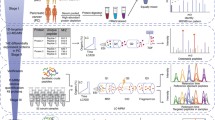
Abstract
Purpose
The detection of pancreatic tumors lacks a sensitive and specific diagnostic tool. Mass spectrometry (MS)-based profiling of serum proteins is a promising approach for discovery of new clinical biomarkers or biomarker signatures.
Methods
Serum samples from pancreatic cancer (PC) patients and control individuals were collected and processed using a standardized protocol. Samples were divided in a calibration set (n = 49 PC and 110 controls) and a validation set (n = 39 PC and 75 controls). Peptide profiles were obtained using a combination of automated solid-phase extraction with reversed-phase C18 paramagnetic beads and matrix-assisted laser desorption ionization time-of-flight MS.
Results
Linear discriminant analysis with double cross-validation resulted in a discriminating peptide signature for PC in the calibration set with a sensitivity of 78 % and a specificity of 91 % [area under the curve (AUC) of 92 %]. Classification was validated with a sensitivity of 93 % and a specificity of 100 % (AUC of 98 %), and the results were compared with carbohydrate antigen 19-9 levels and currently available clinical imaging techniques. The ten most discriminating peptide peaks were identified as fragments of proteins involved in the clotting cascade, acute phase response and immunologic response.
Conclusions
In this study, it is shown that MS-based serum peptide profiles can discriminate between PC and control samples. The approach has great potential for high-throughput analysis in surveillance programs and appears to be most promising for patients with an inherited risk for PC, who benefit from more frequent screening.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
In developed countries, pancreatic cancer (PC) has an annual incidence of 8.2 cases per 100,000 males and of 5.4 cases per 100,000 females and is the fifth (male) and fourth (female) leading cause of cancer death (Jemal et al. 2011). Patients with PC have an extremely poor prognosis with an overall 5-year survival rate of <5 % (Siegel et al. 2011). After surgical resection, 5-year survival rates are approximately 25 % for node-negative and 10 % for node-positive tumors. However, upon diagnosis, most tumors (80 %) are in advanced stage (Sener et al. 1999). There is an urgent need for new strategies that aid in diagnosis for accurate clinical decision making and for use in surveillance programs. Currently, for diagnosis and staging, imaging techniques such as computed tomography (CT) scan, magnetic resonance imaging (MRI) and magnetic resonance cholangiopancreatography (MRCP), positron emission tomography (PET) scan and endoscopic ultrasound (EUS) are available. Reported sensitivities and specificities for CT scan are 91 and 85 %, for MRI 84 and 82 %, respectively, and for EUS sensitivity values of up to 100 % have been reported (Bipat et al. 2005; Dewitt et al. 2006). A CT scan is significantly better than MRI in the detection of PC, but there are no differences for both methods regarding their performance in staging and detection of metastases (Bipat et al. 2005). The choice for a CT scan or MRI often depends on the level of locally available expertise and may therefore vary. Multiple studies have compared EUS with other imaging modalities for initial diagnosis and staging of PC. In a systematic overview of EUS-based studies performed by Dewitt and co-workers, a pooled sensitivity (85 %) and specificity (94 %) were determined, and found to be similar to CT scan results (Dewitt et al. 2006). EUS is a more accurate device for local T (small tumors <2 cm) and N staging and for predicting vascular invasion, while a CT scan is preferred in the evaluation of distant metastasis. EUS-guided fine-needle aspiration biopsy (FNA) is the best modality, with a low chance of tumor-seeding, for obtaining a conclusive diagnosis when the tumor seems irresectable or when neo-adjuvant therapy is planned (Micames et al. 2003). PET scanning with the tracer 18-fluorodeoxyglucose (FDG) relies upon functional activity to differentiate metabolically active proliferative lesions such as cancers from benign masses, most of which do not accumulate FDG with the exception of inflammatory lesions such as chronic pancreatitis. A meta-analysis described the diagnostic value of the PET scan in case of a positive (sensitivity 92 %, specificity 68 %), negative (sensitivity 73 %, specificity 86 %) and non-conclusive (sensitivity 100 %, specificity 68 %) result of the CT scan (Orlando et al. 2004). In summary, a PET scan could be of additional value in the differential diagnosis of PC, but until now this is only be performed on an individual base. Chronic pancreatitis could imitate PC at diagnostics and hinders patient selection for a pancreaticoduodenectomy.
Major risk factors for developing PC include smoking, diabetes, chronic pancreatitis and hereditary predisposition (carriers of a p16-Leiden mutation, Peutz-Jeghers syndrome, familial atypical multiple mole melanoma (FAMMM) of BRCA2 mutation carriers) (Lynch et al. 2009; Vasen et al. 2000). At this moment, screening of such a group with elevated risk factors is performed annually with MRI (Vasen et al. 2011). Furthermore, carbohydrate antigen 19-9 (CA 19-9) is commonly used as a clinical biomarker in assisting differential diagnosis and monitoring of patients with PC. For monitoring purposes, this commercially available immunoassay-based test using the monoclonal antibody 1116-NS-19-9 has a sensitivity of 70–87 % (Hansson and Zopf 1985). However, CA19-9 analysis is not optimal for the detection of small, resectable cancers and only recommended for follow-up (Goonetilleke and Siriwardena 2007). A more sensitive and specific option could be the use of serum protein biomarkers. By comparing peptide- or protein patterns, i.e., signatures, in serum from cancer patients with those obtained from control individuals, proteins that are most discriminating can be determined. The resulting peptide- or protein signature has the potential to identify a person with cancer. Mass spectrometry (MS) is the method of choice for such an analysis of peptides and proteins in serum (Aebersold and Mann 2003). Previously, we have described discriminating serum protein profiles of PC patients obtained from weak-cation exchange paramagnetic bead (MB) fractionation followed by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) MS (Velstra et al. 2013). Since human body fluids (e.g., serum) are highly complex mixtures of salts, metabolites, lipids, peptides and proteins, these require a reproducible and robust “clean-up” procedure before MS analysis (Aebersold and Mann 2003; Albrethsen 2007; Hortin 2006). Often this clean-up is performed by solid-phase extraction (SPE), and to this end, functionalized MBs allow for selective enrichment of subsets of peptides and proteins (Bladergroen et al. 2012; Jimenez et al. 2007). Other studies for diagnosis of PC have been performed using various sample preparation or fractionation techniques and resulted in sensitivities ranging from 77 % up to even 100 % and specificities ranging from 77 % up to 98 % (Deng et al. 2007; Ehmann et al. 2007; Honda et al. 2005; Kakisaka et al. 2007; Koopmann et al. 2004; Navaglia et al. 2009; Sun et al. 2007; Yu et al. 2005). Previously, Koomen and co-workers (Koomen et al. 2005) reported a sensitivity of 88 % and a specificity of 75 % using plasma purified with the so-called C18-disc, whereas Kojima and co-workers used serum, urine and plasma for analysis on a MALDI-TOF MS system in combination with a C18-filter plate resulting in a sensitivity of 88 % and a specificity of 93 % (Kojima et al. 2008). In this study, we have generated RPC18-based serum peptide profiles using an automated, high-throughput platform aiming at discrimination of pancreatic cancer patients from healthy control individuals and providing data complementary to our previous protein profiles from WCX clean-up (Velstra et al. 2013). An additional advantage is that peptides can be characterized more easily than proteins. Furthermore, we have validated the obtained discriminating signatures in an independent case-control group, and the results were evaluated with regard to routinely used imaging modalities and compared to CA19-9 values. Finally, the structures of the most discriminating peptides were elucidated which could allow further investigation into their biological role in cancer progression.
Materials and methods
Patients and serum samples
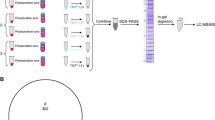
Serum samples for the calibration set were obtained from 49 patients with pancreatic cancer prior to surgery, and from 110 healthy volunteers (“controls”) over a time period ranging from October 2002 until December 2008 at the Leiden University Medical Center (LUMC), in the Netherlands. For the validation set, serum samples were obtained from 39 patients and 75 healthy volunteers over a time period ranging from January 2009 until July 2010. Patients were selected candidates for curative surgery. All surgical specimens were examined according to routine histological evaluation, and the extent of the tumor spread was assessed by TNM (TNM Classification of Malignant Tumors) classification (TNM-atlas, seventh edition, 2009). Tumor localization, preoperative biliary drainage, smoking, and coincidence of diabetes were recorded. The tumor marker CA 19-9 was determined preoperatively for 46 patients using an Elecsys CA 19-9 tumor marker immunoassay based on the monoclonal 116-NS 19-9 antibody (Roche). This tumor marker has a normal reference value of 0.0–37.0 U/ml (99th percentile). Informed consent was obtained from all subjects, and the study was approved by the Medical Ethical Committee of the LUMC. Imaging diagnostic methods (CT-scan, MRI, PET, EUS) were recorded, and the results were classified into malignant, benign or uncertain. Samples from both the calibration set and the validation set were collected and processed following a standardized high-throughput protocol as described (Velstra et al. 2012). All blood samples were drawn by antecubital venapuncture. The samples were collected in an 8.5 cc Serum Separator Vacutainer Tube and centrifuged within maximally 4 h (de Noo et al. 2005). Aliquots were stored at −80 °C until further sample processing.
Sample processing by SPE
The isolation of peptides from serum was performed using RPC18-functionalized MBs (Bladergroen et al. 2012; Jimenez et al. 2007). For this purpose, 10 μl of RPC18 Dynabeads was used for the analysis of 5 μl human serum. The activation, washing and desorption steps of the RPC18 beads were based on the manufacturers protocol and optimized to allow implementation on our 96-channel Hamilton pipetting robot. Two microliter of eluate material (obtained from the final desorption step) was used for quadruplicate MALDI spotting onto a MALDI target plate carried out on the 96-channel pipetting robot.
Mass spectrometry peptide profiling
Isotopically resolved MALDI-TOF mass spectra were obtained using an Ultraflex III TOF/TOF mass spectrometer (Bruker Daltonics, Bremen, Germany) operating in positive reflectron mode in the m/z-range of 600–4,000. The spectra were externally calibrated and baseline-corrected, and alignment was carried out as described previously. For alignment, the following five peptides were selected based on manual inspection of all spectra, namely those at m/z-value 1,465.8, m/z-value 1,778.1, m/z-value 1,865.2, m/z-value 2,602.5 and m/z-value 2,931.5, with a tolerance window of 100 ppm for the m/z-value 1,465.8 peak increasing up to 300 ppm for the highest m/z-value. Forty-two peptide signals were selected and quantified for further analysis based on careful inspection of all profiles. Peak intensities were determined by summing all isotopic peaks of the 42 selected peptides, resulting in a single intensity value for each peptide. MALDI-TOF profiles were exported as DAT (.dat) files, all containing m/z-values with corresponding intensities.
Data processing and statistics
First, the (.dat) files with summarized peak isotope intensities were generated by in-house developed Xtractor for data analysis. Then, in a next step, low-quality profiles as a result of failed sample workup or bad MALDI spotting were excluded from statistical analysis (n = 6, 0.5 %). The mean of the available peak intensities of the quadruplicate spots was used for analysis. For a detailed description of the data processing and statistics, we refer to our previous work (Velstra et al. 2012). The first statistical analysis was based on 42 peptide signals in each profile. A double cross-validatory implementation of linear discriminant analysis for the calibration of a diagnostic rule using principal component-based shrinkage calibration on a single (mean) spectrum per patient was performed, as previously described by our group (Mertens et al. 2006). Each sample was assigned to the group for which the predicted class probability was highest. For each analysis error rate (the total number of observations misclassified, expressed as a fraction of the number of observations (cases and controls) in the sample), sensitivity, specificity and area under the curve (AUC) were calculated. Next, the diagnostic rule was implemented on the validation set, and for each analysis, error, sensitivity, specificity and AUC were calculated. To further investigate the calibrated discriminating rule, a second statistical analysis on the obtained profiles was performed. The original discriminating rule, based on “leaving-one-item-out and finding-the-best-model” principle, was used to select a subset of 10 peptide peaks.
Results
Patient description
All patients were pre- or postoperatively diagnosed with histologically confirmed adenocarcinoma of the pancreas. The calibration set contained 49 preoperative pancreatic cancer patients (n = 23 males, n = 26 females) with a median age of 66 years (range 41–80). The validation set consisted of 39 preoperative pancreatic cancer patients (17 males, 22 females) according to the same criteria as the calibration set with a median age of 63 years (range 38–81). Patient characteristics for both sets are shown in Table 1. The median age of the control group was 63 years (range 44–80) and 46 years (range 21–75) for the calibration set (n = 50 males, n = 60 females) and validation set (n = 27 males, n = 48 females), respectively. Thus, age of cases and controls was matched in the calibration set (p = 0.073); however, in the validation set, the age was significantly different for these two groups (p value of <0.001).
In both sets, the majority of the tumors (n = 75, 85 %) were located in the head of the pancreas. In four cases, it was not known where the tumor was located due to the presence of high infiltration. Other patient characteristics are demonstrated in Table 1.
Tumor differentiation was known for 36 patients (73 %) in the calibration set, and 28 patients (72 %) in the validation set. In total, five patients had an increased risk for developing PC (5.7 %). For 3 out of these 5 patients, a P16 Leiden mutation (also annotated as a 19-base pair deletion of exon 2 of the CDKN2A gene) was found. One patient was never tested but had a 50 % risk of being a carrier because the mother was a carrier, while the fifth patient was a BRCA2 mutation carrier.
Statistical classification of serum profiles
For the whole study, 273 serum samples were processed with RPC18 MBs and MALDI-TOF profiles were obtained in quadruplicate, yielding 1,092 profiles. Six profiles (0.5 %) were excluded from further analysis because of their low quality as a result of non-optimal MALDI spotting. Thus, for both the calibration set and the validation set, high-quality data were obtained for all samples for further statistical analysis, i.e., replicate profiles could be averaged for each serum sample. A typical example of an RPC18-profile of serum peptides is shown in Fig. 1. Performing linear discriminant analysis with double cross-validation on the calibration set resulted in a sensitivity of 78 %, a specificity of 91 % with an AUC of 0.92, and an error of 0.14. The validation data were predicted using the calibration classification rule. In this way, the promising first results could be confirmed with a sensitivity of 93 %, a specificity of 100 %, an AUC of 0.98 and an error rate of 0.027. ROC curves are shown in Fig. 2.
Example of an RPC18 profile including multiple overlays of the most discriminating peaks. On the x-axis, the m/z-values are depicted, and on the y-axis, the intensities (in arbitrary units). Peaks with m/z-values of 1,206.7, 1,518.9, 1,536.9, 1,865.2, 2,271.1, 2,602.5, 3,190.6 and 3,261.7 were statistically evaluated as discriminating cases from controls. Profiles of cases are plotted in blue, profiles of controls in green
ROC curves of the calibration set and validation set after peak selection with standardization. The sensitivities are plotted in function of the 1-specificities for different cut-off points of the classification threshold. The area under the ROC curve is a measure of the between-group separation (diseased/normal)
Discriminating capability compared to TNM stage and grade
A correct classification was obtained for 37 of 49 (76 %) PC patients in the calibration set. In Table 1, these classifications are displayed for TNM stage and grade. For stage, the large majority (73 %) of the patients were diagnosed with early-stage PC (stages I and II). Twenty-nine (81 %) of these 36 patients were correctly classified, 10 out of 12 (83 %) for stage I and 19 out of 24 (79 %) for stage II. Thirteen patients (4 and 9) were diagnosed with late-stage PC (stages III and IV). Eight (62 %) of these patients were correctly classified. In the validation set, a correct classification was achieved for 37 (95 %) of 39 patients. Twenty-eight (72 %) of the patients were operated upon in an early stage. Ninety-three percent of the patients with an early stage were correctly classified, 2 out of 4 (50 %) for stage I and all 24 (100 %) for stage II. All of the eleven patients with late-stage PC were correctly classified. In the calibration set, a correct classification was obtained for eight patients with a grade I tumor (67 %), 10 with a grade II tumor (91 %) and 10 with a grade III tumor (77 %). In the validation set, these correct classifications were 6 (100 %), 13 (87 %) and 7 (100 %), respectively.
Discriminating capability compared to serum biomarker (CA19-9)
A preoperative CA19-9 value was available for 22 patients in the calibration set (45 %) and for 20 patients in the validation set (51 %) (Table 1). Note that CA19-9 levels were not determined for all patients included in this study because of reasons discussed in the “Introduction” section (Goonetilleke and Siriwardena 2007). Comparisons with class probabilities based on RPC18 peptide signatures are plotted in Fig. 3. In the calibration set for 12 patients, the CA19-9 value was elevated, whereas in the other 10 cases, no increase in CA19-9 levels was diagnosed. On the contrary, peptide profiling resulted in a correct classification of all 10 “CA 19-9 negative” patients and 10 out of 12 other patients (83 %) This implies, in a case-based way, that peptide profiling enhanced the sensitivity of detection for these 10 cases. In the validation set, 12 patients had an elevated CA19-9 value of which 11 were correctly classified by peptide profiling. In eight patients, CA19-9 was not increased. Seven of these patients were correctly classified using peptide profiles.
Confounders
Patients that were smokers were correctly classified in the calibration set with 82 %. In the calibration set, 82 % of the patients with diabetes were found positive using peptide profiling versus 74 % of the patients without diabetes. For the validation set, these values were 91 and 96 %, respectively. Patients with a bilirubin level of more than 50 were correctly classified in 18 out of the 21 cases (85 %) in the calibration set versus 11 out of the 17 patients (65 %) with a bilirubin level of less than 50 in the calibration set. In the validation set, these values were 100 and 92 %, respectively.
Discriminating capability compared to imaging methods
In Table 2, the performance of the used imaging methods is displayed. In 79 cases (90 %), imaging could provide a conclusive diagnosis. However, 9 cases (10 %) remained inconclusive after performing the routine diagnostic methods. In four of these cases, RPC18-based peptide profiling could add to correct diagnosis. For all 88 patients, a CT scan was performed and this resulted in 76 positives (sensitivity of 86 %). In 12 cases, malignancy was not diagnosed [initial diagnosis: benign lesion (n = 3), no consensus (n = 4) and no lesion seen (n = 5)]. Seven of these 12 patients (58 %) were correctly classified as malignant using peptide profiling (6 with tumors in the pancreatic head, 1 in the pancreatic tail; 6 females, 1 male; 1 with stage IIA, 5 with stage IIB, 1 with stage IV). In 8 cases, an MRI was performed. In one patient in whom no lesion was found with MRI, peptide profiling also resulted in incorrect classification. Adversely, 6 out of 7 patients with a malignant diagnosis on MRI were classified as positive with peptide profiling. A PET scan was performed for four patients. This resulted in 3 “cases,” with one analysis remaining inconclusive. This latter patient was correctly classified by peptide profiling. In 21 cases, an EUS was part of the diagnostic method. Six patients had an uncertain or negative result, and four of these patients were correctly classified by peptide profiling.
Patients with hereditary risk
All five patients with increased risk were correctly classified with peptide profiling (100 % accuracy). For 3 cases, CA 19-9 values were known, of which two were elevated. In all 5 cases, a CT scan was performed. In 4 cases, this scan resulted in conclusive classification as malignant and the remaining CT scan resulted in an uncertain diagnosis. Two patients received a preoperative MRI; on both scans, a malignant lesion was seen. In one case, an EUS was carried out with inconclusive result.
Structure characterization of discriminating peptide signals
The calibrated discriminating rule was further investigated in an additional, second statistical analysis. The original discriminating rule was used to select a subset of ten peptide peaks which could replicate the above-reported classification results. This resulted in ten peaks with m/z-values of 1,206.7, 1,518.9, 1,536.9, 1,561.9, 1,563.0, 1,865.2, 2,271.1, 2,602.5, 3,190.6 and 3,261.7. The analyses were repeated using only these ten discriminating peaks, which resulted in a sensitivity of 80 % and a specificity of 92 % with an AUC of 95 % in the calibration set. For the validation set, a sensitivity of 92 % and a specificity of 95 % with an AUC of 96 % were found. The structures of the ten discriminating peaks (i.e., peptides) with good separation performance in this study were identified (Table 3), either as fragments of FPA-chain or FGA chain (clotting cascade) (m/z-values at 1,206.7, 1,518.9, 1,536.9, 3,190.6 and 3,261.7—P02671), or as fragments of complement C3 (immunologic response) (m/z-values at 1,563.0 and 1,865.2—P01024), the peak at m/z-value 2,602.5 was identified as Factor XIIIa (clotting cascade) and inter-alpha-trypsin inhibitor (ITIH4) (acute phase reaction) (m/z-values at 2,271.1—Q14624) as overviewed by Tiss and co-workers and others (Erica Gianazza et al. 2012; Shen et al. 2010; Tiss et al. 2010). Note that the signals at m/z-values 1,561.9 and 1,563.0 overlap and can only be resolved by using ultrahigh-resolution Fourier transform ion cyclotron resonance (FTICR) (Nicolardi et al. 2010).
As part of the subsequent exploratory analysis of the calibrated discriminant rule, all ten peaks were tested univariately. In Table 3, the selected peaks from the multivariate analysis are listed together with a two-sample t test (univariate assessment), the corresponding pooled estimate of the population standard deviation, the corresponding p value and the mean confidence interval. In addition, the direction (positive or negative) of the calibrated discriminant weight in the multivariate analysis (expr) is included in Table 3. All peaks have individual univariate significant p values (<0.05). It is important to note the distinction between the univariate and multivariate results, as the univariate measures denote the individual expression differences between cases and controls for each peak separately—without reference to the expression of other peaks, while the multivariate results (discriminant weights) denote the conditional effect associated with each peak, after taking into account the variation in expression across the other peaks selected. Thus, a positive discriminant weight implies a higher case probability for larger values of expression at the corresponding peak—after correction for the variation of other peaks. The reverse applies to controls. As is clear from Table 3, larger expression values for peaks with m/z-values of 1,518.9, 1,536.9, 1,865.2, 2,271.1 and 3,261.7, combined with smaller expression values for peaks with m/z-values of 1,206.7, 1,561.9, 1,563.0, 2,602.5 and 3,190.6, imply greater case probability, and the reverse applies for controls. It is important to note, however, that with respect to the univariate behavior for each peak separately, cases are consistently associated with higher expression as opposed to controls and across all peaks (as may also be seen from the t tests in Table 3 which all have the same sign). The multivariate behavior of the discriminant rule can be better understood by separating the selected peaks—all of which have the said significant univariate effect—into two groups, depending on whether their calibrated discriminant coefficient/weight is positive or negative. As shown above, five of the selected peaks have a positive discriminant weight and five a negative discriminant weight. For each patient, the mean of the expression of peaks with a positive discriminant weight (MEANPOSEXPR) and the mean of the expression of peaks with a negative discriminant weight (MEANNEGEXPR) were calculated. As in the above explanation, it should be expected that samples showing a higher expression of MEANPOSEXPR and comparatively smaller expression on MEANNEGEXPR should be more likely to belong to the group of cases. Likewise, if the situation is the other way around, the probability to belong to the control group increases. This interpretation is confirmed in Fig. 4 which shows a stem and leaf plot in which the difference between MEANPOSEXPR and MEANNEGEXPR is plotted for each sample in the validation set. From this figure, it becomes clear that cases can be separated from controls based on those two summarized measured expressions. To obtain final verification of our interpretation, the discrimination rule was repeated using this MEANPOSEXPR and MEANNEGEXPR summarized expression only. This resulted in a sensitivity of 80 %, a specificity of 82 % and an AUC of 90 % for the calibration set and a sensitivity of 90 %, a specificity of 93 % and an AUC of 97 % for the validation set. The differences between MEANPOSEXPR and MEANNEGEXPR expression were calculated, and a two-sample t test was performed on this contrast that resulted in a t value of 9.76 and a p value of <0.0001 in the calibration set and a t value of 13.87 with a p value of <0.0001 in the validation set.
Stem-and-leave plot of the validation data based on MEANPOSEXPR–MEANNEGEXPR. The blue circles correspond to the cases, the red crosses with the controls. Items with the same expression were plotted vertically. The controls are more located on the left of the horizontal axis; the cases are more located on the right
Discussion
Clinical proteomics approaches have emerged as a powerful tool for the diagnosis of PC and application in surveillance programs (Cecconi et al. 2011; Chen et al. 2009; Liu et al. 2009; Pawa et al. 2010; Zapico-Muniz et al. 2010). In this study, a serum peptide signature is presented for the detection of pancreatic cancer. Serum fractionation was performed with RPC18-MBs, followed by high-resolution MS profiling, in order to develop a discriminating peptide profile for patients with PC, resulting in a high sensitivity of 76 % (95 %) and a specificity of 91 % (100 %) in the calibration and validation sets, respectively. We found an enhanced sensitivity and specificity compared to our previous reported classification results for (small) proteins (molecular weights 3–12 kDa) obtained from a WCX-SPE method, which were 74 and 91 % for the validation set (Velstra et al. 2013). Note that the study population mostly consisted of patients with early-stage PC. Early detection of PC may result in a higher number of patients that might benefit from a pancreaticoduodenectomy and, moreover, in increased survival times. The sensitivity value for the entire set of PC patients obtained from imaging CT scans was 86 %, and from MRI, PET and EUS 75, 75 and 60 %, respectively. Moreover, the MS-based profiling results were compared to CA19-9 readouts, with an overall sensitivity of 65 %. Previously, Goonetilleke and co-workers reviewed CA19-9 as a diagnostic serum marker for PC and found a median sensitivity of 79 % and a specificity of 82 % (Goonetilleke and Siriwardena 2007). In the current study, out of 24 patients with a positive CA19-9 test, 21 patients were classified as “cases” on the basis of peptide profiling results. Out of 18 patients with a negative CA19-9 value, 17 were classified as “cases” based on peptide signatures, thus implying the beneficial aspects of the here presented methodology.
In principle, with current MS-based proteomics approaches, any protein can be detected, but the “real-life” sensitivity of the method is strongly affected by the nature of the sample. Biological samples are typically characterized by a wide range of protein abundances (for proteins in blood, this is more than 10 orders of magnitude), and no analytical method is available that can tackle such dynamic range in one go. Sample fractionation at least in part overcomes this problem and allows a more in-depth analysis of the proteome (Nilsson et al. 2010). In the current study, RPC18-MBs with specific binding characteristics were used. Such paramagnetic beads can well be implemented in a robotic platform for fully automated use, thus ensuring the control of each step in the extraction protocol and minimizing technical variability (Jimenez et al. 2007). Previously, profiles studies were criticized because protein identities were not reported with these “black-box” approaches (Nilsson et al. 2010). As a consequence, emphasis in biomarker research has moved from discovery to structure characterization. We therefore performed additional statistical analyses to further investigate the discriminative rule and to give insight into the identity of the peaks. The classification of PC patient and control samples could be validated even after summarizing the data in the mean of the expression of peaks with a positive discriminant weight (MEANPOSEXPR) and the mean of the expression of peaks with a negative discriminant weight (MEANNEGEXPR). Taking the mean of the profiles with the same direction of expression is only one of the methods that could be used to explore this type of data. It seems logical to first determine the diagnostic power of candidate markers before performing identification studies. Nevertheless, characterization of the discriminating peptides has become a crucial element to allow further investigations into their biological role in disease mechanisms. It should be noted, however, that identification of peptide- or protein signals in a profile is not straightforward. Until now, most reported identifications of serum peptides in profiles have been based on SELDI enrichment chips [i.e., Immobilized Metal Affinity Capture (IMAC) (Koopmann et al. 2004)] or on SPE-based (RPC18) procedures (Koomen et al. 2005; Tiss et al. 2010). Recently, prognostic protein markers of resectable PC were identified by applying MS-based proteomic analysis on tissues (Takadate et al. 2013). The structures of the 10 discriminating peaks, which in this study showed good separation performance, have been elucidated previously (Table 3). The identified peptides were found to originate from proteins that relate to the clotting cascade, or the immunologic response of the acute phase reaction. The clinical value of these peptides requires further investigation. It is known that this profiling method primarily results in the detection of high-abundant proteins and their proteolytic fragments. Fractionation techniques remain crucial for obtaining information on low-abundant peptides and proteins. The here identified proteins could be a result of pancreatitis that accompanies PC development. A very important next step is to analyze the serum proteomic profile in a group of patients with chronic pancreatitis. Any reliable comparison of PC markers requires the use of large number of pancreatitis controls, and we are currently collecting samples.
The most practical and cost-effective clinical advantage would be achieved if patients with an elevated risk to develop pancreatic cancer could benefit from a highly sensitive and specific peptide- or protein profiling serum test (Ghatnekar et al. 2013). With the current peptide-profiling technique, it was possible to correctly classify all five patients with an increased risk for PC. Three of these patients were carrier of a p16-Leiden mutation. A simple screening test to be performed between regular screening moments with MRI might be beneficial for these patients. However, our MS profile has not been tested for such a group with hereditary higher risk. We are now analyzing retrospective series and in addition collect serum samples of individuals belonging to this high-risk group during surveillance. For similar reasons, other risk factors for developing PC such as diabetes and smoking habits will be considered in future analyses. This study can be seen as a second step, following our first step presented earlier (Velstra et al. 2013), in which we have improved the cleaning up procedures. Further technical improvements are ongoing, with promising preliminary results.
In conclusion, the applied MS approach allowed rapid, accurate and reproducible analysis of serum peptides to obtain signatures with a high sensitivity at relatively low cost. Automation ensures a robust peptide isolation protocol. This study demonstrates the potential of a serum peptide signature as a diagnostic method for the detection of pancreatic cancer. This application of this method can be complementary to currently used diagnostic methods in the clinic. Such a test would be beneficial for patients with an inherited risk for PC by allowing more frequent screening.
References
Aebersold R, Mann M (2003) Mass spectrometry-based proteomics. Nature 422:198–207
Albrethsen J (2007) Reproducibility in protein profiling by MALDI-TOF mass spectrometry. Clin Chem 53:852–858
Bipat S, Phoa SS, van Delden OM, Bossuyt PM, Gouma DJ, Lameris JS, Stoker J (2005) Ultrasonography, computed tomography and magnetic resonance imaging for diagnosis and determining resectability of pancreatic adenocarcinoma: a meta-analysis. J Comput Assist Tomogr 29:438–445
Bladergroen MR, Derks RJ, Nicolardi S, de Visser B, van Berloo S, van der Burgt YE, Deelder AM (2012) Standardized and automated solid-phase extraction procedures for high-throughput proteomics of body fluids. J Proteomics 77:144–153
Cecconi D, Palmieri M, Donadelli M (2011) Proteomics in pancreatic cancer research. Proteomics 11:816–828
Chen JH, Ni RZ, Xiao MB, Guo JG, Zhou JW (2009) Comparative proteomic analysis of differentially expressed proteins in human pancreatic cancer tissue. Hepatobiliary Pancreat Dis Int 8:193–200
de Noo ME, Tollenaar RA, Ozalp A, Kuppen PJ, Bladergroen MR, Eilers PH, Deelder AM (2005) Reliability of human serum protein profiles generated with C8 magnetic beads assisted MALDI-TOF mass spectrometry. Anal Chem 77:7232–7241
Deng R, Lu Z, Chen Y, Zhou L, Lu X (2007) Plasma proteomic analysis of pancreatic cancer by 2-dimensional gel electrophoresis. Pancreas 34:310–317
Dewitt J, Devereaux BM, Lehman GA, Sherman S, Imperiale TF (2006) Comparison of endoscopic ultrasound and computed tomography for the preoperative evaluation of pancreatic cancer: a systematic review. Clin Gastroenterol Hepatol 4:717–725
Ehmann M, Felix K, Hartmann D, Schnolzer M, Nees M, Vorderwulbecke S, Bogumil R, Buchler MW, Friess H (2007) Identification of potential markers for the detection of pancreatic cancer through comparative serum protein expression profiling. Pancreas 34:205–214
Ghatnekar O, Andersson R, Svensson M, Persson U, Ringdahl U, Zeilon P, Borrebaeck CA (2013) Modelling the benefits of early diagnosis of pancreatic cancer using a biomarker signature. Int J Cancer 133:2392–2397
Gianazza E, Chinello C, Mainini V, Cazzaniga M, Squeo V, Albo G, Signorini S, Di Pierro SS, Ferrero S, Nicolardi S, van der Burgt YEM, Deelder AM, Magni F (2012) Alterations of the serum peptidome in renal cell carcinoma discriminating benign and malignant kidney tumors. J Proteomics 5:125–140
Goonetilleke KS, Siriwardena AK (2007) Systematic review of carbohydrate antigen (CA 19-9) as a biochemical marker in the diagnosis of pancreatic cancer. Eur J Surg Oncol 33:266–270
Hansson GC, Zopf D (1985) Biosynthesis of the cancer-associated sialyl-Lea antigen. J Biol Chem 260:9388–9392
Honda K, Hayashida Y, Umaki T, Okusaka T, Kosuge T, Kikuchi S, Endo M, Tsuchida A, Aoki T, Itoi T, Moriyasu F, Hirohashi S, Yamada T (2005) Possible detection of pancreatic cancer by plasma protein profiling. Cancer Res 65:10613–10622
Hortin GL (2006) The MALDI-TOF mass spectrometric view of the plasma proteome and peptidome. Clin Chem 52:1223–1237
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011) Global cancer statistics. CA Cancer J Clin 61:69–90
Jimenez CR, El FZ, Knol JC, Hoekman K, Kruyt FA, Giaccone G, Smit AB, Li KW (2007) Automated serum peptide profiling using novel magnetic C18 beads off-line coupled to MALDI-TOF-MS. Proteomics Clin Appl 1:598–604
Kakisaka T, Kondo T, Okano T, Fujii K, Honda K, Endo M, Tsuchida A, Aoki T, Itoi T, Moriyasu F, Yamada T, Kato H, Nishimura T, Todo S, Hirohashi S (2007) Plasma proteomics of pancreatic cancer patients by multi-dimensional liquid chromatography and two-dimensional difference gel electrophoresis (2D-DIGE): up-regulation of leucine-rich alpha-2-glycoprotein in pancreatic cancer. J Chromatogr B Analyt Technol Biomed Life Sci 852:257–267
Kojima K, Asmellash S, Klug CA, Grizzle WE, Mobley JA, Christein JD (2008) Applying proteomic-based biomarker tools for the accurate diagnosis of pancreatic cancer. J Gastrointest Surg 12:1683–1690
Koomen JM, Shih LN, Coombes KR, Li D, Xiao LC, Fidler IJ, Abbruzzese JL, Kobayashi R (2005) Plasma protein profiling for diagnosis of pancreatic cancer reveals the presence of host response proteins. Clin Cancer Res 11:1110–1118
Koopmann J, Zhang Z, White N, Rosenzweig J, Fedarko N, Jagannath S, Canto MI, Yeo CJ, Chan DW, Goggins M (2004) Serum diagnosis of pancreatic adenocarcinoma using surface-enhanced laser desorption and ionization mass spectrometry. Clin Cancer Res 10:860–868
Liu D, Cao L, Yu J, Que R, Jiang W, Zhou Y, Zhu L (2009) Diagnosis of pancreatic adenocarcinoma using protein chip technology. Pancreatology 9:127–135
Lynch SM, Vrieling A, Lubin JH, Kraft P, Mendelsohn JB, Hartge P, Canzian F, Steplowski E, Arslan AA, Gross M, Helzlsouer K, Jacobs EJ, LaCroix A, Petersen G, Zheng W, Albanes D, Amundadottir L, Bingham SA, Boffetta P, Boutron-Ruault MC, Chanock SJ, Clipp S, Hoover RN, Jacobs K, Johnson KC, Kooperberg C, Luo J, Messina C, Palli D, Patel AV, Riboli E, Shu XO, Rodriguez SL, Thomas G, Tjonneland A, Tobias GS, Tong E, Trichopoulos D, Virtamo J, Ye W, Yu K, Zeleniuch-Jacquette A, Bueno-de-Mesquita HB, Stolzenberg-Solomon RZ (2009) Cigarette smoking and pancreatic cancer: a pooled analysis from the pancreatic cancer cohort consortium. Am J Epidemiol 170:403–413
Mertens BJ, de Noo ME, Tollenaar RA, Deelder AM (2006) Mass spectrometry proteomic diagnosis: enacting the double cross-validatory paradigm. J Comput Biol 13:1591–1605
Micames C, Jowell PS, White R, Paulson E, Nelson R, Morse M, Hurwitz H, Pappas T, Tyler D, McGrath K (2003) Lower frequency of peritoneal carcinomatosis in patients with pancreatic cancer diagnosed by EUS-guided FNA vs. percutaneous FNA. Gastrointest Endosc 58:690–695
Navaglia F, Fogar P, Basso D, Greco E, Padoan A, Tonidandel L, Fadi E, Zambon CF, Bozzato D, Moz S, Seraglia R, Pedrazzoli S, Plebani M (2009) Pancreatic cancer biomarkers discovery by surface-enhanced laser desorption and ionization time-of-flight mass spectrometry. Clin Chem Lab Med 47:713–723
Nicolardi S, Palmblad M, Dalebout H, Bladergroen M, Tollenaar RA, Deelder AM, van der Burgt YE (2010) Quality control based on isotopic distributions for high-throughput MALDI-TOF and MALDI-FTICR serum peptide profiling. J Am Soc Mass Spectrom 21:1515–1525
Nilsson T, Mann M, Aebersold R, Yates JR III, Bairoch A, Bergeron JJ (2010) Mass spectrometry in high-throughput proteomics: ready for the big time. Nat Methods 7:681–685
Orlando LA, Kulasingam SL, Matchar DB (2004) Meta-analysis: the detection of pancreatic malignancy with positron emission tomography. Aliment Pharmacol Ther 20:1063–1070
Pawa N, Wright JM, Arulampalam TH (2010) Mass spectrometry based proteomic profiling for pancreatic cancer. JOP 11:423–426
Sener SF, Fremgen A, Menck HR, Winchester DP (1999) Pancreatic cancer: a report of treatment and survival trends for 100,313 patients diagnosed from 1985-1995, using the National Cancer Database. J Am Coll Surg 189:1–7
Shen Y, Tolic N, Liu T, Zhao R, Petritis BO, Gritsenko MA, Camp DG, Moore RJ, Purvine SO, Esteva FJ, Smith RD (2010) Blood peptidome-degradome profile of breast cancer. PLoS ONE 5:e13133
Siegel R, Ward E, Brawley O, Jemal A (2011) Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin 61:212–236
Sun ZL, Zhu Y, Wang FQ, Chen R, Peng T, Fan ZN, Xu ZK, Miao Y (2007) Serum proteomic-based analysis of pancreatic carcinoma for the identification of potential cancer biomarkers. Biochim Biophys Acta 1774:764–771
Takadate T, Onogawa T, Fukuda T, Motoi F, Suzuki T, Fujii K, Kihara M, Mikami S, Bando Y, Maeda S, Ishida K, Minowa T, Hanagata N, Ohtsuka H, Katayose Y, Egawa S, Nishimura T, Unno M (2013) Novel prognostic protein markers of resectable pancreatic cancer identified by coupled shotgun and targeted proteomics using formalin-fixed paraffin-embedded tissues. Int J Cancer 132:1368–1382
Tiss A, Smith C, Menon U, Jacobs I, Timms JF, Cramer R (2010) A well-characterised peak identification list of MALDI MS profile peaks for human blood serum. Proteomics 10:3388–3392
Vasen HF, Gruis NA, Frants RR, van Der Velden PA, Hille ET, Bergman W (2000) Risk of developing pancreatic cancer in families with familial atypical multiple mole melanoma associated with a specific 19 deletion of p16 (p16-Leiden). Int J Cancer 87:809–811
Vasen HF, Wasser M, van MA, Tollenaar RA, Konstantinovski M, Gruis NA, Bergman W, Hes FJ, Hommes DW, Offerhaus GJ, Morreau H, Bonsing BA, De Vostot Nederveen Cappel WH (2011) Magnetic resonance imaging surveillance detects early-stage pancreatic cancer in carriers of a p16-Leiden mutation. Gastroenterology 140:850–856
Velstra B, van der Burgt YE, Mertens BJ, Mesker WE, Deelder AM, Tollenaar RA (2012) Improved classification of breast cancer peptide and protein profiles by combining two serum workup procedures. J Cancer Res Clin Oncol 138:1983–1992
Velstra B, Bonsing BA, Mertens BJ, van der Burgt YE, Huijbers A, Vasen H, Mesker WE, Deelder AM, Tollenaar RA (2013) Detection of pancreatic cancer using serum protein profiling. HPB (Oxford) 15:602–610
Yu KH, Rustgi AK, Blair IA (2005) Characterization of proteins in human pancreatic cancer serum using differential gel electrophoresis and tandem mass spectrometry. J Proteome Res 4:1742–1751
Zapico-Muniz E, Farre-Viladrich A, Rico-Santana N, Gonzalez-Sastre F, Mora-Brugues J (2010) Standardized peptidome profiling of human serum for the detection of pancreatic cancer. Pancreas 39:1293–1298
Acknowledgements
We greatly appreciate the technical assistance from Marco Bladergroen and Hans Dalebout (Center for Proteomics and Metabolomics), Rob Keyzer, Gabi van Pelt and Ronald van Vlierberghe (Department of Surgery), and Linda Verhoeff, Arienne Loor, Annemarie Voet-van den Brink and Richard Zwaan from the Datacenter of the Department of Surgery. Lisa Waller Hayes Research Grant 2011.
Conflict of interest
The authors declare that they have no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Velstra, B., Vonk, M.A., Bonsing, B.A. et al. Serum peptide signatures for pancreatic cancer based on mass spectrometry: a comparison to CA19-9 levels and routine imaging techniques. J Cancer Res Clin Oncol 141, 531–541 (2015). https://doi.org/10.1007/s00432-014-1812-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-014-1812-2