Abstract
Purpose
Cancer stem cells (CSC) and epithelial–mesenchymal transition (EMT) pathways are crucial for cancer progression. However, synergistic interactions between CSC and EMT are not clear in non-small cell lung cancer (NSCLC). The objective of this study was to investigate CSC markers such as CD44, NANOG, and ALDH1 expression and its correlation with EMT markers in NSCLC patients. Its association with survival was also determined.
Methods
CD44, NANOG, and ALDH1 protein expression was evaluated in 267 resected NSCLC and its correlation with e-cadherin, β-catenin, p120 catenin, vimentin, SNAIL, and TWIST expressions was determined based on immunohistochemical and mRNA expression data from The Cancer Genome Atlas (TCGA) database. Survival analyses also were performed based on immunohistochemistry and mRNA expression data from Gene Expression Omnibus dataset.
Results
ALDH1 expression in lung adenocarcinoma was positively correlated with the epithelial-like phenotype, low vimentin and low TWIST in immunohistochemical and mRNA expression data. NANOG and ALDH1 expressions measured by immunohistochemical and mRNA expression profiling data of adenocarcinomas were associated with a favorable prognosis. ALDH1 was an independent favorable prognostic marker for overall survival or recurrence-free survival in adenocarcinoma (P = 0.026 and P = 0.033, respectively). The epithelial-like phenotype expressing P120-catenin and beta-catenin was associated with a favorable prognosis; however, the TWIST-expressing mesenchymal-like phenotype was correlated with an unfavorable prognosis.
Conclusions
NANOG and ALDH1 protein or mRNA expression showed improved prognosis in adenocarcinoma alone. ALDH1 expression correlated with an epithelial-like phenotype.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lung cancer, especially non-small cell lung cancer (NSCLC), is the most common cancer in the world (Bray et al. 2018). Despite recent advances in surgical techniques, chemotherapy, and radiotherapy over the past decades, the prognosis of NSCLC remains poor (Ettinger et al. 2017). Even when surgical resections were performed in the early stage of NSCLC, the 5-year survival rate varied from 73% in stage IA to 9% in stage IIIB (Goldstraw et al. 2007). In addition, despite adjuvant chemotherapy, the risk of recurrence is still relatively high (Tanaka and Yoneda 2016). To reduce lung cancer mortality, it is important to identify biomarkers that can effectively predict the progression of resected NSCLC patients.
Cancer stem cells (CSCs) are a subpopulation of tumor cells that possess stem or progenitor cell-like characteristics. These cells exhibit the dual properties of self-renewal and pluripotency and generate a heterogeneous population of tumor cells (Clarke and Fuller 2006). CSCs correlate with the properties of tumor initiation, migration, recurrence, and drug resistance (Simple et al. 2015). Several CSC markers, including CD133, CD44, aldehyde dehydrogenase 1 (ALDH1), and NANOG have been examined in patients with NSCLC (Bertolini et al. 2009; Dimou et al. 2012; Leung et al. 2010; Park et al. 2016). Those studies have yielded mixed results. Although two earlier reports found a negative prognostic effect of NANOG (Park et al. 2016) and CD133 (Wang et al. 2018), other studies suggested a beneficial prognostic effect of ALDH1 (Dimou et al. 2012) and CD44 (Leung et al. 2010), in patients with NSCLC.
Epithelial–mesenchymal transition (EMT) is a mechanism by which epithelial cells lose their cell polarity and cell–cell junction, and acquire migratory and invasive properties to become mesenchymal cells (Kalluri and Weinberg 2009). Previous studies showed that the EMT state was associated with CSC properties, including expression of CD44 in breast cancer (Mani et al. 2008), CD133 in hepatocellular cancer (Zhang et al. 2017), NANOG in ovarian cancer (Qin et al. 2017), and ALDH1 in prostate cancer (Nastaly et al. 2018). In lung adenocarcinoma cell line research, ALDH1 expression was correlated with an epithelial-like phenotype marker (Tiran et al. 2017). In lung adenocarcinoma, the expression of NANOG was positively associated with the expression of mesenchymal marker SNAIL (Park et al. 2016). The relationship between EMT and CSC has not been thoroughly studied in NSCLC, and little is known about the underlying mechanisms of their processes.
Therefore, the objective of this study was to investigate expression of CSC markers, including CD44, NANOG, and ALDH1 in NSCLC patients, using immunohistochemistry. We also analyzed the correlation between expression of CSC markers and expression of EMT markers, including e-cadherin, β-catenin, p120 catenin, vimentin, SNAIL, and TWIST, and conducted survival analysis of patients with NSCLC in immunohistochemical and web-based mRNA expression data (Gyorffy et al. 2013; Network 2012; Network. 2014).
Materials and methods
Patients
Formalin-fixed, paraffin-embedded lung samples were obtained from patients who underwent surgical resection for NSCLC at Ajou University Hospital from January 2000 to December 2015. A total of 267 patients with NSCLC were selected. This retrospective study was approved by the Institutional Review Board of Ajou University School of Medicine. Informed consent was waived due to the retrospective nature of this study.
Histopathological analysis and immunohistochemistry
All hematoxylin and eosin stained slides were carefully reviewed by two pathologists (YWK and JHH) to determine tumor subtype, according to the 2015 World Health Organization Classification of Lung Tumors (Travis et al. 2015). Pathological staging was recorded according to the eighth edition of the TNM classification. We used tissue microarrays for immunohistochemical (IHC) analysis. Tissue microarray blocks were constructed from a representative paraffin block of a tumor section using a trephine apparatus. TNM stage was based on the 8th edition of the AJCC Cancer Staging Manual. Tissue microarray sections were arranged in a Benchmark XT automatic IHC staining device (Ventana Medical Systems, Tucson, AZ, USA). Samples were incubated with antibodies against CD44 (monoclonal, clone MRQ-13, Cell Marque), NANOG (polyclonal, Abcam), ALDH1 (monoclonal, clone 44/ALDH, BD biosciences), e-cadherin (monoclonal, clone 36B5, Novocastra), β-catenin (monoclonal, clone β-catenin1, DAKO), p120 catenin (monoclonal, clone MRQ-5, Cell Marque), vimentin (monoclonal, clone V9, Novocastra), SNAIL (polyclonal, Abcam), and TWIST (polyclonal, Abcam).
Various tissues were used as positive controls for each antibody (Supplementary Figure 1). Normal breast tissue was used as positive control of p120 catenin, E-cadherin. Normal gastric tissue was used as positive control of ALDH1. Normal placental tissue was used as positive control of SNAIL and TWIST. Embryonal carcinoma tissue was used as positive control of NANOG. Synovial sarcoma tissue was used as a positive control for vimentin. Phosphate-buffered saline (PBS) was used instead of primary antibody as a negative control.
Intensity of CD44, NANOG, ALDH1, e-cadherin, β-catenin, p120 catenin, vimentin, SNAIL, and TWIST was evaluated on a four-point intensity scale: 0 (no staining), 1 (faint staining = light yellow), 2 (moderate staining = yellow–brown), and 3 (strong staining = brown) (Fig. 1 and Supplementary Figure 2). Percentages (0–100%) of cytoplasmic expression of NANOG, ALDH1, vimentin, SNAIL, and TWIST; and membranous expression of CD44, e-cadherin, β-catenin, and p120 catenin were also evaluated. We used the H-score for interpretation of IHC stain (McCarty et al. 1986). H-score = [1 × (% cells 1 +) + 2 × (% cells 2 +) + 3 × (% cells 3 +)]. H-scores (0–300) were obtained by multiplying the intensity and percentage of positive cells. Based on the mean value of the H-score, patients were divided into low- and high-expression groups.
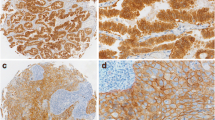
Cancer stem cell marker expression in lung adenocarcinoma. a Positive CD44 expression on tumor cells. b Negative CD44 expression on tumor cells. c Positive NANOG expression on tumor cells. d Negative NANOG expression on tumor cells. e Positive ALDH1 expression on tumor cells. f Negative ALDH1 expression on tumor cells
Web-based mRNA profiling
We obtained mRNA expression data of 230 lung adenocarcinoma patients and 178 lung squamous cell carcinoma patients of The Cancer Genome Atlas (TCGA) database from cBioPortal for Cancer Genomics (http://cbioportal.org) (Network 2012; Network 2014). We performed correlation analyses between mRNA expression of CSC markers and EMT markers in TCGA data.
The online Kaplan–Meier plotter tool has been used to assess the effect of 54,675 genes on survival, using 10,461 cancer samples including lung cancer (Gyorffy et al. 2013). mRNA expression profiling and overall survival (OS) information were downloaded from GEO datasets (GSE14814, GSE19188, GSE29013, GSE30219, GSE31210, GSE3141, GSE31908, GSE37745, GSE50081). These datasets were handled by the PostgreSQL server, which integrated mRNA expression and clinical data at the same time. To analyze the prognostic information of a particular gene, patient samples were divided into two groups, according to the median mRNA expression of the proposed biomarkers, and compared using the Kaplan–Meier survival plot.
Statistical analyses
OS or recurrence-free survival (RFS) was analyzed by Kaplan–Meier curve and compared by the log-rank test. Cox proportional hazards regression model was used in multivariate prognostic analysis of OS or RFS. In the univariate analysis, significant predictors with a P value < 0.05 were included in the final multivariate analysis. The enter method was used to determine the final Cox model for multivariate analysis. Categorical variables were compared, using Chi squared test. Spearman correlation analysis was used to describe the correlation between quantitative variables. SPSS statistical software version 18.0 (SPSS; Chicago, IL, USA) was used in all analyses and a p value of less than 0.05 was considered statistically significant.
Results
Patient demographics
Demographic data of patients included in this study are provided in Table 1. Patient age ranged from 35 to 86 years (median 64 years). There were 110 (42.3%), 71 (27.3%), and 79 (30.4%) stage I, II, and III patients, respectively. The median OS was 71.06 months and the estimated 5-year OS was 56.4%. The median RFS was 63 months and the estimated 5-year RFS was 51.1 percent. The median follow-up time was 37.6 months (range 0.5–123 months).
Correlation between stem cell-related marker and epithelial–mesenchymal transition marker expression in IHC data
We performed correlation analyses between CSC marker and EMT marker expressions in IHC data (Table 2). In adenocarcinoma, CD44 expression was positively correlated with P120-catenin expression (Spearman’s rho = 0.154 and P = 0.044). There was no correlation between NANOG and EMT marker expression. ALDH1 expression was positively correlated with P120-catenin expression (Spearman’s rho = 0.262 and P < 0.001) and negatively correlated with vimentin (Spearman’s rho = -0.037 and P < 0.001) and TWIST (Spearman’s rho = − 0.150 and P = 0.049). In squamous cell carcinoma, CD44 expression was not correlated with EMT markers. NANOG was negatively correlated with E-cadherin (Spearman’s rho = − 0.244 and P = 0.018), and ALDH1 was positively correlated with E-cadherin (Spearman’s rho = 0.485 and P < 0.001).
Correlation between stem cell-related marker and epithelial–mesenchymal transition marker expression in mRNA expression data
Next, we performed correlation analyses between CSC marker and EMT marker expressions in mRNA expression data (Table 3). In adenocarcinoma, CD44 expression was positively correlated with vimentin (Spearman’s rho = 0.531 and P < 0.001), SNAIL (Spearman’s rho = 0.195 and P < 0.001), and TWIST expression (Spearman’s rho = 0.282 and P < 0.001). NANOG expression was negatively correlated with SNAIL (Spearman’s rho = − 0.183 and P < 0.001) and TWIST expression (Spearman’s rho = − 0.224 and P < 0.001). ALDH1 expression was negatively correlated with β-catenin (Spearman’s rho = − 0.224 and P < 0.001), vimentin expression (Spearman’s rho = − 0.14 and P = 0.02), and TWIST expression (Spearman’s rho = − 0.200 and P < 0.001).
In squamous cell carcinoma, CD44 expression was positively correlated with β-catenin (Spearman’s rho = 0.178 and P = 0.01) and p120-catenin (Spearman’s rho = 0.191 and P = 0.01). NANOG was negatively correlated with β-catenin (Spearman’s rho = − 0.152 and P = 0.042). ALDH1 was positively correlated with E-cadherin (Spearman’s rho = 0.277 and P < 0.001) and β-catenin (Spearman’s rho = 0.156 and P < 0.001) and negatively correlated with vimentin (Spearman’s rho = − 0.329 and P < 0.001), SNAIL (Spearman’s rho = − 0.356 and P < 0.001), and SNAIL (Spearman’s rho = − 0.242 and P < 0.001).
Prognostic significance of stem cell-related marker and EMT marker expression
Mean values of CD44 (55.4), NANOG (30.4), ALDH1 (93.85), e-cadherin (201.5), β-catenin (279.7), p120-catenin (242.1), vimentin (20.5), SNAIL (29.3), and TWIST (43.6) protein expressions were used as cut-offs.
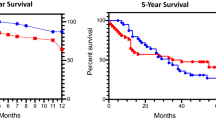
In adenocarcinoma, IHC CD44 expression was not associated with OS or RFS rates (P = 0.456; Fig. 2a and P = 0.73; Fig. 2b, respectively). However, in Kaplan–Meier plotter analysis, patients with high CD44 expression had higher OS rates than patients with low CD44 [Hazard ratio (HR) = 0.58 and P < 0.01, Fig. 2c]. Higher expression of IHC NANOG was associated with a favorable prognosis for OS and was not correlated with RFS rate (P = 0.016; Fig. 2d and P = 0.174; Fig. 2e, respectively). Higher expression of NANOG was also correlated with a favorable prognosis for OS by Kaplan–Meier plotter analysis (HR = 0.49 and P < 0.01, Fig. 2f]. Higher expression of IHC ALDH1 was associated with a favorable prognosis for OS and RFS rates (P = 0.003; Fig. 2g and P = 0.007; Fig. 2h, respectively) and was also correlated with a favorable prognosis for OS rate by Kaplan–Meier plotter analysis (HR = 0.71 and P < 0.01, Fig. 2i].
Comparison of survival rates, according to CD44, NANOG, and ALDH1 immunohistochemistry (IHC) and mRNA expression in patients with adenocarcinoma. a Overall survival (OS) and CD44 by IHC. b Recurrence free survival (RFS) and CD44 IHC. c OS and CD44 mRNA. d OS and NANOG IHC. e RFS and NANOG IHC. f OS and NANOG mRNA. g OS and ALDH1 IHC. h RFS and ALDH1 IHC. i OS and ALDH1 mRNA
In squamous cell carcinoma, IHC CD44 expression was not associated with OS or RFS rates (P = 0.55 and P = 0.742, respectively) and was not correlated with OS rate by Kaplan–Meier plotter analysis (P = 0.25). IHC NANOG expression was also not associated with OS or RFS rates (P = 0.446 and P = 0.506, respectively) and was not correlated with OS rate by Kaplan–Meier plotter analysis (P = 0.75). IHC ALDH1 expression was also not associated with OS or RFS rates (P = 0.575 and P = 0.755, respectively) and was not correlated with OS rate by Kaplan–Meier plotter analysis (P = 0.5).
Because NANOG and ALDH1 expressions were associated with favorable clinical outcomes, and NANOG and ALDH1 expressions were associated with epithelial-like phenotypes in adenocarcinoma, we performed survival analysis of EMT markers in adenocarcinoma. Higher expression of P120-catenin detected by IHC correlated with a favorable prognosis for both OS and RFS rates (P = 0.008 and P = 0.069, respectively). Higher expression of IHC beta-catenin was associated with a favorable prognosis for OS and RFS rates (P = 0.002 and P < 0.001, respectively). Higher expression of IHC TWIST correlated with a worse prognosis for OS and RFS rates (P = 0.01 and P = 0.054, respectively). However, other EMT markers did not correlate with prognosis.
Univariate analysis revealed that OS was associated with sex, stage, NANOG, and ALDH1 expression; and RFS was associated with stage, solid pattern histology, lymphovascular invasion, and ALDH1 expression (Table 4). In multivariate analysis, higher expression of ALDH1 was an independent favorable prognostic marker for OS and RFS (HR = 0.428, P = 0.026 and HR = 0.505, P = 0.033, respectively; Table 4).
Correlation between stem cell-related marker and clinicopathologic variables
In adenocarcinoma, solid pattern histology was more common in the higher CD44 expression group, but acinar pattern histology was relatively rare (P < 0.001) (Supplementary Table 1). High expression of NANOG was more frequent in female patients and non- smokers (56.9% vs. 34.4%, P = 0.007; and 60% vs. 38.3, P = 0.015, respectively). There was no correlation between ALDH1 expression and clinicopathologic variables.
In squamous cell carcinoma, stem cell-related markers were not correlated with clinicopathologic variables (Supplementary Table 2).
Discussion
Our study made several novel findings. First, NANOG and ALDH1 expressions were associated with a favorable prognosis and ALDH1 was an independent favorable prognostic marker for OS and RFS in adenocarcinoma. Second, ALDH1 expression was positively correlated with an epithelial-like phenotype: low vimentin and low TWIST in IHC and mRNA expression data. Third, the epithelial-like phenotype expressing P120-catenin and beta-catenin was associated with a favorable prognosis; however, the mesenchymal-like phenotype expressing TWIST was correlated with an unfavorable prognosis. These results suggest that ALDH1 expression may improve lung adenocarcinoma clinical outcomes by enhancing the epithelial-like phenotype.
CSC has been characterized by lower proliferation rates and higher expression of DNA repair than normal cells, which can contribute to failure of cancer chemotherapy (Jiang et al. 2012). Another characteristic of CSC is their ability to metastasize (Balic et al. 2006). EMT is crucial for the invasion of cancer cells through dissolution of epithelial-cell junctions and changes in cytoskeletal organization (Shibue and Weinberg 2017). EMT is also one of the mechanisms that promote metastasis (Shibue and Weinberg 2017). According to previous studies, these EMT functions are closely related to CSC (Aktas et al. 2009). In contrast to the expected association between the CSC and EMT, our study found that ALDH1 expression was positively correlated with an epithelial-like marker, P120-catenin expression and negatively correlated with mesenchymal markers vimentin and TWIST. Previous studies have rarely examined the relationship between ALDH1 and EMT phenotypes in lung adenocarcinoma. Tiran et al. found that lung adenocarcinoma cells expressing ALDH1 were strongly associated with epithelial-like phenotype markers, including e-cadherin, EpCAM, and pan-cytokeratin in primary patient-derived cancer cell culture (Tiran et al. 2017). Park et al. reported no correlation between ALDH1 and EMT phenotypes in lung adenocarcinoma, using e-cadherin and SNAIL1 (Park et al. 2016). Sung et al. also reported no correlation between ALDH1 and EMT phenotype in lung adenocarcinoma, using e-cadherin and vimentin (Sung et al. 2015). In our study, epithelial-like markers, including P120-catenin and beta-catenin were associated with a favorable prognosis; however, the TWIST mesenchymal marker was correlated with a worse prognosis. Therefore, we suggest that the epithelial-like phenotype is important to a favorable prognosis in tumors expressing ALDH1.
In our study, NANOG and ALDH1 expressions were associated with a favorable prognosis. Previous studies have also reported on the association of NANOG and ALDH1 with the prognosis of NSCLC. Li et al. and Park et al. reported that NANOG expression was significantly correlated with a poor prognosis in NSCLC (Li et al. 2013; Park et al. 2016). We used Abcam’s NANOG antibody; however, Li et al. used Cell Signaling Technology’s NANOG antibody, and Park et al. used Epitomics’s NANOG antibody. We used the H-score for interpretation of IHC data, but Li et al. and Park et al. used different scoring systems and cut-offs for IHC data than our study. Sung et al. and Dimou et al. showed that ALDH1 expression defined a group with a better prognosis in NSCLC (Dimou et al. 2012; Sung et al. 2015). Okudela et al. and Zhou et al. found a negative prognostic impact of ALDH1 in NSCLC (Okudela et al. 2012; Zhou et al. 2016). We used the same ALDH1 antibody (clone name 44/ALDH) as the Dimou study, but Okudela et al. and Zhou et al. used a different antibody (Abcam antibody). Additionally, Okudela et al. and Zhou et al. used different scoring systems and cut-off values for IHC data than our study. Most previous studies performed survival analyses using only immunohistochemistry. Web-based biomarker assessment using mRNA expression data showed that CD44, NANOG, and ALDH1 had a significantly beneficial prognostic effect in lung adenocarcinoma. These results are consistent with the results of our NANOG and ALDH1 immunochemical staining.
Previous studies found relationships between clinicopathologic features and CSC phenotypes. NANOG expression has been shown to correlate significantly with large tumor size and presence of lymphatic permeation (Park et al. 2016). Park et al. reported that ALDH1 expression was significantly associated with the absence of lymphatic permeation and low pathologic stage in lung adenocarcinoma (Park et al. 2016). These features of ALDH1, including absence of lymphatic permeation and low pathologic stage suggest a good prognosis. In our study, NANOG expression in lung adenocarcinoma correlated with female gender and absence of smoking. However, there was no correlation between clinicopathologic features and ALDH1 expression.
Targeted agents have been developed that inhibit ALDH1 function, such as dimethylaminobenzaldehyde and disulfiram (MacDonagh et al. 2017; Yue et al. 2015). MacDonagh et al. reported that dimethylaminobenzaldehyde and disulfiram significantly re-sensitized resistant NSCLC cells to the cytotoxic effects of cisplatin (MacDonagh et al. 2017). Addition of diethylaminobenzaldehyde to dacarbazine chemotherapy reduced growth and metastasis in a human melanoma xenograft model (Yue et al. 2015). However, in our study, ALDH1 expression correlated with better prognosis and an epithelial-like phenotype. In order to better target ALDH1 for treatment, further studies are needed to understand the functional role of ALDH1 in lung adenocarcinoma.
In our study, ALDH1 protein expression was positively correlated with p120-catein protein expression and was not correlated with β-catenin. However, ALDH1 mRNA expression was not correlated with p120-catein mRNA expression and was negatively correlated with β-catenin mRNA expression. Discrepancies were also found in correlation analysis between immunohistochemistry and mRNA expression data in other genes. Protein expression levels are influenced by a variety of factors. Alternative splicing produces proteomic diversity and plays an important role in the function and expression of NANOG and CD44 (Das et al. 2011; Prochazka et al. 2014). MicroRNAs are well-known regulators of mRNA stability and microRNAs can also reduce the rate of translation initiation (Barrett et al. 2012). Autophagy and ubiquitin–proteasome systems are two important pathways for intracellular proteolysis and can affect protein expression regardless of transcription level (Tang and Amon 2013).
This study had some limitations. First, we did not evaluate protein expression and mRNA expression in the same sample. We used a web-based biomarker assessment for mRNA profiling. Therefore, further studies of mRNA and IHC data should be performed on the same specimen. Second, we used tissue microarray for IHC expression. Unfortunately, tissue microarrays cannot reflect the entire tumor section, unlike whole tissue sections. Third, our study had a relatively small sample size.
In conclusion, CSC and EMT pathways are crucial for cancer progression. However, synergistic interactions between CSC and EMT in NSCLC are not yet fully understood. In this study, we found a clinical correlation of ALDH1 expression in adenocarcinoma with an epithelial-like phenotype in IHC and mRNA data. mRNA and IHC data for NANOG and ALDH1 expression were correlated with improved survival outcome. Epithelial-like phenotypes, such as P120-catenin and beta-catenin were associated with a favorable prognosis. Unlike previous studies on CSC and EMT, we obtained new results, which are consistent with mRNA data. We believe that our data provide important clinical evidence challenging the current model of EMT and CSC in lung adenocarcinoma progression.
Abbreviations
- ALDH1:
-
Aldehyde dehydrogenase 1
- CSC:
-
Cancer stem cell
- EMT:
-
Epithelial–mesenchymal transition
- GEO:
-
Gene expression omnibus
- HR:
-
Hazard ratio
- IHC:
-
Immunohistochemical
- NSCLC:
-
Non-small cell lung cancer
- OS:
-
Overall survival
- RFS:
-
Recurrence free survival
References
Aktas B, Tewes M, Fehm T, Hauch S, Kimmig R, Kasimir-Bauer S (2009) Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res 11:R46. https://doi.org/10.1186/bcr2333
Balic M et al (2006) Most early disseminated cancer cells detected in bone marrow of breast cancer patients have a putative breast cancer stem cell phenotype Clinical cancer research: an official journal of the American Association for. Can Res 12:5615–5621. https://doi.org/10.1158/1078-0432.ccr-06-0169
Barrett LW, Fletcher S, Wilton SD (2012) Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell Mol Life Sci 69:3613–3634. https://doi.org/10.1007/s00018-012-0990-9
Bertolini G et al (2009) Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proc Natl Acad Sci USA 106:16281–16286. https://doi.org/10.1073/pnas.0905653106
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. https://doi.org/10.3322/caac.21492
Clarke MF, Fuller M (2006) Stem cells and cancer: two faces of eve. Cell 124:1111–1115. https://doi.org/10.1016/j.cell.2006.03.011
Das S, Jena S, Levasseur DN (2011) Alternative splicing produces Nanog protein variants with different capacities for self-renewal and pluripotency in embryonic stem cells. J Biol Chem 286:42690–42703. https://doi.org/10.1074/jbc.M111.290189
Dimou A, Neumeister V, Agarwal S, Anagnostou V, Syrigos K, Rimm DL (2012) Measurement of aldehyde dehydrogenase 1 expression defines a group with better prognosis in patients with non-small cell lung cancer. Am J Pathol. 181:1436–1442. https://doi.org/10.1016/j.ajpath.2012.06.037
Ettinger DS et al (2017) Non-Small Cell Lung Cancer, Version 5.2017, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Cancer Netw 15:504–535
Goldstraw P et al (2007) The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol. 2:706–714. https://doi.org/10.1097/jto.0b013e31812f3c1a
Gyorffy B, Surowiak P, Budczies J, Lanczky A (2013) Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One 8:e82241. https://doi.org/10.1371/journal.pone.0082241
Jiang W, Peng J, Zhang Y, Cho WC, Jin K (2012) The implications of cancer stem cells for cancer therapy. Int J Mol Sci 13:16636–16657. https://doi.org/10.3390/ijms131216636
Kalluri R, Weinberg RA (2009) The basics of epithelial-mesenchymal transition. J Clin Investig 119:1420–1428. https://doi.org/10.1172/jci39104
Leung EL et al (2010) Non-small cell lung cancer cells expressing CD44 are enriched for stem cell-like properties. PLoS ONE. 5:14062. https://doi.org/10.1371/journal.pone.0014062
Li XQ et al (2013) Nuclear beta-catenin accumulation is associated with increased expression of Nanog protein and predicts poor prognosis of non-small cell lung cancer. J Transl Med 11:114. https://doi.org/10.1186/1479-5876-11-114
MacDonagh L et al (2017) Targeting the cancer stem cell marker, aldehyde dehydrogenase 1, to circumvent cisplatin resistance in NSCLC. Oncotarget 8:72544–72563. https://doi.org/10.18632/oncotarget.19881
Mani SA et al (2008) The epithelial–mesenchymal transition generates cells with properties of stem cells. Cell 133:704–715. https://doi.org/10.1016/j.cell.2008.03.027
McCarty KS Jr et al (1986) Use of a monoclonal anti-estrogen receptor antibody in the immunohistochemical evaluation of human tumors. Can Res 46:4244s–4248s
Nastaly P et al (2018) ALDH1-positive intratumoral stromal cells indicate differentiated epithelial-like phenotype and good prognosis in prostate cancer. Transl Res. 1:1. https://doi.org/10.1016/j.trsl.2018.08.007
Network CGAR (2012) Comprehensive genomic characterization of squamous cell lung cancers. Nature 489:519–525. https://doi.org/10.1038/nature11404
Network CGAR (2014) Comprehensive molecular profiling of lung adenocarcinoma. Nature 511:543–550. https://doi.org/10.1038/nature13385
Okudela K, Woo T, Mitsui H, Tajiri M, Masuda M, Ohashi K (2012) Expression of the potential cancer stem cell markers, CD133, CD44, ALDH1, and beta-catenin, in primary lung adenocarcinoma–their prognostic significance. Pathol Int. 62:792–801. https://doi.org/10.1111/pin.12019
Park E et al (2016) Prognostic significance of stem cell-related marker expression and its correlation with histologic subtypes in lung adenocarcinoma. Oncotarget 7:42502–42512. https://doi.org/10.18632/oncotarget.9894
Prochazka L, Tesarik R, Turanek J (2014) Regulation of alternative splicing of CD44 in cancer. Cell Signal. 26:2234–2239. https://doi.org/10.1016/j.cellsig.2014.07.011
Qin S, Li Y, Cao X, Du J, Huang X (2017) NANOG regulates epithelial-mesenchymal transition and chemoresistance in ovarian cancer. Biosci Rep. https://doi.org/10.1042/bsr20160247
Shibue T, Weinberg RA (2017) EMT, CSCs, and drug resistance: the mechanistic link and clinical implications Nature reviews. Clin Oncol. 14:611–629. https://doi.org/10.1038/nrclinonc.2017.44
Simple M, Suresh A, Das D, Kuriakose MA (2015) Cancer stem cells and field cancerization of oral squamous cell carcinoma. Oral Oncol 51:643–651. https://doi.org/10.1016/j.oraloncology.2015.04.006
Sung WJ, Park KS, Kwak SG, Hyun DS, Jang JS, Park KK (2015) Epithelial-mesenchymal transition in patients of pulmonary adenocarcinoma: correlation with cancer stem cell markers and prognosis. Int J Clin Exp Pathol 8:8997–9009
Tanaka F, Yoneda K (2016) Adjuvant therapy following surgery in non-small cell lung cancer (NSCLC). Surg Today. 46:25–37. https://doi.org/10.1007/s00595-015-1174-7
Tang YC, Amon A (2013) Gene copy-number alterations: a cost-benefit analysis Cell 152:394–405. https://doi.org/10.1016/j.cell.2012.11.043
Tiran V et al (2017) Primary patient-derived lung adenocarcinoma cell culture challenges the association of cancer stem cells with epithelial-to-mesenchymal transition. Sci Rep 7:10040. https://doi.org/10.1038/s41598-017-09929-0
Travis WD et al (2015) The 2015 World Health Organization Classification of Lung Tumors: impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol 10:1243–1260. https://doi.org/10.1097/jto.0000000000000630
Wang D, Wen GM, Hou W, Xia P (2018) The roles of CD133 expression in the patients with non-small cell lung cancer Cancer biomarkers: section A of Disease markers 22:385-394 https://doi.org/10.3233/cbm-170835
Yue L et al (2015) Targeting ALDH1 to decrease tumorigenicity, growth and metastasis of human melanoma. Melanoma Res. 25:138–148. https://doi.org/10.1097/cmr.0000000000000144
Zhang Z, Wang Q, Bu X, Zhang C, Chen H, Sha W, Liu W (2017) Overexpression of Bmi1 promotes epithelial mesenchymal transition in CD133 + Hep G2 cells. Mol Med Rep. 16:6156–6161. https://doi.org/10.3892/mmr.2017.7347
Zhou L, Yu L, Zhu B, Wu S, Song W, Gong X, Wang D (2016) Metastasis-associated in colon cancer-1 and aldehyde dehydrogenase 1 are metastatic and prognostic biomarker for non-small cell lung cancer. BMC Cancer. 16:876. https://doi.org/10.1186/s12885-016-2903-z
Funding
This research was supported by a Grant (NRF-2017R1C1B5076342 for Young Wha Koh) of the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning.
Author information
Authors and Affiliations
Contributions
YWK, JHH, SH and JJ collected the data, YWK, SH and JJ processed the data, YWK, JHH, SH and JJ designed the study and wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have stated that they have no conflicts of interest.
Ethical approval
All procedures of this study were in accordance with the ethical standards of the Institutional Research Committee and with the 1964 Helsinki declaration and its later amendments.
Informed consent
This retrospective study was approved by the Institutional Review Board of Ajou University School of Medicine. Informed consent was waived owing to the retrospective nature of this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
432_2019_2906_MOESM1_ESM.tif
Supplementary material 1. Supplementary Figure 1. Positive control tissue for antibodies. (A) Positive CD44 expression on epidermis of normal skin. (B) Positive NANOG expression on embryonal carcinoma cells. (C) Positive ALDH1 expression on parietal cells of normal gastric tissue. (D) Positive E-cadherin expression on ductal cells of normal breast tissue. (E) Positive beta-catenin expression on epidermis of normal skin. (F) Positive P120 catenin expression on ductal cells of normal breast tissue. (G) Positive vimentin expression on synovial carcinoma cells. (H) Positive SNAIL expression on trophoblast of normal placenta tissue. (I) Positive TWIST expression on trophoblast of normal placenta tissue
432_2019_2906_MOESM2_ESM.tif
Supplementary material 2. Supplementary Figure 2. Epithelial mesenchymal transition marker expression in lung adenocarcinoma. (A) Positive E-cadherin expression on tumor cells. (B) Negative E-cadherin expression on tumor cells. (C) Positive beta-catenin expression on tumor cells. (D) Negative beta-catenin expression on tumor cells. (E) Positive P120 catenin expression on tumor cells. (F) Negative P120 catenin expression on tumor cells. (G) Positive vimentin expression on tumor cells. (H) Negative vimentin expression on tumor cells. (I) Positive SNAIL expression on tumor cells. (J) Negative SNAIL expression on tumor cells. (K) Positive TWIST expression on tumor cells. (L) Negative TWIST expression on tumor cells
Rights and permissions
About this article
Cite this article
Koh, Y.W., Han, JH., Haam, S. et al. ALDH1 expression correlates with an epithelial-like phenotype and favorable prognosis in lung adenocarcinoma: a study based on immunohistochemistry and mRNA expression data. J Cancer Res Clin Oncol 145, 1427–1436 (2019). https://doi.org/10.1007/s00432-019-02906-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-019-02906-2