Abstract
Purpose
Altered expression of serine protease inhibitor peptidase inhibitor clade E member 2 (SERPINE2) associates with human cancer development and progression; thus, this study investigated SERPINE2 expression in gastric cancer tissues for association with clinicopathological and survival data from the patients and then investigated the role of SERPINE2 in gastric cancer cells in vitro.
Methods
The levels of SERPINE2 mRNA in 243 gastric cancer tissues and paired non-cancerous mucosa were determined using quantitative PCR. Inhibition of SERPINE2 expression by small interfering RNA (siRNA) was detected by Western blotting. tetrazolium, soft agar, and transwell assays were performed to evaluate the proliferation, anchorage-independent growth, and motility of gastric cancer SGC7901 cells transfected with SERPINE2 siRNA.
Results
Compared with the normal mucosa, SERPINE2 mRNA was increased in gastric cancer tissues and cells. Analysis of the 243 matched specimens showed that high SERPINE2 levels were associated with lymph node metastasis, distant metastasis, and clinical stage. Patients with high SERPINE2 mRNA levels had poorer survival compared with patients with low SERPINE2 mRNA levels. In vitro, SERPINE2 inhibited anchorage-independent growth, migration, and invasion of SGC7901 cells, but not proliferation.
Conclusions
Our findings indicate that upregulated SERPINE2 may contribute to the aggressive phenotype of gastric cancer and suggest that SERPINE2 can be used as a novel prognostic factor and anticancer target in patients with gastric cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In developing countries, gastric cancer is the second most common cancer in men and the fourth most common cancer in women, accounting for over 70 % of new cases and death following gastric cancer diagnosis worldwide (Jemal et al. 2011). At present, although surgical resection combined with chemotherapy or radiotherapy is utilized in gastric cancer treatment, the outcomes for patients with metastasis are relatively poor. Therefore, it remains an extremely urgent matter to identify the novel factors for early diagnostic and prognostic assessment.
More and more intracellular signal molecules have been discovered as biomarkers of gastric cancer at present. It is worth noting that various proteolytic enzymes, including matrix metalloproteinases (MMPs, Wieczorek et al. 2014) and serine proteases (Dorn et al. 2014), also play crucial roles in gastric cancer progression due to the degradation of extracellular matrix components.
Serine protease inhibitor (serpin) peptidase inhibitor clade E member 2 (SERPINE2, also known as prostate nexin-1 [PN-1]), a member of the serpin protein family, mainly modulates thrombin, urokinase, plasmin, and trypsin activity. SERPINE2 was first discovered as a susceptibility factor for chronic obstructive pulmonary disease and panlobular-type emphysema (DeMeo et al. 2006; Kukkonen et al. 2011). Recent studies have shown that SERPINE2 is closely associated with tumorigenesis. Altered SERPINE2 expression has been reported in several types of tumors, such as breast cancer (Candia et al. 2006), colon cancer (Selzer-Plon et al. 2009), and oral squamous cell carcinoma (Gao et al. 2008). SERPINE2 is also a tumor suppressor gene that decreases cell proliferation and vessel formation in prostate cancer (McKee et al. 2013). In contrast, another report demonstrated that SERPINE2 promoted lymph node metastasis (LNM) in a xenograft model of testicular cancer, suggesting its function as an oncogene (Nagahara et al. 2010). However, SERPINE2 expression and its relationship with gastric cancer, as well as its biological function on gastric cancer cells, remain unclear.
In the present study, the expression levels of the SERPINE2 gene in 243 gastric cancer tissues and matched adjacent normal mucosa were measured by quantitative PCR. We aimed to establish whether SERPINE2 expression levels correlate with clinicopathological parameters and prognosis for patients with gastric cancer. Moreover, the biological function of SERPINE2 was investigated in the gastric cancer cell line SGC7901.
Materials and methods
Patient information and data collection
The present study involved 243 patients with gastric cancer who underwent surgery at QiLu Hospital and Qianfoshan Hospital from 2006 to 2008. Gastric cancer and paired non-cancerous gastric mucosa (>5 cm away from the tumor) tissue were obtained and frozen immediately in liquid nitrogen after surgical resection. Pathological and clinical data were collected from medical records (Table 1). There were 172 men and 71 women; the median age was 65 years (range 27–89 years). The 5-year follow-up patient records were available. All patients were added to overall survival (OS) rate analysis, and 216 cases were included in disease-free survival (DFS) rate analysis, excluding 27 cases with distant metastasis before surgery. We obtained written informed consent from all patients involved in this study. All human studies have been approved by Ethics Committee of Shandong University and followed the principals outlined in the 1964 Declaration of Helsinki.
RNA extraction and real-time quantitative PCR
Total RNA was extracted from tumor and paired normal gastric mucosa using TRIzol reagent (Invitrogen, USA) according to the manufacturer’s instructions. Total RNA (1 μg) was quantified for reverse transcription, and PCR was performed using SYBR reagent (Genecopeia, USA) in a 7900HT Fast PCR System (Applied Biosystems, USA). The primers for SERPINE2 were 5′-AATGAAACCAGGGATATGATTGAC-3′ (forward) and 5′-TTGCAAGATATGAGAAACATGGAG-3′ (reverse). The primers for the endogenous control β-actin were 5′-CTCCTTAATGTCACGCACGAT-3′ (forward) and 5′-CATGTACGTTGCTATCCAGGC-3′ (reverse). The PCR was carried out at 95 °C for 10 min, then 40 cycles of 95 °C for 10 s, 60 °C for 20 s, and 72 °C for 20 s. The relative expression of SERPINE2 was calculated using the formula 2−ΔCt. Each reaction was conducted in duplicate.
SERPINE2 small interfering RNA and cell transfection
SERPINE2 small interfering RNA (siRNA) (AGGAACCATGAATTGGCAT, synthesized by Shanghai Shenggong Biological, China) and negative control were separately transfected into SGC7901 cells using 2 μL X-tremeGENE (Roche Applied Science, USA) transfection reagent according to the manufacturer’s instructions. After transfection, cells were cultured in RPMI 1640 medium supplemented with 10 % fetal bovine serum (FBS; Gibco, USA) at 37 °C for an additional 24 h.
Western blotting
To test whether SERPINE2 was silenced by siRNA, sodium dodecyl sulfate–polyacrylamide gel electrophoresis and immunoblot analyses were performed. Specimens were incubated with anti-SERPINE2 (1:1000; Abcam, UK) and anti-β-actin (1:1000; Santa Cruz Biotechnology, USA) antibodies at 4 °C overnight. Reactive bands were detected using ECL Plus™ (Millipore, Germany) and X-ray exposure and then analyzed using ImageJ (Broken Symmetry Software, USA). SERPINE2 expression was quantified against β-actin.
Cell proliferation assay
The proliferation ability of cells transfected with SERPINE2 siRNA was measured using CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega, USA). After 24-h transfection, 6 × 103 SGC7901 cells were seeded in 96-well plates. Tetrazolium (MTS) solution (20 μL) was added to each well and incubated at 37 °C for 2 h. Then, the absorbance at 490 nm was detected at 24, 48, and 72 h using a microplate reader (Bio-Rad, Singapore). Each experiment was performed in triplicate.
Soft agar formation assay
Agar culture medium (0.5 %, 2 mL; 1:9 5 % agar and warm RPMI 1640 medium mixed at 40 °C) was used to coat the bottom of a 60-mm plate. Cells transfected with SERPINE2 siRNA (2 × 104/mL) were suspended at 1:1 with agar culture medium (0.6 %) at 40 °C. Then, 1 mL cell–agar mixture was immediately seeded on the surface of the solidified agar layer. After 2-week incubation, colonies with >50 cells were counted under a microscope (Olympus, Japan).
Migration and invasion assays
At 24 h after transfection, the migration ability of cells was assessed by transwell assay in 24-well plates containing polycarbonate membranes with 8.0-μm pores (Corning, USA). SGC7901 cells (1 × 105) in 100 μL serum-free medium were added to the top chamber; medium containing 10 % FBS was placed in the bottom chamber. The cell invasion assay was performed using membranes coated with Matrigel matrix (BD Biosciences, USA) and a method similar to that described above. After 24-h incubation, cells that had migrated to the lower surface of the membrane were fixed and stained. Data were obtained from three independent experiments.
Statistical analysis
Data analysis was performed using Statistical Product and Service Solution version 13.0 (SPSS, USA). Fisher’s exact test was used to analyze the association between SERPINE2 expression and clinicopathological variables. OS or DFS curves were plotted using the Kaplan–Meier method, and the differences were compared with the log-rank test. Independent risk factors were determined using a Cox proportional hazard model. Differences between two groups were evaluated with Student’s t test. The P value was considered statistically significant at <0.05.
Results
SERPINE2 was overexpressed in gastric cancer tissues and cells compared with normal gastric mucosa
SERPINE2 mRNA levels in gastric cancer tissues, cells, and normal mucosa were determined by reverse transcription–quantitative PCR. According to the presence or absence of LNM, we divided the gastric cancer specimens into two further groups: tumor with LNM (156/243, 64.2 %) and tumor without LNM (87/243, 35.8 %). The data revealed a stepwise upregulation of SERPINE2 mRNA in normal mucosa, tumor, and tumor with LNM (Fig. 1). SERPINE2 mRNA was upregulated 5.09-fold in gastric cancer tissues and 4.18-fold in SGC7901 cells compared with the normal mucosa (P = 0.0089, 0.0015, respectively). Compared with the normal mucosa, SERPINE2 mRNA in the tumor with LNM group was upregulated 6.79-fold (P = 0.004). SERPINE2 mRNA in the tumor without LNM group was 2.24-fold higher than that in the normal mucosa, although no statistical difference was found between the two groups (P = 0.0682). It is worth noting that compared with the tumor without LNM group, SERPINE2 mRNA was increased 3.02-fold in the tumor with LNM group (P = 0.023). These results demonstrate that SERPINE2 is overexpressed in gastric cancer tissues and cells, especially in cases with LNM.
Comparison of SERPINE2 expression in gastric cancer tissues, cancer cells, and normal mucosa. Relative SERPINE2 expression is higher in human gastric cancer tissues and cells than in the normal mucosa (P = 0.0089, 0.0015, respectively). SERPINE2 mRNA is significantly increased in tumor with LNM compared with normal mucosa and tumor without LNM (P = 0.004, 0.0023). *P < 0.05; **P < 0.01
Association between SERPINE2 mRNA level and clinicopathological parameters in gastric cancer
Of the 243 paired specimens, we considered a >1.25-fold increase (Fayard et al. 2009) as the cutoff for SERPINE2 overexpression in gastric cancer: 157 cases (64.4 %) had high SERPINE2 expression; expression was low in the remaining 86 cases (35.6 %). To determine the potential clinical implications of SERPINE2, the associations between clinicopathological factors and SERPINE2 expression level were analyzed. High SERPINE2 mRNA expression was significantly correlated with LNM (N classification, P = 0.01), distant metastasis (M classification, P = 0.018), and clinical stage (International Union Against Cancer [UICC], P = 0.001). However, no correlations were found between SERPINE2 mRNA level and age, sex, tumor size, tumor location, differentiation, or Lauren classification (P > 0.05, Table 1).
SERPINE2 may serve as a predictive prognostic biomarker in patients with gastric cancer
To further explore the prognostic value of high SERPINE2, the correlation between SERPINE2 expression and OS or DFS was analyzed using Kaplan–Meier survival curves. Patients with high SERPINE2 expression had significantly worse OS and DFS than patients with low SERPINE2 expression (both P < 0.001, Fig. 2). Univariate analysis (Table 2) revealed that both decreased OS and DFS were associated with T classification, LNM, distant metastasis, clinical stage, poor differentiation, and SERPINE2 expression. Multivariate analysis further demonstrated that high SERPINE2 expression was an independent prognostic factor for patients with gastric cancer (Table 2, P = 0.000, hazard ratio [HR] = 3.05, 95 % confidence interval [95 % CI] = 1.78–4.96; P = 0.002, HR = 2.30, 95 % CI = 1.57–3.55).
SERPINE2 siRNA had no effect on cell proliferation in vitro
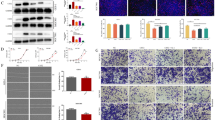
To test the effect of increased SERPINE2 on cell biological behavior in vitro, the proliferation ability of SGC7901 cells treated with SERPINE2 siRNA or negative control was detected by MTS assay. After transfection, the interference efficiency of SERPINE2 was first confirmed by Western blotting. Normalized to the negative control, SERPINE2 expression was downregulated 56.83 ± 11.80 % in the SERPINE2 siRNA group (P = 0.0039, Fig. 3a). Then, the cell proliferation ability was monitored at 24, 48, and 72 h. However, as illustrated in Fig. 3b, no difference in cell proliferation rate was observed between the two groups (P > 0.05).
Effect of SERPINE2 siRNA on gastric cancer SGC7901 cells. a SERPINE2 expression was decreased in cells transfected with SERPINE2 siRNA compared with the negative control (P = 0.0039). b MTS assay revealing no significant difference between the cell proliferation ability of SERPINE2 siRNA and negative control groups (P > 0.05). c Transwell assay revealing markedly decreased cell migration and invasion ability of the SERPINE2 siRNA group compared with the negative control group (P = 0.0158, 0.008, respectively). d Significant reduction in colonies formed by SERPINE2 siRNA-transfected cells compared with the negative control group (P = 0.001). *P < 0.05; **P < 0.01
SERPINE2 siRNA decreased cell migration and invasion and inhibited soft agar colony formation in vitro
As SERPINE2 expression was associated with LNM and distant metastasis, we were interested in determining whether SERPINE2 affected cell migration and invasion ability in vitro. The migration and invasion activity of SGC7901 cells transfected with SERPINE2 siRNA were assessed by transwell assay. In contrast to the negative control group, a significantly lower number of cells in the SERPINE2 siRNA group migrated to the lower surface of the membrane in the migration and invasion assays (Fig. 3c, P = 0.0158, 0.008, respectively). Furthermore, to verify whether decreased SERPINE2 expression would alter colony formation ability, a soft agar assay was performed after SERPINE2 siRNA transfection. As expected, there was a significant reduction in colony formation by SERPINE2 siRNA-transfected cells on soft agar compared with the negative control group (Fig. 3d, P = 0.001). These data imply that SERPINE2 controls the migration, invasion, and anchorage-independent growth capability of SGC7901 cells.
Discussion
Although SERPINE2 has been investigated in several tumors, its possible role in gastric cancer is poorly understood to date. In the present work, we report for the first time that upregulated SERPINE2 gene expression in gastric cancer is associated with cancer progression, such as LNM, distant metastasis, and clinical stage. The data also support the premise that SERPINE2 serves as a novel prognostic indicator of gastric cancer outcomes independent of clinical and tumor–node–metastasis stage. Furthermore, the effect of SERPINE2 in inhibiting SGC7901 cell migration, invasion, and anchorage-independent growth supports the idea that SERPINE2 may contribute to gastric carcinogenesis and progression.
In fact, using Northern blotting, Buchholz and colleagues were the first to report that SERPINE2 was overexpressed in three of four gastric cancers, four of four colorectal carcinoma, and 22 of 27 pancreatic cancer samples (Buchholz et al. 2003). SERPINE2 expression was also increased 1.5- to 3.5-fold in 21 of 26 breast cancer specimens compared with normal mammary tissues (Candia et al. 2006). In concordance with the above results, we observed a 5.09-fold increase in SERPINE2 mRNA in 243 cases, namely a larger cohort of gastric cancer specimens. Through oligonucleotide microarray analysis, Bergeron and others reported that SERPINE2 gene expression is mainly reduced by mitogen-activated protein kinase/extracellular signal-regulated kinase (MEK) signal activation (Bergeron et al. 2010). In addition, upregulated SERPINE2 expression is mediated by phosphatase and tensin homolog (PTEN) deletion in MEF cells (Li et al. 2006). Considering these findings, we speculate that SERPINE2 may have oncogenic properties in gastric cancer development.
More importantly, SERPINE2 mRNA was obviously increased in tumors from patients with LNM: The increase was 6.79-fold higher than that of the normal mucosa and 3.02-fold higher than that of patients without LNM. This result also agreed well with the published findings on oral squamous cell carcinoma. Gao et al. reported a 13-fold increase of SERPINE2 in oral squamous cell carcinoma and a 3.27-fold difference between cases with and without LNM (Gao et al. 2008). Moreover, we observed a significant correlation between SERPINE2 expression and both N classification and distant metastasis (Table 1). Cases with high SERPINE2 expression tended to involve LNM or distant metastasis. Using a xenograft model, Buchholz et al. (2003) presented evidence that the local invasion potential of pancreatic tumors is enhanced by SERPINE2 overexpression, accompanied by abundant extracellular matrix deposition. Another study on breast cancer reported that elevated SERPINE2 expression was markedly associated with the probability of lung metastasis (Fayard et al. 2009). These data strongly suggest that SERPINE2 upregulation might promote tumor invasion and metastasis. In the future, SERPINE2 might be used as a novel biomarker for identifying the aggressive phenotype of malignant tumors.
It is worth noting that due to different outcomes among patients with the same clinical stage, investigating novel risk factors for recurrence assessment and individualized therapy remains urgent. To date, no reports have been published on SERPINE2 expression and gastric cancer prognostic analysis. Here, we are the first to demonstrate that SERPINE2 upregulation is connected with decreased OS and DFS and that it can be an independent prognostic factor for patients with gastric cancer.
To assess the effect of SERPINE2 in human gastric cancer SGC7901 cells, proliferation, soft agar, and transwell assays were performed to monitor the growth and motility of cells transfected with SERPINE2 siRNA. Targeting SERPINE2 with siRNA decreased anchorage-independent growth, migration, and invasion, but not proliferation. The mechanism by which SERPINE2 promotes tumorigenesis has not been fully described. As an extracellular matrix-secreted component, SERPINE2 plays an important role in tumor cell and stroma interaction in pancreatic cancer (Neesse et al. 2007). SERPINE2 is a direct target of MMP-9 in prostate cancer, and MMP-9 promotes cell invasion by cleaving SERPINE2 and inhibiting urokinase plasminogen activation (Xu et al. 2010). However, there appears to be a paradox in that SERPINE2 suppresses breast cancer metastasis through low-density lipoprotein receptor-1 (LRP1)-mediated MMP-9 expression (Fayard et al. 2009). Researchers also disagree on the suppressive effect of SERPINE2 on cell growth. For example, McKee et al. (2012) found that SERPINE2 regulated Hedgehog (Hh) signaling by decreasing the Hh ligand Sonic protein expression to inhibit prostate adenocarcinoma tumor growth in mice. Contradicting this, downregulating SERPINE2 expression in murine breast cancer 4T1 cells had no effect on tumor growth in BALB/c mice (Fayard et al. 2009). Taken together, we presume that the different functions of SERPINE2 in different tumors might be partly ascribed to its tissue and cell specificity. Further investigation is also required to define the molecular mechanisms that govern SERPINE2 expression in gastric cancer development and progression.
The present study demonstrated the potential function of SERPINE2 in gastric cancer cells and the possibility of assessing SERPINE2 expression for predicting patients’ prognosis. These data could help clinical evaluation and contribute to target therapeutics to patients with a higher likehood of a poor prognosis. The limitations of this study include the small number of patients with relatively shorter follow-up time. As increased SERPINE2 expression in gastric cancer cells indicates an aggressive phenotypic change, it would be interesting to detect SERPINE2 expression in atypical hyperplasia, gastric ulcer, and gastric adenoma and to clarify whether there is any correlation between the two factors. And a larger, controlled, and clinical study also needed to validate our results in the future.
In summary, this work provides new insights into the biological function of SERPINE2 in gastric cancer. SERPINE2 is overexpressed in gastric cancer tissues and correlates with poor survival. SERPINE2 is involved in gastric carcinogenesis by promoting cell migration, invasion, and anchorage-independent growth. We propose that SERPINE2 is a useful prognostic indicator and a potential target of gene therapy for gastric cancer.
References
Bergeron S, Lemieux E, Durand V, Cagnol S, Carrier JC, Lussier JG, Boucher MJ, Rivard N (2010) The serine protease inhibitor serpinE2 is a novel target of ERK signaling involved in human colorectal tumorigenesis. Mol Cancer 9:271
Buchholz M, Biebl A, Neesse A, Wagner M, Iwamura T, Leder G, Adler G, Gress TM (2003) SERPINE2 (protease nexin I) promotes extracellular matrix production and local invasion of pancreatic tumors in vivo. Cancer Res 63:4945–4951
Candia BJ, Hines WC, Heaphy CM, Griffith JK, Orlando RA (2006) Protease nexin-1 expression is altered in human breast cancer. Cancer Cell Int 6:16
DeMeo D, Mariani T, Lange C, Lake S, Litonjua A, Celedon J, Reilly J, Chapman HA, Sparrow D, Spira A et al (2006) The SERPINE2 gene is associated with chronic obstructive pulmonary disease. Proc Am Thorac Soc 3:502
Dorn J, Beaufort N, Schmitt M, Diamandis EP, Goettig P, Magdolen V (2014) Function and clinical relevance of kallikrein-related peptidases and other serine proteases in gynecological cancers. Crit Rev Clin Lab Sci 51:63–84
Fayard B, Bianchi F, Dey J, Moreno E, Djaffer S, Hynes NE, Monard D (2009) The serine protease inhibitor protease nexin-1 controls mammary cancer metastasis through LRP-1-mediated MMP-9 expression. Cancer Res 69:5690–5698
Gao S, Krogdahl A, Sorensen JA, Kousted TM, Dabelsteen E, Andreasen PA (2008) Overexpression of protease nexin-1 mRNA and protein in oral squamous cell carcinomas. Oral Oncol 44:309–313
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011) Global cancer statistics. CA Cancer J Clin 61:69–90
Kukkonen MK, Tiili E, Hämäläinen S, Vehmas T, Oksa P, Piirilä P, Hirvonen A (2011) SERPINE2 haplotype as a risk factor for panlobular type of emphysema. BMC Med Genet 12:157
Li G, Hu Y, Huo Y, Liu M, Freeman D, Gao J, Liu X, Wu DC, Wu H (2006) PTEN deletion leads to up-regulation of a secreted growth factor pleiotrophin. J Biol Chem 281:10663–10668
McKee CM, Xu D, Cao Y, Kabraji S, Allen D, Kersemans V, Beech J, Smart S, Hamdy F, Ishkanian A et al (2012) Protease nexin 1 inhibits hedgehog signaling in prostate adenocarcinoma. J Clin Investig 122:4025–4036
McKee CM, Xu D, Muschel RJ (2013) Protease nexin 1: a novel regulator of prostate cancer cell growth and neo-angiogenesis. Oncotarget 4:1–2
Nagahara A, Nakayama M, Oka D, Tsuchiya M, Kawashima A, Mukai M, Nakai Y, Takayama H, Nishimura K, Jo Y et al (2010) SERPINE2 is a possible candidate promotor for lymph node metastasis in testicular cancer. Biochem Biophys Res Commun 391:1641–1646
Neesse A, Wagner M, Ellenrieder V, Bachem M, Gress TM, Buchholz M (2007) Pancreatic stellate cells potentiate proinvasive effects of SERPINE2 expression in pancreatic cancer xenograft tumors. Pancreatology 7:380–385
Selzer-Plon J, Bornholdt J, Friis S, Bisgaard HC, Lothe IM, Tveit KM, Kure EH, Vogel U, Vogel LK (2009) Expression of prostasin and its inhibitors during colorectal cancer carcinogenesis. BMC Cancer 9:201
Wieczorek E, Wasowicz W, Gromadzinska J, Reszka E (2014) Functional polymorphisms in the matrix metalloproteinase genes and their association with bladder cancer risk and recurrence: A mini-review. Int J Urol. doi:10.1111/iju.12431
Xu D, McKee CM, Cao Y, Ding Y, Kessler BM, Muschel RJ (2010) Matrix metalloproteinase-9 regulates tumor cell invasion through cleavage of protease nexin-1. Cancer Res 70:6988–6998
Acknowledgments
This study is supported by Shandong Province Health Science and Technology Development Program (2009HZ067).
Conflict of interest
All authors promise that there is no conflict to disclose.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Kun Wang and Bin Wang have contributed equally to this article.
Rights and permissions
About this article
Cite this article
Wang, K., Wang, B., Xing, A.Y. et al. Prognostic significance of SERPINE2 in gastric cancer and its biological function in SGC7901 cells. J Cancer Res Clin Oncol 141, 805–812 (2015). https://doi.org/10.1007/s00432-014-1858-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-014-1858-1