Glioblastoma. Curr Treat Options Oncol 20(3):24
Monti S, Tamayo P, Mesirov J, Golub T (2003) Consensus clustering: a resampling-based method for class discovery and visualization of gene expression microarray data. Mach LEARN 52:91–118
Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y et al (2015a) Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 12:453–457
Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y et al (2015b) Robust enumeration of cell subsets from tissue expression profles. Nat Methods 12(5):453–7
Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, Louis DN, Rozenblatt-Rosen O, Suvà ML, Regev A, Bernstein BE (2014) Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344(6190):1396–1401
Saltz J, Gupta R, Hou L, Kurc T, Singh P, Nguyen V et al (2018) Spatial organization and molecular correlation of tumor-infltrating lymphocytes using deep learning on pathology images. Cell Rep 23(1):181–93
Sanders S, Debinski W (2020) Challenges to successful implementation of the immune checkpoint inhibitors for treatment of glioblastoma. Int J Mol Sci 21:2759
Sener U, Ruff MW, Campian JL (2022) Immunotherapy in Glioblastoma: current approaches and future perspectives. Int J Mol Sci 23(13):7046
Sia D, Jiao Y, Martinez-Quetglas I, Kuchuk O, Villacora-Martin C, Castro de Moura M et al (2017) Identification of an immune-specific class of hepatocellular carcinoma, based on molecular features. Gastroenterology 153(3):812–26
Sia D, Jiao Y, Martinez-Quetglas I, Kuchuk O, Villacorta-Martin C, Castro de Moura M et al (2017) Identifcation of an immune-specifc class of hepatocellular carcinoma based on molecular features. Gastroenterology 153(3):812–26
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profles. Proc Natl Acad Sci U S A 102(43):15545–50
Tay RE, Richardson EK, Toh HC (2021) Revisiting the role of CD4(+) T cells in cancer immunotherapy-new insights into old paradigms. Cancer Gene Ther 28(1–2):5–17
Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH et al (2018) The immune landscape of cancer. Immunity 48(4):812–30
Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O’Kelly M, Tamayo P, Weir BA, Gabriel S, Winckler W, Gupta S, Jakkula L, Feiler HS, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN (2010) Cancer genome atlas research network integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1,EGFR, and NF1. Cancer Cell 17(1):98–110. https://doi.org/10.1016/j.ccr.2009.12.020
Wang H, Xu T, Huang Q et al (2020) Immunotherapy for malignant glioma: current status and future directions. Trends Pharmacol Sci 41(2):123–138
Wesseling P, Capper D (2018) WHO 2016 classifcation of gliomas. Neuropathol Appl Neurobiol 44:139–150
Yang M, Lin C, Wang Y, Chen K, Zhang H, Li W (2022) Identification of a cytokine-dominated immunosuppressive class in squamous cell lung carcinoma with implications for immunotherapy resistance. Genome Med 14(1):72
Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, TorresGarcia W et al (2013) Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 4:2612
Yu MW, Quail DF (2021) Immunotherapy for Glioblastoma: current progress and challenges. Front Immunol 13(12):676301
Zhao Z, Zhang KN, Wang Q et al (2021) Chinese Glioma genome atlas (CGGA): a comprehensive resource with functional genomic data from chinese glioma patients. Genomics Proteomics Bioinformatics 19(1):1–12
Acknowledgements
The authors thank all the participants for their cooperation.
Funding
The Scientific Research Planning Project of the Education Department of Jilin Province (Grant Nos. JJKH20181256KJ).
Author information
Authors and Affiliations
Contributions
ZT, RD. designed the study, performed the major data analysis, and drafted the manuscript; ZT. collected a part of data, performed a part of data analysis, and help to generated figures. ZY. performed a part of data analysis. MJ. helped to generate figures. WZ. provided funding source, designed, oversaw, and supervised the project and edited, reviewed, and finalized the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher. The authors declare no conflicts of interest.
Ethical approval
The only data used in this study are the original data of public databases, so no ethical approval is involved.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tian, Z., Yang, Z., Jin, M. et al. Identification of cytokine-predominant immunosuppressive class and prognostic risk signatures in glioma. J Cancer Res Clin Oncol 149, 13185–13200 (2023). https://doi.org/10.1007/s00432-023-05173-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-023-05173-4