Abstract
Purpose
Aberrant DNA methylation could regulate the expression of tumor suppressor gene DLEC1 and oncogene PBX3 and was related to the occurrence and prognosis of gastric cancer (GC). In this study, the associations between DLEC1 and PBX3 promoter methylation in peripheral blood leukocytes (PBLs) and the risk and prognosis of GC were investigated.
Methods
The methylation status of DLEC1 and PBX3 promoter in PBLs of 368 GC cases and 382 controls was detected by the methylation-sensitive high-resolution melting (MS-HRM) method. Logistic and Cox regression were adopted to analyze the associations of DLEC1 and PBX3 methylation with GC risk and prognosis, respectively. Confounding biases were controlled by propensity score (PS).
Results
Compared with negative methylation (Nm), DLEC1-positive methylation (Pm) was associated with increased GC risk in PS (OR 2.083, 95% CI 1.220–3.558, P = 0.007), but PBX3 Pm was not associated with GC risk. In the elderly group (≥ 60 years), DLEC1 Pm was associated with increased GC risk (OR 2.951, 95% CI 1.426–6.104, P = 0.004). The combined effects between DLEC1 methylation and consumption of dairy products, fried food intake and Helicobacter pylori (H. pylori) infection on GC risk were discovered (ORc 3.461, 95% CI 1.847–6.486, P < 0.001, ORc 3.246, 95% CI 1.708–6.170, P < 0.001 and ORc 2.964, 95% CI 1.690–5.197, P < 0.001, respectively). Furthermore, DLEC1 and PBX3 methylation were not associated with GC prognosis.
Conclusion
DLEC1 methylation in PBLs and the combined effects of gene–environment can influence GC risk.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gastric cancer (GC) is a common malignant tumor and is responsible for 1,034,000 new cases and 783,000 deaths in 2018, making it the fifth most commonly diagnosed cancer and the third major reason of cancer death all over the world (Bray et al. 2018). China is a country with high incidence of GC, with estimates of 679,000 new cases and 498,000 deaths in 2015 according to the China Cancer Data Report (Chen et al. 2016).
Genetic and epigenetic alterations are considered to be the two main factors involved in gastric carcinogenesis. There is plenty of evidence to suggest that gene polymorphisms and mutations are associated with the occurrence of GC (He et al. 2012, 2018). Beside the genetic change, research has shown that epigenetic dysregulation plays a crucial role in GC development (Nakamura et al. 2014). Epigenetics is defined as heritable changes in gene expression that are not due to alterations in gene sequence (Guo and Yan 2015). As one of the most widely studied epigenetic modifications, DNA methylation is closely related to the occurrence and prognosis of GC. Abnormal DNA methylation often occurs at the promoter regions of genes, particularly tumor-suppressor genes, which are related to tumor cell cycle, apoptosis, proliferation, differentiation, and invasion (Li and Chen 2013). To date, more and more researches have revealed that dietary factors and lifestyle could contribute to cancer development by inducing both epigenetic and genetic changes (Herceg 2007).
Deleted in lung and esophageal cancer 1 (DLEC1), located at the commonly deleted locus 3p22.3, has been demonstrated to act as a tumor suppressor gene in multiple cancers (Kwong et al. 2006; Pastuszak-Lewandoska et al. 2016; Zhang et al. 2015) which can suppress tumor growth or reduce the invasiveness of cancer cells (Ye et al. 2014). It was reported that the promoter methylation of DLEC1 could result in the downregulation or silence of its own expression in most gastric cell lines (Ying et al. 2009) and was associated with GC risk and prognosis in tissue samples (Ye et al. 2014; Zhang et al. 2010).
Pre-leukemia transcription factor 3 (PBX3) is a member of the PBX family that belongs to the three amino acid loop extension family with a highly conserved homologous domain (Han et al. 2014). As an oncogene, PBX3 is over-expressed in GC and its overexpression can accelerate cell proliferation and colony formation. In addition, PBX3 is closely associated with invasion depth, clinical stage, and differentiation of GC (Li et al. 2014; Wang et al. 2016a). A recent research reported that PBX3 hypermethylation in PBLs was associated with prognosis in colorectal cancer (Sun et al. 2019). However, the relationship between PBX3 methylation and GC is unclear.
In recent years, increasing studies focused on the relationship between DNA methylation in tissues and the incidence and prognosis of tumors. Compared with the acquisition of tissues, blood sampling is convenient and minimally invasive, making it adaptive for the population-based study (Tahara and Arisawa 2015). Furthermore, DNA from peripheral blood can dynamically monitor tumors in real-time, so it can provide more information for the early detection and prognosis of tumors than tissues (Hu et al. 2019). Therefore, we conducted this case–control study to detect the methylation levels of DLEC1 and PBX3 in PBLs, explore the relationship between environmental factors and gene methylation and investigate the relationship between environmental factors, gene methylation and their interactions with GC risk. We also conducted a follow-up study of GC patients to assess the association of gene methylation in PBLs with GC prognosis.
Materials and methods
Study samples
368 GC cases and 382 controls were enrolled in a hospital-based case–control study. Patients of GC diagnosed by pathological examination of the Third Affiliated Hospital of Harbin Medical University in 2010 and 2012 were selected as cases. The vast majority of these patients (> 98%) had undergone surgery. Patients from the Department of Orthopaedics and Ophthalmology of the Second Affiliated Hospital and the Department of Neurology of the Fourth Affiliated Hospital of Harbin Medical University, and healthy people who participated in physical examination at the Harbin Xiangfang Center for Disease Control and Prevention between 2010 and 2013 were selected as controls. The control individuals with the history of malignant tumors such as gastric cancer and gastrointestinal diseases were excluded. After obtaining the patient's informed consent, face-to-face investigations were conducted in both cases and controls, and 5 ml blood samples were collected from each subject. The overall response rate for cases and controls were approximately 90%. All cases were included in the follow-up study. Finally, a total of 347 GC patients were included in the analysis, 21 cases were lost to follow-up due to death or withdrawal. Demographic, clinical, and treatment information of each patient was extracted from the electronic medical record system.
H. pylori serologic tests by ELISA
The enzyme immunoassay kit (IBL, German) was used to detect the infection status of H. pylori. The criterion is that lower than 8 units/ml was represented negative, from 8 to 12 units/ml was represented suspicious, upper to 12 units/ml was represented positive.
Methylation assay
The DNA was extracted using QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany), and then modified by bisulfite using EpiTect Fast DNA Bisulfite Kit (Qiagen). The procedure was referred to the kit instructions. Nanodrop 2000c (Thermo Scientific) was adopted to measure the DNA quantity. The bisulfite-modified DNA was stored at – 80 ℃. Primers of DLEC1 and PBX3 gene were designed using Primer Premier 5.0 software. PCR amplification and MS-HRM assay were implemented on the LightCycler480 (Roche Applied Science, Mannheim, Germany) equipped with Gene Scanning software (version 2.0) to identify and analyze the methylation status of genes. The 5 µl reaction system contained 2.5 µl LightCycler480 High-Resolution Melting Master Mix (Roche), 0.5 µl sodium bisulfite-modified template DNA, 0.1 µl forward primer, 0.1 µl reverse primer, 0.6 µl MgCl2 and 1.2 µl PCR-grade water. The primer sequences and reaction conditions are listed in Table S1.
Methylated DNA standards with different levels, including 100%, 5%, 2%, 1%, 0.5%, and 0% methylated DNA, were constructed by mixing 100% methylated and 0% methylated human whole genomic DNA (Zymo Research). Normalized melting curves and melting peaks of the MS-HRM assay for two genes are shown in Figs. 1 and 2. The methylation levels of genes in samples were confirmed by contrasting with the standard curves. In Figs. S1 and S2, the distribution of DNA methylation levels of samples was showed. Based on the area under the curve (AUC), 0% and 2% methylated DNA acted as the cut-off values to divide Nm and Pm of DLEC1 and PBX3, respectively (Fig. S3). Replicate measurements of some samples were performed at different times for DLEC1 and PBX3. The consistency rates of DLEC1 and PBX3 were 96.0% and 95.2%, respectively. Rank sum test of paired samples was used for consistency analysis, and the results showed no difference (P > 0.05) (Tables S2). PCR-grade water was employed as negative (no-template) control in each batch, and second experiments were performed for the equivocal results.
Statistical analysis
The Chi-square (χ2) test was used for categorical variables and t test was used for continuous variables. Multiple imputation method was adopted for variables with less than 30%. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated by logistic regression and adjusted by propensity score (PS). Univariate and multivariate logistic regression analyses were adopted to evaluate the relationships between gene methylation, environmental factors and GC risk, as well as the relationships between gene methylation and environmental factors. The interactions of gene methylation and environmental factors on GC risk were estimated on a multiplicative scale with a product-term coefficient using multivariate logistic regression. The combined effects of gene methylation and environmental factors on GC risk were calculated by crossover analysis. Kaplan–Meier analysis was adopted to acquire the survival curve of GC patients. Univariate and multivariate Cox regression analyses were adopted to calculate hazard ratios (HRs) and 95% CIs for the relationship between gene methylation and clinical characteristics and prognosis of GC patients, and these results were adjusted by PS. All statistical analyses were performed using SPSS software version 23.0. PS was executed using R-3.1.3 for Windows with PS matching 3.04 software packages. P values < 0.05 were considered statistically significant.
Results
Demographic characteristics of all subjects
368 cases and 382 controls were carried out in our study. The PS value calculated by all 28 covariates was an adjustment factor. The demographic characteristics of all subjects are listed in Table 1.
The results showed no difference between cases and controls in the aspect of sex and age (P = 0.878 and P = 0.301, respectively). The distribution of body mass index (BMI) and monthly income between GC cases and controls were statistically different (P < 0.05). Compared with controls (3.1%), the percentage of cases with GC family history (13.9%) was higher (P < 0.001).
Associations between environmental factors and GC risk
Multivariate logistic regression analysis was adopted to assess associations between environmental factors and GC risk, and the results are shown in Table S3. Then backward conditional selection method was adopted for multivariate analysis. Finally, thirteen environmental factors were integrated into the regression model. As shown in Table S4, alcohol consumption, H. pylori infection, salty food intake, food left overnight intake, dairy products intake, eat fried food and freshwater fish intake significantly increased GC risk (adjusted by PS, P < 0.05). On the contrary, regular diet, drinking tap water and mineral water, refrigerated food, beef and mutton intake, garlic intake and green vegetable intake significantly decreased GC risk (P < 0.05).
Associations between the methylation of DLEC1 and PBX3 and GC risk
As shown in Table 2, DLEC1 Pm was associated with increased GC risk compared with Nm (OR 2.083, 95% CI 1.220–3.558, P = 0.007). However, PBX3 methylation was not associated with GC risk.
Associations between the methylation of DLEC1 and PBX3 and GC risk by stratified analysis
In the elderly group (≥ 60 years), the results displayed that DLEC1 Pm was associated with increased GC risk (OR 2.951, 95% CI 1.426–6.104, P = 0.004), but no correlation was found in the younger group (< 60 years). When the individuals were classified by H. pylori infection, DLEC1 Pm was marginally associated with increased GC risk in H. pylori-positive and -negative individuals (OR 1.875, 95% CI 0.944–3.727, P = 0.073 and OR 2.179, 95% CI 0.935–5.080, P = 0.071, respectively). But PBX3 Pm was still not associated with GC risk by stratified analysis (Tables 3, 4).
Associations between the methylation of DLEC1 and PBX3 and environmental factors
As shown in Table S5, consumption of garlic was associated with decreased risk of DLEC1 methylation (OR 0.615, 95% CI 0.397–0.953, P = 0.030), and refrigerated food was marginally associated with DLEC1 methylation (OR 0.664, 95% CI 0.441–1.000, P = 0.050). Alcohol consumption was associated with decreased risk of PBX3 methylation (OR 0.728, 95% CI 0.539–0.984, P = 0.039) (Table S6).
The interactions between the methylation of DLEC1 and PBX3 and their interactions with environmental factors on GC risk
The results showed that the combined effects between DLEC1 methylation and H. pylori infection, consumption of dairy products (≥ 1 times/week) and fried food intake (≥ 1 times/month) on GC risk existed (ORc 2.964, 95% CI 1.690–5.197, P < 0.001, ORc 3.461, 95% CI 1.847–6.486, P < 0.001 and ORc 3.246, 95% CI 1.708–6.170, P < 0.001, respectively), whereas no interactions between DLEC1 methylation and environmental factors on the GC risk were found (Table S7). As for PBX3, its methylation and some environmental factors have both interactions and combined effects on GC risk (P < 0.05) (Table S8). In addition, the results in Table S9 showed that DLEC1 methylation did not interact with PBX3 methylation on GC risk.
Demographic characteristics of GC patients
A total of 347 GC patients were included in this 5-year follow-up study. The association between demographic characteristics and prognosis of GC patients was analyzed, as shown in Table S10.
Although the relationship between each demographic characteristic and GC prognosis was not statistically significant, age, gender and BMI were still used as the adjustment factors in analyzing the relationship between clinical characteristics and GC prognosis since they were common confounding factors. The results of multivariate Cox analysis revealed that tumor–node–metastasis (TNM) stage, differentiation, tumor size, carbohydrate antigen 19–9 (CA 19–9) level and carcinoembryonic antigen (CEA) level were significantly associated with GC prognosis (all P values < 0.05) (Table 5).
Backward conditional selection results showed that GC patients with TNM stage III had marginally poorer prognosis (HR 1.887, 95% CI 0.959–3.715, P = 0.066). GC patients with stage IV had obviously poorer prognosis (HR 4.178, 95% CI 2.296–7.603, P < 0.001). In addition, tumor size was also associated with poorer GC prognosis (HR 1.689, 95% CI 1.229–2.320, P = 0.001) (Table S11).
Associations between methylation of DLEC1 and PBX3 and GC prognosis
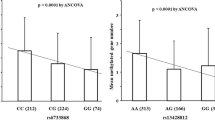
As shown in Table 6, compared with Nm, PBX3 Pm was marginally associated with GC prognosis only by multivariate adjustment (HR 1.349, 95% CI 0.981–1.856, P = 0.065). DLEC1 Pm had no association with GC prognosis by both multivariate and PS adjustment. The Kaplan–Meier survival curves for the relationships between methylation of DLEC1 and PBX3 and GC prognosis are shown in Fig. 3.
Associations between methylation of DLEC1 and PBX3 and GC prognosis by stratified analysis
Stratified analyses were conducted in prognostic analysis by age, gender, H. pylori infection, TNM stage, and tumor size. The results indicated PBX3 Pm was associated with poorer GC prognosis only in the elderly group (HR 1.678, 95% CI 1.046–2.693, P = 0.032) and female group (HR 2.058, 95% CI 1.024–4.137, P = 0.043), but DLEC1 Pm was not associated with GC prognosis in each age and gender subgroup (Tables S12 and S13). In addition, stratified analysis by H. pylori infection indicated that DLEC1 Pm was associated with poorer GC prognosis in H. pylori-negative individuals (HR 2.040, 95% CI 1.104–3.769, P = 0.023), but PBX3 Pm was not associated with GC prognosis in both H. pylori-positive and -negative individuals (Table S14). No significant relationships of DLEC1 Pm and PBX3 Pm with GC prognosis were found in each TNM stage and tumor size subgroup (Tables S15 and S16).
Discussion
Gastric cancer is a common malignant tumor of the gastrointestinal tract. In recent decades, more and more evidence indicates that epigenetics plays an important role during cancer progression, including GC (Heyn and Esteller 2012). Abnormal DNA methylation, a common event of epigenetics, could regulate the expression of tumor suppressor genes and oncogenes (Puneet et al. 2018; Qu et al. 2013). It is reported that tumors do not develop as an isolated phenomenon in their target tissue; other organ systems including the immune system (such as PBLs) also participate in tumor initiation and prognosis (Marsit and Christensen 2013). Moreover, since peripheral blood is relatively easy to obtain, PBLs are the most commonly used alternative to studying the risk of epigenome induction and the epigenetic response to disease-associated stress (Hohos et al. 2016). Therefore, this study was performed to evaluate the effect of the promoter methylation of genes derived from PBLs on the risk and prognosis of GC.
Compared with Nm, DLEC1 Pm increased GC risk (OR 2.083). Wang et al. found that the hypermethylation levels of DLEC1 were significantly associated with GC risk by quantitative methylation-specific PCR in the serum of 82 GC patients, 46 chronic atrophic gastritis subjects, and 40 healthy controls (Wang et al. 2015). Our finding verified above result in larger populations by MS-HRM.
It is reported that DNA methylation changes during aging are closely correlated to the occurrence of cancer (Wang et al. 2016b). The result of age-stratified analysis declared that DLEC1 Pm individuals had a higher GC risk than Nm individuals in the elderly group. Fuke et al. measured 5-methyldeoxycytidine ((met)C) content by HPLC in PBLs obtained from 76 healthy individuals and found that the age-dependent decrease of (met)C was statistically highly significant in the aged group compared with the young group (Fuke et al. 2004), which indicated that methylation differences were remarkable in older individuals. Previous studies had revealed that H. pylori infection could cause an intensive inflammatory response in the gastric mucosa, leading to upregulation of certain inflammatory cytokines such as IL-1β which in turn caused abnormal levels of DNA methylation (Lamb and Chen 2013). The result of H. pylori infection-stratified analysis declared that DLEC1 Pm was marginally associated with GC risk in both H. pylori-positive and -negative individuals. Considering DLEC1 Pm was significantly associated with GC in unstratified analyses, so more researches were needed to determine marginal associations in H. pylori infection-stratified analysis.
In this study, a combined effect between methylation status of DLEC1 and H. pylori infection on GC risk was found. Helicobacter pylori infection could contribute to GC due to its role in increasing chronic inflammation and cell proliferation (Yousefi et al. 2019). Cell proliferation has been recognized as a contributing factor for de novo DNA methylation (Issa et al. 2001; Velicescu et al. 2002). On the other hand, expression of many genes is inhibited in the processes of inflammatory and low expression of these genes can promote de novo methylation (De Smet et al. 2004; Song et al. 2002; Ushijima and Okochi-Takada 2005). Research showed that H. pylori infection enhances abnormal DNA methylation in the gastric mucosa, which further promotes GC by inducing abnormal methylation of gene promoters (Xie et al. 2017). After eradication of H. pylori, the methylation levels of genes were reduced, and H. pylori-mediated gastric tumorigenesis could be postponed or even reversed (Perri et al. 2007; Zhou et al. 2019). In this study, no interaction was found between H. pylori infection and DLEC1 methylation.
Our results also observed that there was a combined effect between DLEC1 methylation and consumption of dairy products on GC risk. Dairy products contain high amounts of methionine (Finkelstein 1990), which serves as the precursor of S-adenosylmethionine, the universal methyl donor for DNA methylation in the hepatic one-carbon metabolism (Zhang 2018). Moreover, we found that DLEC1 methylation also showed the combined effect with fried food intake on GC risk. During the frying process, protein-rich foods produce heterocyclic amine carcinogens, and starchy foods produce acrylamide carcinogen, which causes human tumors to occur. Acrylamide is metabolized to glycidyl amide in the human body, and it can directly bind to hydrazine, altering DNA structure and causing DNA methylation (de Conti et al. 2019; Guo et al. 2018).
It is well known that dietary and lifestyle are convertible factors and profoundly influence the occurrence and development of GC. Among these influence factors, regular diet, refrigerated food, consumption of vegetables, garlic and beef and mutton and drinking tap and mineral water were recognized as protective factors for GC; however, alcohol consumption, salty food intake and consumption of food left overnight, dairy products and fried food were identified as risk factors for GC (den Hoed and Kuipers 2016; Ghaffari et al. 2019; Rastaghi et al. 2019; Zhao et al. 2011), and our results were consistent with these findings. In this study, we also found that consumption of freshwater fish was recognized as risk factor for GC, which was somewhat unexpected. Freshwater fish may increase tumor susceptibility. The reason may be that large amounts of heavy metals and toxic chemicals accumulate in freshwater fish (Hou et al. 1988). In addition, due to the habit of high-temperature cooking in China, cooking freshwater fish could produce high levels of carcinogenic compounds, which may be a major reason of the increased GC risk (Felton et al. 1997; Sugimura and Terada 1998).
The relationship between methylation status of DLEC1 and PBX3 and GC prognosis was also explored in this study. The results found that TNM stage and tumor size were factors influencing the GC prognosis. The previous research published by our department had been reported that PBX3 hypermethylation in PBLs was associated with better prognosis in colorectal cancer (Sun et al. 2019). This was the first time to explore the association between PBX3 methylation and GC prognosis. Compared with Nm, PBX3 Pm was marginally associated with worse prognosis of GC only in multivariate Cox regression analysis. To be conservative, the result of this study preferred to prompt that PBX3 Pm was not associated with GC prognosis. However, PBX3 Pm subjects had a worse GC prognosis than Nm subjects in elder and female groups.
There were three limitations to this study. First, recall bias might remain unavoidable at the time of gathering information on environmental factor although we attempted to reduce this bias. Second, the causality between gene methylation and GC risk remains unclear, and prospective cohort studies are needed to determine in the future. Third, the intake of dietary was not explicitly quantified, which might affect the results of the gene–environment interaction analysis.
Conclusions
In conclusion, this study indicated that DLEC1 methylation and the combined effects between environmental factors and its methylation in PBLs were associated with the GC risk. As a new biomarker, DLEC1 methylation can predict the risk of GC.
References
Bray F, Ferlay J, Soerjomataram I et al (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68(6):394–424. https://doi.org/10.3322/caac.21492
Chen W, Zheng R, Baade PD et al (2016) Cancer statistics in China, 2015. CA Cancer J Clin 66(2):115–132. https://doi.org/10.3322/caac.21338
de Conti A, Tryndyak V, VonTungeln LS et al (2019) Genotoxic and epigenotoxic alterations in the lung and liver of mice induced by acrylamide: a 28 days drinking water study. Chem Res Toxicol 32(5):869–877. https://doi.org/10.1021/acs.chemrestox.9b00020
De Smet C, Loriot A, Boon T (2004) Promoter-dependent mechanism leading to selective hypomethylation within the 5′ region of gene MAGE-A1 in tumor cells. Mol Cell Biol 24(11):4781–4790. https://doi.org/10.1128/MCB.24.11.4781-4790.2004
den Hoed CM, Kuipers EJ (2016) Gastric cancer: how can we reduce the incidence of this disease? Curr Gastroenterol Rep 18(7):34. https://doi.org/10.1007/s11894-016-0506-0
Felton JS, Malfatti MA, Knize MG et al (1997) Health risks of heterocyclic amines. Mutat Res 376(1–2):37–41. https://doi.org/10.1016/s0027-5107(97)00023-7
Finkelstein JD (1990) Methionine metabolism in mammals. J Nutr Biochem 1(5):228–237
Fuke C, Shimabukuro M, Petronis A et al (2004) Age related changes in 5-methylcytosine content in human peripheral leukocytes and placentas: an HPLC-based study. Ann Hum Genet 68(Pt 3):196–204. https://doi.org/10.1046/j.1529-8817.2004.00081.x
Ghaffari HR, Yunesian M, Nabizadeh R et al (2019) Environmental etiology of gastric cancer in Iran: a systematic review focusing on drinking water, soil, food, radiation, and geographical conditions. Environ Sci Pollut Res Int 26(11):10487–10495. https://doi.org/10.1007/s11356-019-04493-8
Guo LW, Liu SZ, Zhang M et al (2018) Multivariate analysis of the association between consumption of fried food and gastric cancer and precancerous lesions. Zhonghua Yu Fang Yi Xue Za Zhi 52(2):170–174. https://doi.org/10.3760/cma.j.issn.0253-9624.2018.02.010
Guo M, Yan W (2015) Epigenetics of gastric cancer. Methods Mol Biol 1238:783–799. https://doi.org/10.1007/978-1-4939-1804-1_41
Han HB, Gu J, Ji DB et al (2014) PBX3 promotes migration and invasion of colorectal cancer cells via activation of MAPK/ERK signaling pathway. World J Gastroenterol 20(48):18260–18270. https://doi.org/10.3748/wjg.v20.i48.18260
He J, Qiu LX, Wang MY et al (2012) Polymorphisms in the XPG gene and risk of gastric cancer in Chinese populations. Hum Genet 131(7):1235–1244. https://doi.org/10.1007/s00439-012-1152-8
He J, Zhuo ZJ, Zhang A et al (2018) Genetic variants in the nucleotide excision repair pathway genes and gastric cancer susceptibility in a southern Chinese population. Cancer Manag Res 10:765–774. https://doi.org/10.2147/CMAR.S160080
Herceg Z (2007) Epigenetics and cancer: towards an evaluation of the impact of environmental and dietary factors. Mutagenesis 22(2):91–103. https://doi.org/10.1093/mutage/gel068
Heyn H, Esteller M (2012) DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet 13(10):679–692. https://doi.org/10.1038/nrg3270
Hohos NM, Lee K, Ji L et al (2016) DNA cytosine hydroxymethylation levels are distinct among non-overlapping classes of peripheral blood leukocytes. J Immunol Methods 436:1–15. https://doi.org/10.1016/j.jim.2016.05.003
Hou H, She Y, Ma Y et al (1988) Investigations on methyl mercury contamination of fishes in the Second Songhua River. Biomed Environ Sci 1(1):79–82
Hu Y, Ma P, Feng Y et al (2019) Predictive value of the serum RASSF10 promoter methylation status in gastric cancer. J Int Med Res 47(7):2890–2900. https://doi.org/10.1177/0300060519848924
Issa JP, Ahuja N, Toyota M et al (2001) Accelerated age-related CpG island methylation in ulcerative colitis. Cancer Res 61(9):3573–3577
Kwong J, Lee JY, Wong KK et al (2006) Candidate tumor-suppressor gene DLEC1 is frequently downregulated by promoter hypermethylation and histone hypoacetylation in human epithelial ovarian cancer. Neoplasia 8(4):268–278. https://doi.org/10.1593/neo.05502
Lamb A, Chen LF (2013) Role of the Helicobacter pylori-induced inflammatory response in the development of gastric cancer. J Cell Biochem 114(3):491–497. https://doi.org/10.1002/jcb.24389
Li W, Chen BF (2013) Aberrant DNA methylation in human cancers. J Huazhong Univ Sci Technol Med Sci 33(6):798–804. https://doi.org/10.1007/s11596-013-1201-0
Li Y, Sun Z, Zhu Z et al (2014) PBX3 is overexpressed in gastric cancer and regulates cell proliferation. Tumour Biol 35(5):4363–4368. https://doi.org/10.1007/s13277-013-1573-6
Marsit C, Christensen B (2013) Blood-derived DNA methylation markers of cancer risk. Adv Exp Med Biol 754:233–252. https://doi.org/10.1007/978-1-4419-9967-2_12
Nakamura J, Tanaka T, Kitajima Y et al (2014) Methylation-mediated gene silencing as biomarkers of gastric cancer: a review. World J Gastroenterol 20(34):11991–12006. https://doi.org/10.3748/wjg.v20.i34.11991
Pastuszak-Lewandoska D, Kordiak J, Antczak A et al (2016) Expression level and methylation status of three tumor suppressor genes, DLEC1, ITGA9 and MLH1, in non-small cell lung cancer. Med Oncol 33(7):75. https://doi.org/10.1007/s12032-016-0791-3
Perri F, Cotugno R, Piepoli A et al (2007) Aberrant DNA methylation in non-neoplastic gastric mucosa of H. pylori infected patients and effect of eradication. Am J Gastroenterol 102(7):1361–1371. https://doi.org/10.1111/j.1572-0241.2007.01284.x
Puneet KHR, Kumari S et al (2018) Epigenetic mechanisms and events in gastric cancer-emerging novel biomarkers. Pathol Oncol Res 24(4):757–770. https://doi.org/10.1007/s12253-018-0410-z
Qu Y, Dang S, Hou P (2013) Gene methylation in gastric cancer. Clin Chim Acta 424:53–65. https://doi.org/10.1016/j.cca.2013.05.002
Rastaghi S, Jafari-Koshki T, Mahaki B et al (2019) Trends and risk factors of gastric cancer in Iran (2005–2010). Int J Prev Med 10:79. https://doi.org/10.4103/ijpvm.IJPVM_188_17
Song JZ, Stirzaker C, Harrison J et al (2002) Hypermethylation trigger of the glutathione-S-transferase gene (GSTP1) in prostate cancer cells. Oncogene 21(7):1048–1061. https://doi.org/10.1038/sj.onc.1205153
Sugimura T, Terada M (1998) Experimental chemical carcinogenesis in the stomach and colon. Jpn J Clin Oncol 28(3):163–167. https://doi.org/10.1093/jjco/28.3.163
Sun H, Huang H, Li D et al (2019) PBX3 hypermethylation in peripheral blood leukocytes predicts better prognosis in colorectal cancer: a propensity score analysis. Cancer Med 8(8):4001–4011. https://doi.org/10.1002/cam4.2321
Tahara T, Arisawa T (2015) DNA methylation as a molecular biomarker in gastric cancer. Epigenomics 7(3):475–486. https://doi.org/10.2217/epi.15.4
Ushijima T, Okochi-Takada E (2005) Aberrant methylations in cancer cells: where do they come from? Cancer Sci 96(4):206–211. https://doi.org/10.1111/j.1349-7006.2005.00035.x
Velicescu M, Weisenberger DJ, Gonzales FA et al (2002) Cell division is required for de novo methylation of CpG islands in bladder cancer cells. Cancer Res 62(8):2378–2384
Wang G, Zhang W, Zhou B et al (2015) The diagnosis value of promoter methylation of UCHL1 in the serum for progression of gastric cancer. Biomed Res Int 2015:741030. https://doi.org/10.1155/2015/741030
Wang S, Li C, Wang W, Xing C (2016a) PBX3 promotes gastric cancer invasion and metastasis by inducing epithelial–mesenchymal transition. Oncol Lett 12(5):3485–3491. https://doi.org/10.3892/ol.2016.5305
Wang Y, Zhang J, Xiao X et al (2016b) The identification of age-associated cancer markers by an integrative analysis of dynamic DNA methylation changes. Sci Rep 6:22722. https://doi.org/10.1038/srep22722
Xie Y, Zhou JJ, Zhao Y et al (2017) H. pylori modifies methylation of global genomic DNA and the gastrin gene promoter in gastric mucosal cells and gastric cancer cells. Microb Pathog 108:129–136. https://doi.org/10.1016/j.micpath.2017.05.003
Ye X, Feng G, Jiao N et al (2014) Methylation of DLEC1 promoter is a predictor for recurrence in Chinese patients with gastric cancer. Dis Markers 2014:804023. https://doi.org/10.1155/2014/804023
Ying J, Poon FF, Yu J et al (2009) DLEC1 is a functional 3p22.3 tumour suppressor silenced by promoter CpG methylation in colon and gastric cancers. Br J Cancer 100(4):663–669. https://doi.org/10.1038/sj.bjc.6604888
Yousefi B, Mohammadlou M, Abdollahi M et al (2019) Epigenetic changes in gastric cancer induction by Helicobacter pylori. J Cell Physiol 234(12):21770–21784. https://doi.org/10.1002/jcp.28925
Zhang L, Zhang Q, Li L et al (2015) DLEC1, a 3p tumor suppressor, represses NF-kappaB signaling and is methylated in prostate cancer. J Mol Med 93(6):691–701. https://doi.org/10.1007/s00109-015-1255-5
Zhang N (2018) Role of methionine on epigenetic modification of DNA methylation and gene expression in animals. Anim Nutr 4(1):11–16. https://doi.org/10.1016/j.aninu.2017.08.009
Zhang Y, Ye X, Geng J, Chen L (2010) Epigenetic inactivation of deleted in lung and esophageal cancer 1 gene by promoter methylation in gastric and colorectal adenocarcinoma. Hepatogastroenterology 57(104):1614–1619
Zhao DL, Chen WQ, Yu TT et al (2011) A population-based matched case–control study on the risk factors of gastric cardia cancer. Zhonghua Zhong Liu Za Zhi 33(10):775–778
Zhou H, Sun H, Liu X et al (2019) Combined effect between WT1 methylation and Helicobacter pylori infection, smoking, and alcohol consumption on the risk of gastric cancer. Helicobacter. https://doi.org/10.1111/hel.12650
Funding
This research was supported by grant from National Natural Science Foundation of China (2016–2019, Grant no. 81573219).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The study was approved by the Human Research and Ethics Committee of Harbin Medical University.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xie, W., Zhou, H., Han, Q. et al. Relationship between DLEC1 and PBX3 promoter methylation and the risk and prognosis of gastric cancer in peripheral blood leukocytes. J Cancer Res Clin Oncol 146, 1115–1124 (2020). https://doi.org/10.1007/s00432-020-03171-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-020-03171-4