Abstract
Purpose
To investigate the effects of TRF2 depletion on radiosensitivity in both the telomerase-positive cell lines (A549) and alternative lengthening of telomere (ALT) cell lines (U2OS).
Methods
X-ray irradiation was used to establish two radioresistant cancer models (A549R and U2OSR) from A549 and U2OS. Colony formation assay was applied to examine the radiosensitivity of radioresistant A549R and U2OSR cells and TRF2 low-expression cells. Real-time PCR and TeloTAGGG Telomerase PCR ELISA Kit were performed to examine telomere length and telomerase activity separately. γ-H2AX was detected by immunofluorescence to assess the radiation-induced DSBs.
Results
Radioresistant cancer models were established, in which TRF2 was significantly over-expressed. Low expression of TRF2 protein could enhance the radiosensitivity and induce telomere length of A549 and U2OS cell shortening. In A549 cells with TRF2 down-regulated, the telomerase activity was inhibited, too. TRF2 deficiency increases γ-H2AX foci and fails to protect telomere from radiation.
Conclusion
The data suggest that TRF2 is a radioresistant protein in A549 and U2OS cells, and could potentially be a target for radiosensitization of both telomerase-positive and ALT cells in radiotherapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Radiation is the main treatment for most cancers, but patients receiving radiation therapy might encounter radioresistance, which results in the invariable local recurrence or distant metastasis of tumors. This unresolved issue has encouraged the development of targeted molecular therapy seeking to sensitize tumor cells to radiation and predict tumor radioresistance.
Telomere length was considered as a useful predictor of tumor radiosensitivity (Ayouaz et al. 2008). The inverse relationship between telomere length and radiation sensitivity in mouse (McIlrath et al. 2001) and human cancer cells (Slijepcevic 2004) has been proven. Our previous study confirmed the negative correlation between telomere length and radiosensitivity in human cancer cells as the telomerase-dependent telomere mechanism played an important role in DNA repair. A number of telomere-associated proteins were involved in the process (Zhong et al. 2008; Zhou et al. 2007).
It is well known that TRF2 (telomeric repeat binding factor 2) protects the telomeres from end-to-end fusion and inhibits the activation of DNA damage response, thereby decreasing the telomere dysfunction-induced DNA damage and serving as a survival factor in telomere maintenance (Denchi and de Lange 2007; Okamoto et al. 2013). While in response to DNA damage, TRF2 is rapidly and transiently phosphorylated and this DNA damage-induced phosphorylation is essential for DNA double-strand break repair, including the repair of telomere double-strand breaks (Huda et al. 2012). However, it has not yet been reported whether knockdown of TRF2 would enhance the radiosensitivity of telomerase-positive cells and ALT cells by modulating telomere homeostasis. Further study of the relationship between TRF2 and radiosensitivity is needed.
The aim of this study is to investigate whether the depletion of TRF2 could improve radiosensitivity in both telomerase-positive and ALT cells. This hopefully furthers our understanding of the distinct or unidentified roles of TRF2 in telomeric protection and provides a new target for cancer radiotherapy.
Materials and methods
Cell culture
The human lung carcinoma (A549) and osteosarcoma (U2OS) cell lines were obtained from China Center for Type Culture Collection. The cells were cultured with RPMI 1640 medium (Sigma chemical Co., St Louis, MO, USA) in a humidified atmosphere (5 % CO2) at 37 °C.
X-ray irradiation
The human lung carcinoma (A549) and osteosarcoma (U2OS) cell lines were firstly grown to approximately 90 % confluence and then irradiated at an average dose rate of 2.0 Gy/min (6 Mv X-ray) by a Siemens Primus accelerator. The total dose was 64 Gy, and the new radioresistant cell lines were named A549R and U2OSR.
Small interfering RNA and transfection
TRF2 small interfering RNA (siRNA) sequences were designed as: 5′-CCA GAA GGA UCU GGU UCU UTT-3′ (sense) and 5′-AAG AAC CAG AUC CUU CUG GTT-3′ (antisense). The target sequences were all located within the 3′-UTR region of the TRF2 cDNA (GenBank accession number: XM_005256124). The sequences of control were designed as: 5′-UUC UCC GAA CGU GUC ACG UTT-3′ (sense) and 5′-ACG UGA CAC GUU CGG AGA ATT-3′ (antisense). All of the RNA oligonucleotides and the mock siRNAs were synthesized by the Genepharma Co. (Shanghai, China). The siRNAs were transfected with the Oligofectamine reagent (invitrogen) according to the manufacturer’s instruction.
RNA extraction and RT-PCR
Total RNA was isolated using TRIzol reagent (invitrogen) according to the manufacturer’s instructions. The first strand of cDNA was obtained using revert aidTM first strand cDNA synthesis kit (Fermentas). The semiquantitative reverse transcription-PCR (RT-PCR) for TRF2 mRNA was performed with the TaKaRa RT-PCR Kit (TaKaRa biotechnology Co., Ltd). TRF2 primers were 5′-TCC CAA AGT ACC CAA AGG C-3′ (sense) and 5′-ACT CCA GCC TTG ACC CAC TC-3′ (antisense). Glyceraldehyde-3-phosphate dehydrogenase was used as the loading control with the primers of 5′-TGG AAG GAC TCA TGA CCA CA-3′(sense), 5′-TTC AGC TCA GGG ATG ACC TT-3′ (antisense) or GAA ATC CCA TCA CCA TCT TCC A-3′(sense), 5′-CAT GAG TCC TTC CAC GAT ACC AA-3′(antisense). The RT-PCR program was run as follows: for the reverse transcription, 42 °C for 60 min and 70 °C for 5 min; and for PCR, hot start 94 °C for 2 min followed by 37 cycles of 94 °C for 30 s, 62 °C for 30 s and 72 °C for 45 s, and then 72 °C for 10 min. The PCR products were resolved by 1.5 % agarose electrophoresis. All experiments were repeated at least three times.
Western blot analysis
Cultured cells were rinsed twice with phosphate-buffered saline (PBS) and mixed with 200 μl of lysis buffer (Beyotime biotechnology). The cells in lysis buffer in the dish were removed using a scraper and transferred to an Eppendorf tube. The cells were homogenized and centrifuged at 12,000 rpm for 5 min, and the supernatant was stored at −20 °C. The protein concentration of the whole cell was determined using a BCA protein assay kit (Beyotime biotechnology). The protein extracts (50 μg) were incubated in loading buffer (60 mmol/L Tris–Hcl, 25 % Glycerol, 2 % SDS, 14.4 mmol/L Mercaptoethanol, 0.1 % Bromophenol blue) and boiled for 5 min. These samples were electrophoresed on a 10 % SDS-PAGE. After electrophoresis, proteins were transferred to a PVDF membrane and processed for immunoblotting. For the detection of TRF2 and β-actin, blots were incubated with 1:1,000 dilution mouse monoclonal antibodies (Abcam ab13579) and further incubated with Horseradish peroxidase-conjugated secondary antibody diluted at 1:5,000 and specific bands were visualized by ECL (Beyotime biotechnology). Autoradiographs were recorded onto X-Omat AR film (Eastman kodak Co). The density of bands in the resulting film was quantified using the ImageJ analysis program.
Colony formation assay
An appropriate number of cells transfected with siRNA–TRF2 and control cells were plated into 6-well plates (numbers of A549/A549-siTRF2 cells: 100, 100, 200, 400, 800, 1,000, 2,000; numbers of U2OS/U2OS-siTRF2 cells transfected with siRNAs: 200, 200, 400, 800, 1,600, 2,000, 4,000). Each group was irradiated at the dose point of 0, 1, 2, 4, 6, 8, 10 Gy, respectively. After 14 days of incubation, the colonies were fixed and stained with crystal violet (1 % in absolute alcohol). Those colonies containing ≥50 cells were scored as viable colonies. These data were fit into the single-hit multi-target model, and survival curves for each group were demonstrated using GraphPad prism 5.0 software. Radiobiological parameters, such as D 0, D q and SF2, were calculated according to the survival curves.
Telomere length
Telomere length was detected in the A549 and U2OS cells transfected with either siRNA–TRF2 or siRNA-NC by using real-time PCR. The dissociation curve showing a unique peak from the PCR amplification of telomere and single copy gene (36B4) attested to the specificity of the amplification. Our previous research showed that there was a linear relationship between Ct values and DNA copy number, suggesting that real-time PCR approach would be useful for quantification. The cells were divided into four groups: cells transfected with siRNA-NC (A549-NC), cells transfected with siRNA-TRF2 (A549-siTRF2), cells received irradiation (A549-R) and cells transfected with siRNA-TRF2 and received R(A549-siTRF2-R). Cells were exposed to X-rays at a dose rate of 61.3 cGy/min. The total dose was 4 Gy. Cells were seeded in six wells and incubated at 37 °C under humidified 5 % CO2, 95 % air in culture medium.
Telomerase activity
Telomerase activity was detected by using the TeloTAGGG Telomerase PCR ELISA Kit (Roche, Switzerland) as recommended by the manufacturer. Sample absorbance was measured with Model 550 Microplate Reader (Bio-Rad, USA) at the wavelength of 450/690 nm within 30 min after addition of the stop reagent.
Phosphorylated H2AX foci
A549 and U2OS cells plated on coverslips in 6-well plates were used to study the subcellular localization of γ-H2AX. Cells were air-dried, fixed for 20 min with 4 % para formaldehyde in PBS, pH 7.4, and washed twice with PBS. Then, the cells were incubated for 15 min with 0.2 % Triton X-100 and washed with PBS. After that, the cells were incubated in blocking buffer (3 % bovine serum albumin in PBS) for 45 min before being incubated for 1 h with primary antibodies against γ-H2AX (Millipore Ser139). After three washes (5 min each) with PBS, the cells were incubated for an additional 45 min with the Alexa FITC-conjugated secondary antibody (FITC-labeled goat anti-mouse IgG; Beyotime). The cells were washed three times and stained with 20 μl (0.1 mg/ml) DAPI for 10 min in the dark. Images were taken using an Olympus BX51 fluorescence microscope equipped with an Olympus DP72 camera (Olympus Optical Co., Ltd., Tokyo, Japan).
Statistical analyses
Statistical analyses were performed with SPSS software (version 17.0, SPSS Institute, Chicago, IL, USA). All the data are represented as mean values ±SD. The significant differences between means were assessed by Student’s t test, with P ≤ 0.05 considered statistically significant.
Results
TRF2 expression is up-regulated in radioresistant cancer modes
To examine whether TRF2 affects radiosensitivity, we established two radioresistant cancer models (Fig. 1a, b; Table 1). TRF2 was significantly over-expressed in A549R and U2OSR than in A549 and U2OS. TRF2 protein has a 2.8-fold and 3.8-fold increase in A549R and U2OSR (Fig. 1c, d) (P < 0.05).
Establishment of radioresistant cancer modes. a The survival fraction of A549 and A549R. Each point of A549R was significantly higher than the A549 cell line. b The changes of cell survival curves of clonogenic assay of U2OS and U2OSR cell lines, and the irradiated group was significantly higher than the parent cell. c The expression of TRF2 in mRNA level was significantly over-expressed in the radioresistant cell lines of A549R and U2OSR than the parent cell lines (*P < 0.05). d The expression of TRF2 in protein level was significantly over-expressed in the radioresistant cell lines of A549R and U2OSR compared with the parent cell lines (*P < 0.05). Three experiments were done
Efficient knockdown of TRF2 increases radiosensitivity
To examine the impact of down-regulation of TRF2 on cancer cells, we used siRNA to block the expression of TRF2 and examined the effect on radiosensitivity. RT-PCR and Western blot were used to analyze the TRF2 expression. Compared with the negative control group, the expression of TRF2 transcripts (Fig. 2a) and TRF2 proteins (Fig. 2b) was reduced in siRNA-TRF2 group (P < 0.05). Expression of TRF2 protein was declined by 45 and 78 % in A549-siTRF2 and U2OS-siTRF2. However, negative control cells and blank control cells showed no apparent change of TRF2 protein expression. The results indicated that the siRNA-TRF2 could down-regulate the expression of TRF2 efficiently in gene and protein levels. The survival curves describe the radiobiological parameters of each group (Fig. 2c, d). Compared with the negative control group, the survival fractions of siRNA-TRF2 group were much lower at each point. The radiobiological parameters calculated are shown in Table 2. D 0, D q and SF2 values in siRNA-TRF2 group are significantly lower than the control groups (P < 0.05). Negative control and blank control groups showed no significant changes. The results suggest that down-regulating TRF2 protein expression could enhance the radiosensitivity of U2OS cells.
TRF2 knockdown increases radiosensitivity of A549 and U2OS cells. a RT-PCR was used to detect the TRF2 gene expression in the different groups. The PCR products were semiquantified for relative levels of mRNA using image analysis by comparing TRF2 with GAPDH. b TRF2 protein expression was detected by Western blot in different groups. The bar chart shows the semiquantitative analysis of TRF2 protein expression (*P < 0.05). c Survival curves in A549 treated with siRNA-TRF2 and negative control group (*P < 0.05). d Survival curves in U2OS treated with siRNA-TRF2 and negative control group (*P < 0.05). Three experiments were done
Knockdown of TRF2 induced telomere length shortening and telomerase activity inhibition
Telomere length was detected in A549 and U2OS cell lines transfected with siRNA-TRF2 or siRNA-NC by using real-time PCR. The dissociation curve showing a unique peak from the PCR amplification of telomere and single copy gene (36B4) attested to the specificity of the amplification. The standard curve derived from the amplification of various concentrations indicated that there was a linear relationship between Ct values and DNA copy number, suggesting that real-time PCR approach would be useful for quantification purposes. The result suggested that down-regulation of TRF2 protein expression could significantly shorten the telomere length (Fig. 3a, b). The relative of telomere length of A549-NC, A549-R, A549-siTRF2 and A549-siTRF2-R was 2.02 ± 0.08, 2.42 ± 0.04, 1.57 ± 0.16 and 1.89 ± 0.14. The relative of telomere length of U2OS-NC, U2OS-R, U2OS-siTRF2 and U2OS-siTRF2-R was 3.64 ± 0.18, 3.00 ± 0.33, 2.77 ± 0.12 and 2.04 ± 0.10. Telomerase activity was detected from A549, A549-NC and A549-siTRF2, negative control and positive control (Fig. 3c). Telomerase activity of A549, A549-NC and A549-siTRF2 was 2.3554 ± 0.1003, 2.1880 ± 0.15212 and 1.6960 ± 0.3071.
Effect of suppression of TRF2 on telomere length and telomerase activity. a The relative telomere length among the A549 cells treated with siRNA-NC, siRNA-TRF2, R and siRNA-TRF2+R was detected to be 2.02 ± 0.08, 1.57 ± 0.16, 2.42 ± 0.04 and 1.89 ± 0.14, respectively (*P < 0.05). b The relative telomere length among the U2OS cells treated with siRNA-NC, siRNA-TRF2, R and siRNA-TRF2+R was detected to be 3.64 ± 0.18, 2.77 ± 0.12, 3.00 ± 0.33 and 2.04 ± 0.10, respectively (*P < 0.05). c Telomerase activity was significantly increased in A549 cells compared with A549-siTRF2 cells, while A549-NC and A549 cells showed no apparent change of telomerase activity (**P < 0.05). Three experiments were done
TRF2 deficiency increased γ-H2AX foci in TRF2 down-regulated cells
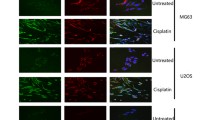
We examined the number of γ-H2AX foci in non-transfected (A549 and U2OS) cells and transfected cells (A549-siTRF2 and U2OS-siTRF2). The γ-H2AX foci increased significantly in A549-siTRF2 (23.32 ± 2.07) and U2OS-siTRF2 (56.34 ± 3.61) cells compared with A549 (9.16 ± 0.81) and U2OS (19.08 ± 1.13) (Fig. 4).
γ-H2AX foci. Immunofluorescence staining of A549, A549-siTRF2, U2OS and U2OS-siTRF2. Exponentially growing cells were transfected with si-TRF2 and then cultured on coverslips for 24 h, followed by immunofluorescence staining for phosphorylated H2AX (green). Nuclei were counterstained with DAPI (blue). At least more than 50 cells were assessed per group. Three experiments were done
Discussion
We have demonstrated that down-regulation of TRF2 was associated with increased radiosensitivity in telomerase-positive and ALT cell lines. Previous researches showed that telomeres play crucial roles in the maintenance of chromosome integrity and stability, and shortened telomere length could increase the risk of cancer (Ma et al. 2011). It has been verified that there is a significant negative correlation between telomere length and radiosensitivity (McIlrath et al. 2001; Zhong et al 2008; Cabuy et al. 2005). Indeed, TRF2 plays an important role in the regulation of telomere integrity and stability. Herein, we found that ectopic low expression of TRF2 leads to radiosensitization and telomere shortening in A549 and U2OS cells, which suggest that TRF2 might regulate radiosensitization through telomere length and DNA damage response regardless of telomerase status.
X-ray is widely used in CEM cells, breast cancer cells, SCLC and so on, to establish a radioresistant model (Shi et al. 2006; Li et al. 2014; Mistuhashi et al. 1996). In our study, human lung carcinoma (A549) and osteosarcoma cell (U2OS) represented telomerase-positive cell line and ALT cell line, respectively. Their radioresistance results revealed that the irradiated groups were much more radioresistant than their parent cell lines after 64 Gy irradiation. In A549 and U2OS, the expression of TRF2 was much lower than the radioresistant models (A549R and U2OSR). Thus, TRF2 may be a potential anticancer target for therapeutic strategies.
Many studies have shown that telomere homeostasis serves as a potential target in cancer treatment, especially in radiotherapy (Ayouaz et al. 2008). As an important part of shelterin which maintains telomere homeostasis, TRF2 has a close relationship with the length and function of telomere. TRF2 is required to prevent activation of the DNA damage response pathway at chromosome ends. TRF2-depleted telomeres as well as critically short telomeres elicit DDR (Okamoto et al. 2013). Loss of TRF2 results in a loss of the single-strand TTAGGG repeat at the end of the telomere, which could induce apoptosis or senescence, and create a substrate for telomere–telomere fusions (Bombarde et al. 2010; Takai et al. 2003; Palm and de Lange 2008). Radiosensitization of A549 and U2OS with TRF2 depletion reconfirmed the importance of telomere homeostasis in radiotherapy.
As expected, telomere length analysis showed significant telomere shortening in A549 and U2OS cells with TRF2 low expression compared with control group, which indicated that TRF2 acts as an important regulator of telomere length. Insufficient TRF2 induces telomere shortening. Several lines of evidence indicate that telomerase inhibition sensitizes human cancer cells to radiation (Zongaro et al. 2008; Gomez-Millan et al. 2007; Ji et al. 2006; Nakamura et al. 2005). So we examined the telomerase activity in telomerase-positive cells (A549). Consistent with the shortening of telomeres, telomerase activity was inhibited, which might be due to the nuclear translocation of hTERT or localized activation of telomerase at the telomere resulting in the inhibition of the telomere lengthening (Yang et al. 2013). Further studies are needed to determine the mechanism by which TRF2 affects the expression of telomerase. As a telomerase-negative cell line, U2OS with TRF2 low expression showed higher radiosensitivity, too. But the mechanism of TRF2 controlling telomere length in ALT cells is still unclear. The results above suggest that both A549 cell lines and U2OS cell lines show radiosensitization upon telomere shortening by down-regulation of TRF2.
Studies have demonstrated that TRF2 plays important roles in DNA damage response and the suppression of TRF2 in cancer cells could result in telomere dysfunction (Okamoto et al. 2013; Kaul et al. 2011). γ-H2AX is involved in the process that TRF2 preventing the initial steps of the DNA damage response pathway (Okamoto et al. 2013). The visualization of discrete foci of γ-H2AX (phosphorylated histone H2AX) in response to ionizing radiation was used to quantify radiation-induced DSBs. To understand more about the mechanism of TRF2 modulating the DNA damage response, we numerated the γ-H2AX foci here. We obtained the same result that TRF2 low expression results in increased recruitment of DNA damage response factors-γ-H2AX, which revealed that the DNA damage that results from radiation was increased by TRF2 low expression. Our study demonstrates that TRF2 could protect telomeres from radiation-induced DSBs in both A549 and U2OS cells. However, the study had certain limitations. We studied two cell lines only; more telomerase-positive and ALT cells need to be included in future researches.
In conclusion, this study revealed that lower TRF2 levels increase the risk of DNA damage and confer radiosensitization in telomerase-positive and ALT cells. Giving certain doses of irradiation to cancer cells is a good way to establish radioresistant cancer models, which showed advantages in studying proteins related with radiosensitization. In addition, the evidence of the correlation between TRF2 expression and telomere length advanced the understanding of the relation between telomere homeostasis and radiosensitization. In summary, our study for the first time indicates that TRF2 may be a potential target in the radiotherapy of lung and osteosarcoma cancer.
References
Ayouaz A, Raynaud C, Heride C, Revaud D, Sabatier L (2008) Telomeres: hallmarks of radiosensitivity. Biochimie 90:60–72
Bombarde O, Boby C, Gomez D et al (2010) TRF2/RAP1 and DNA–PK mediate a double protection against joining at telomeric ends. EMBO J 29:1573–1584
Cabuy E, Newton C, Joksic G et al (2005) Accelerated telomere shortening and telomere abnormalities in radiosensitive cell lines. Radiat Res 164:53–62
Denchi EL, de Lange T (2007) Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 448:1068–1071
Gomez-Millan J, Goldblatt EM, Gryaznov SM, Mendonca MS, Herbert BS (2007) Specific telomere dysfunction induced by GRN163L increases radiation sensitivity in breast cancer cells. Int J Radiat Oncol Biol Phys 67:897–905
Huda N, Abe S, Gu L, Mendonca MS, Mohanty S, Gilley D (2012) Recruitment of TRF2 to laser-induced DNA damage sites. Free Radic Biol Med 53:1192–1197
Ji XM, Xie CH, Fang MH, Zhou FX, Zhang WJ, Zhang MS, Zhou YF (2006) Efficient inhibition of human telomerase activity by antisense oligonucleotides sensitizes cancer cells to radiotherapy. Acta Pharmacol Sin 27:1185–1191
Kaul Z, Cesare AJ, Huschtscha LI, Neumann AA, Reddel RR (2011) Five dysfunctional telomeres predict onset of senescence in human cells. EMBO Rep 13:52–59
Li Z, Yang XX, Xia NX, Yang L, Yu HJ, Zhou FX, Xie CH, Zhou YF (2014) PTOP and TRF1 help enhance the radio resistance in breast cancer cell. Cancer Cell Int 14:7
Ma H, Zhou Z, Wei S et al (2011) Shortened telomere length is associated with increased risk of cancer a meta-analysis. PLoS One 6:e20466
McIlrath J, Bouffler SD, Samper E et al (2001) Telomere length abnormalities in mammalian radiosensitive cells. Cancer Res 61:912–915
Mistuhashi N, Takahashi T, Sakurai H et al (1996) A radioresistant variant cell line isolated from a radiosensitive rat yolk sac tumor cell line: different of early radiation-induced morphological changes, especially apoptosis. Int J Radiat Biol 69:329–336
Nakamura M, Masutomi K, Kyo S, Hashimoto M, Maida Y, Kanaya T, Tanaka M, Hahn WC, Inoue M (2005) Efficient inhibition of human telomerase reverse transcriptase expression by RNA interference sensitizes cancer cells to ionizing radiation and chemotherapy. Hum Gene Ther 16:859–868
Okamoto K, Bartocci C, Ouzounov I, Diedrich JK, Yates JR, Denchi EL (2013) A two-step mechanism for TRF2-mediated chromosome-end protection. Nature 494:502–505
Palm W, de Lange T (2008) How shelterin protects mammalian telomeres. Annu Rev Genet 42:301–334
Shi DG, Shi GM, Huang G (2006) Chemosensitivity of irradiated resistant cells of multicellular spheroids in A549 lung adenocarcinoma. Chin J Radiol Med Prot 28:155–158
Slijepcevic P (2004) Is there a link between telomere maintenance and radiosensitivity? Radiat Res 161:82–86
Takai H, Smogorzewska A, de Lange T (2003) DNA damage foci at dysfunctional telomeres. Curr Biol 13:1549–1556
Yang L, Wang W, Hu L, Yang X, Zhong J, Li Z, Yang H, Lei H, Yu H, Liao Z, Zhou F, Xie C, Zhou Y (2013) Telomere-binding protein TPP1 modulates telomere homeostasis and confers radioresistance to human colorectal cancer cells. PLoS One 8:e81034
Zhong YH, Liao ZK, Zhou FX, Xie CH, Xiao CY, Pan DF, Luo ZG, Liu SQ, Zhou YF (2008) Telomere length inversely correlates with radiosensitivity in human carcinoma cells with the same tissue background. Biochem Biophys Res Commun 367:84–89
Zhou FX, Liao ZK, Dai J, Xiong J, Xie CH, Luo ZG, Liu SQ, Zhou YF (2007) Radiosensitization effect of zidovudine on human malignant glioma cells. Biochem. Biophys. Res Commun 354:351–356
Zongaro S, Verri A, Giulotto E, Mondello C (2008) Telomere length and radiosensitivity in human fibroblast clones immortalized by ectopic telomerase expression. Oncol Rep 19:1605–1609
Acknowledgments
This research was supported by the National Natural Science Foundation of China (No. 81071825, 81201755), the Doctoral Fund of Ministry of Education of China (No. 20120141130010) and the Fundamental Research Funds for the Central Universities. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Xiaoxi Yang and Zheng Li have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Yang, X., Li, Z., Yang, L. et al. Knockdown of telomeric repeat binding factor 2 enhances tumor radiosensitivity regardless of telomerase status. J Cancer Res Clin Oncol 141, 1545–1552 (2015). https://doi.org/10.1007/s00432-015-1911-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-015-1911-8