Abstract
Term born infants are predisposed to human rhinovirus (HRV) lower respiratory tract infections (LRTI) by reduced neonatal lung function and genetic susceptibility. Our aim was to investigate whether prematurely born infants were similarly predisposed to HRV LRTIs or any other viral LRTIs. Infants born less than 36 weeks of gestational age were recruited. Prior to neonatal/maternity unit discharge, lung function (functional residual capacity by helium gas dilution and multiple breath washout, lung clearance index and compliance (Crs), and resistance (Rrs) of the respiratory system) was assessed and DNA samples assessed for eight single nucleotide polymorphisms (SNPs) in seven genes: ADAM33, IL10, MMP16 NFκB1A,SFTPC, VDR, and NOS2A. Infants were prospectively followed until 1 year corrected age. Nasopharyngeal aspirates (NPAs) were sent whenever an infant developed a LRTI and tested for 13 viruses. One hundred and thirty-nine infants were included in the analysis. Infants who developed HRV LRTIs had reduced Crs (1.6 versus 1.2 mL/cmH2O/kg, p = 0.044) at 36 weeks postmenstrual age. A SNP in the gene coding for the vitamin D receptor was associated with the development of HRV LRTIs and any viral LRTIs (p = 0.02).
Conclusion: Prematurely born infants may have both a functional and genetic predisposition to HRV LRTIs.
What is Known: • Term born infants are predisposed to rhinovirus lower respiratory tract (HRV LRTIs) infection by reduced neonatal lung function. • Term born infants requiring hospitalisation due to HRV bronchiolitis were more likely to have single nucleotide polymorphism (SNP) in the IL-10 gene. |
What is New: • Prematurely born infants who developed a HRV LRTI had lower C rs before maternity unit discharge. • A SNP in the gene coding for the vitamin D receptor was associated with the development of HRV LRTIs and overall respiratory viral LRTIs in prematurely born infants. |
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Human rhinoviruses (HRV) are the most common cause of respiratory tract infection in infants, with almost all infants developing at least one HRV infection in the first year after birth [14, 23]. Both term and prematurely born infants are susceptible to developing LRTIs caused by HRV [3, 11, 13, 21, 24]. Some term born infants may be predisposed to wheezy HRV LRTIs by reduced neonatal lung function [22]. The adjusted risk of developing a wheezy HRV LRTI in the first year of life was 1.8 times higher for each standard deviation increase of airway resistance (Rrs) measured at 2 months of age [22]. In addition, some term born infants may be genetically predisposed to HRV infection. Infants developing HRV bronchiolitis requiring hospitalisation at less than 6 months of age were more likely to have a single nucleotide polymorphism (SNP) in the IL-10 gene compared to unselected blood donors [9]. Other SNPs in genes coding for IL-18, TLR4, and IFN-γ did not confer susceptibility to hospitalisation for HRV infection [9]. The aim of this study was to determine whether prematurely born infants were functionally and genetically predisposed to HRV LRTIs. An additional aim was to determine whether prematurely born infants were functionally and genetically predisposed overall to respiratory viral LRTIs.
Materials and methods
Analysis was undertaken of the results of infants entered into a study investigating the risk factors for viral LRTIs in prematurely born infants [7]. Infants were eligible for recruitment into the study if they were born prior to the onset of the RSV season (1st October to 31st March in the UK) in 2008 or 2009 and were born at less than 36 weeks of completed gestation. Ethical approval was obtained from King’s College Hospital NHS Foundation Trust Research Ethics Committee.
Prior to neonatal/maternity unit discharge, either blood or buccal swabs were obtained from infants and stored at −20 °C until tested. The samples were then sent on dry ice to the National Institute for Public Health and the Environment (RIVM) in Bilthoven, The Netherlands for testing. DNA was isolated from the blood samples or buccal swabs and then stored at −20 °C at the RIVM until analysed [6]. Eight single nucleotide polymorphisms (SNPs) were chosen to be tested. The chosen SNPs had previously been associated with HRV infection in term born infants less than 6 months old [9]. Nuclear factor-κ-B activity has been associated with steroid resistant airway epithelium in HRV infection in vitro and thus the SNP NFκB1A rs2233409 was also included [15]. In addition, we have studied SNPs associated with reduced lung function in previously healthy children at 3 and 5 years of age [20], RSV infection in prematurely born infants [19], prematurity [10], or bronchopulmonary dysplasia (BPD) [8]. We also included SNPs associated with RSV infection in prematurely born infants as they may be associated with other viral causes of bronchiolitis (i.e., HRV) in prematurely born infants.
The extracted DNA samples were diluted with TE Buffer to 7 ng/μL and sent to KBioscience (Herts, UK) for genotyping. Six SNPs (ADAM33 rs2787094, IL10 rs1800872, MMP16 rs2664349, MMP16 rs2664352, NFκB1A rs2233409, and SFTPC rs1124) were tested at KBioscience with the KASPar technology and two further SNPs (vitamin D receptor [VDR rs10735810] and nitric oxide synthase 2 A [NOS2A rs1060826]) were tested at the RIVM in the Netherlands. Genotyping of VDR rs10735810 was performed by a custom TaqMan SNP genotyping assay (Applied Biosystems, Carlsbad, USA), and genotyping of NOS2A rs1060826 was performed by using TaqMan SNP genotyping assay C_9458082_10. Genotyping of both SNPs tested at the RIVM was carried out on a 7500 Fast Real-Time PCR system (Applied Biosystems) as previously described [6]. The genotype distributions of the eight SNPs were in Hardy-Weinberg equilibrium [6].
Lung function was assessed at 36 weeks postmenstrual age (PMA) whilst infants were still inpatients on the neonatal or maternity unit. Infants were not sedated or ventilated during lung function testing. Lung volume was assessed by the measurement of functional residual capacity (FRCHe), using a commercially available helium gas dilution system (EBS 2615, Equilibrated Bio Systems, New York) as previously described [5]. Lung volume was also assessed by the measurement of FRC (FBCMBW) using the commercially available open circuit multiple breath wash-in/out system (Exhalyzer D, Ecomedics, Duernten, Switzerland) and using sulphur hexafluoride as a tracer gas as previously described [6]. The MBW technique also measures ventilation inhomogeneity (VI), measured as lung clearance index (LCI) as previously described [6]. Compliance (Crs) and resistance (Rrs) of the respiratory system were measured using the single breath occlusion technique as previously described [7].
Following neonatal or maternity unit discharge, infants were followed prospectively until 1 year corrected age. Whenever an infant developed an LRTI, regardless of whether the child remained at home or required hospitalisation, a nasopharyngeal aspirate (NPA) was taken. An infant was diagnosed with a viral LRTI if they had coryzal symptoms together with a respiratory examination demonstrating either a raised respiratory rate for their age, crackles or wheeze or respiratory distress (e.g., tracheal tug or intercostal or subcostal recession). NPAs were tested for 11 viruses (rhinovirus, RSV A and B, human metapneumovirus, influenza A and B, parainfluenza 1–3, enterovirus and parechovirus) using real-time reverse transcription polymerase chain reaction (PCR) and for adenovirus and bocavirus using real time PCR as previously described [5].
The neonatal notes were reviewed to document demographic and clinical data and to document the duration of the infants’ admission on the neonatal and/or maternity unit. Antenatal, perinatal, and postnatal data collected included that on maternal infections, antenatal steroid use, use of surfactant, duration of respiratory support, development of bronchopulmonary dysplasia (BPD), postnatal infant sepsis, breast/formula feeding, and use of palivizumab [7].
Statistical analysis
The infants were divided into two groups depending on their HRV LRTI status. The “no LRTI group” consisted of infants who did not develop a viral LRTI throughout the study period and the “HRV LRTI group” consisted of infants who developed at least one HRV LRTI during the study period. The infants in the HRV LRTI group may also have had other viral LRTIs. We also undertook a subsidiary analysis of all infants who had LRTIs with NPAs positive for respiratory viruses and compared their outcomes to infants who had no LRTI. Infants who had LRTIs but no virus was detected from the NPA were excluded from the analysis.
Data were tested for normality using the Shapiro-Wilk test. Data were analysed using either the independent T test, the Mann-Whitney U test, the Chi-squared test, or the Fisher’s exact test as appropriate. A multivariable regression model was used to examine whether lung function at 36 weeks PMA was a predictor of HRV LRTI or respiratory viral LRTIs, independent of other variables which in the univariate analysis were significant at p ≤ 0.1. Statistical analysis was carried out with IBM SPSS Statistics (version 19, New York, USA).
Sample size
A sample size of 28 infants in each group allowed the detection of a difference in the premorbid lung function results equivalent to one standard deviation, with 90 % power and two-sided 5 % significance. A previous study [2] demonstrated a significant difference in lung function (Rrs) equivalent to one standard deviation between the groups.
Results
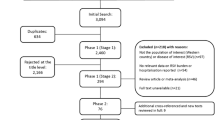
During the study period, 251 infants met the eligibility criteria for recruitment into the study (Fig. 1). One hundred and thirty-nine infants were included in the overall analysis. Their median gestational age (GA) was 34 (range 23–35) weeks and median birth weight 1904 (range 610–3610) g. Four infants received palivizumab of which one was admitted to hospital due to an RSV LRTI. There were significant differences when comparing the demographic data of the HRV group and the no LRTI group. The HRV group were more immature and lighter at birth, more received surfactant, had a longer duration of supplemental oxygen, developed BPD, received palivizumab, developed postnatal sepsis, and had a longer duration of neonatal/maternity unit stay (Table 1). Comparison of those infants who developed any respiratory virus LRTI compared to no LRTI is shown in Appendix Table 1. Some infants developed more than one viral LRTI or had more than one virus detected from an NPA during a HRV LRTI (Table 2).
Eight (25 %) infants in the HRV LRTI group required hospitalisation (six due to a viral LRTI [two HRV]), one due to a minor head injury and one due to gastroenteritis. Nine (12 %) infants in the no LRTI group required hospitalisation (all due to non-respiratory causes).
The HRV LRTI group were more immature (36 versus 37 weeks PMA, p = 0.031) and of lower weight (1908 versus 2113 g, p = 0.007) when their lung function was measured. The HRV LRTI group had a smaller FRCHe uncorrected for weight (p = 0.004), although this was no longer significantly different after correcting for weight (p = 0.13), a smaller FRCMBW uncorrected for weight (p = 0.001) which remained significantly different when corrected for weight (p = 0.042), a lower Crs uncorrected for weight (p = 0.001) which remained significantly different when corrected for weight (p = 0.005) and a higher Rrs (p = 0.028) (Table 3). Multivariate analysis revealed that after correcting for significant differences in the demographic data the only difference in lung function between the groups that remained significant was in the Crs corrected for weight (Table 3). There were no significant differences in the lung function results of the infants who had any respiratory virus LRTI compared to those who had no LRTI after correcting for differences in their demographics (Appendix Table 2).
There were no significant differences at the genotype level in any of the SNPs between the HRV LRTI and no LRTI groups (data not shown). There was a significant difference in the SNP (rs10735810) in the VDR gene at the allele level. Infants with the G allele were significantly more likely (OR 2.07, 95 % CI 0.98–3.13, p = 0.047) to develop HRV LRTIs than those with the A allele (Table 4). Similarly, there was a significant difference in the SNP in the VDR gene at the allele level between infants who did and did not develop a respiratory viral LRTI (p = 0.02) (Appendix Table 3).
Discussion
We have demonstrated that prematurely born infants who developed HRV LRTIs had reduced premorbid lung function that is they had significantly lower Crs than those who did not develop an HRV LRTI. In addition, a SNP in the G allele of the vitamin D receptor gene was associated with an increased risk of developing HRV LRTIs and respiratory viral LRTIs overall.
Term born infants have been shown to have reduced lung function prior to developing HRV LRTIs [22]. Although, in that study overall, there were no significant differences in lung function between the infants who did and did not develop an HRV infection, those infants who wheezed with an HRV infection had significantly reduced lung function (Crs and Rrs) compared with those infants who had HRV infections but did not wheeze [22]. In this study, initial analysis demonstrated several differences in lung function between infants who did and did not develop an HRV LRTI. After adjusting for differences in the demographic data, however, the only significant difference that remained was in Crs corrected for weight. A possible explanation is that infants with a low Crs may have less lung distensibility leading to poorer clearance of respiratory secretions. In term born infants, a reduced Crs was associated with an increased susceptibility to hospitalisation with RSV LRTIs as well as post RSV bronchiolitis wheezing [25]. Although there were significant differences in lung function between the all virus LRTI and the no LRTI group, these disappeared after adjusting for confounding factors.
Vitamin D deficiency has been associated with an increased risk of developing viral LRTIs in infants, in particular RSV LRTIs [1, 18]. In addition, SNPs in the VDR gene have been associated with severe RSV bronchiolitis and other viral LRTIs in infants [12, 17] but no previous study has investigated the role of the VDR in HRV infection. In this study, a SNP in the gene coding for VDR was associated with the development of HRV LRTIs in prematurely born infants and, in addition, in infants overall with respiratory viral LRTIs. Vitamin D has an important role in innate immunity [4]. It is thus plausible that defects in the VDR will increase an infant’s susceptibility to HRV infections. Only one previous study [9] has investigated the genetic susceptibility of infants to HRV infection. In that study [9], term born infants with the A allele of a SNP (at −1082) in the gene coding for IL-10 were more likely to be hospitalised for HRV bronchiolitis at less than 6 months of age than those with the G allele. In this study, a different SNP (rs10735810) in the IL-10 gene was not associated with HRV LRTI. The difference in those results suggests that genetic susceptibility to HRV infection is different in term and prematurely born infants. The other SNPs tested in this study have been associated with severe RSV infection, prematurity, or BPD in prematurely born infants but not HRV infection and did not appear to influence the development of HRV LRTIs, suggesting they may not have a role in prematurely born infants’ susceptibility to HRV LRTI. We also did not find any significant association between the SNPs tested and respiratory viral infections overall. The numbers of infants with each viral infection precluded subanalysis at that level.
The current study has strengths and some weaknesses. A large cohort of prematurely born infants from a variety of ethnic backgrounds was prospectively followed. Lung function was assessed before neonatal or maternity unit discharge that is prior to any of the infants being infected with any viral infection. The wide range of ethnicities in the study may have affected the results, as genotype differences in various ethnic groups may increase the likelihood of associations occurring by chance [16]. No correction was made for multiple testing of the genetics data; it is, therefore, possible the significant differences we demonstrate with respect to VDR could be attributable to chance. Nevertheless, we demonstrate a significant relationship not only with HRV LRTIs but also to any respiratory viral LRTIs. Although infants born at less than 36 weeks GA were eligible for entry into the study, most of the infants recruited were born moderately prematurely (median gestational age 34 weeks) and thus the results of this study may not be generalisable to all infants born extremely prematurely.
In conclusion, prematurely born infants may be predisposed to HRV LRTIs by both reduced premorbid lung function and genetic susceptibility. A SNP in the gene coding for VDR was associated with the development overall of respiratory viral LRTIs.
Authors’ Contributions
AG, SLJ, and LB designed the study. MS and MZ undertook the virological analyses. SBD undertook the lung function assessments. MA, TW, and SBD were responsible for the follow up of the patients. SBD, HMH, RJ, and LB undertook the genetic analyses. All authors were involved in the preparation of the manuscript.
Abbreviations
- BPD:
-
Bronchopulmonary dysplasia
- Crs :
-
Compliance of the respiratory system
- FRCHE :
-
Functional residual capacity (by helium gas)
- HRV:
-
Rhinovirus
- LCI:
-
Lung clearance index
- LRTI:
-
Lower respiratory tract infection
- NPA:
-
Nasopharyngeal aspirates
- PCR:
-
Polymerase chain reaction
- PMA:
-
Postmenstrual age
- Rrs :
-
Resistance of the respiratory system
- RSV:
-
Respiratory syncytial virus
- SNP:
-
Single nucleotide polymorphisms
- VDR:
-
Vitamin D receptor
References
Belderbos ME, Houben ML, Wilbrink B, Lentjes E, Bloemen EM, Kimpen JL, Rovers M, Bont L (2011) Cord blood vitamin D deficiency is associated with respiratory syncytial virus bronchiolitis. Pediatrics 127:e1513–e1520
Broughton S, Bhat R, Roberts A, Zuckerman M, Rafferty GF, Greenough A (2006) Diminished lung function, RSV infection, and respiratory morbidity in prematurely born infants. Arch Dis Child 91:26–30
Chidekel AS, Rosen CL, Bazzy AR (1997) Rhinovirus infection associated with serious lower respiratory illness in patients with bronchopulmonary dysplasia. Pediatr Infect Dis J 16:43–47
Chun RF, Liu PT, Modlin RL, Adams JS, Hewison M (2014) Impact of vitamin D on immune function: lessons learned from genome-wide analysis. Front Physiol 5:151
Drysdale SB, Lo J, Prendergast M, Alcazar M, Wilson T, Zuckerman M, Smith M, Broughton S, Rafferty GF, Peacock JL, Johnston SL, Greenough A (2014) Lung function of preterm infants before and after viral infections. Eur J Pediatr 173:1497–1504
Drysdale SB, Prendergast M, Alcazar M, Wilson T, Smith M, Zuckerman M, Broughton S, Rafferty GF, Johnston SL, Hodemaekers HM, Janssen R, Bont L, Greenough A (2014) Genetic predisposition of RSV infection-related respiratory morbidity in preterm infants. Eur J Pediatr 173:905–912
Drysdale SB, Wilson T, Alcazar M, Broughton S, Zuckerman M, Smith M, Rafferty GF, Johnston SL, Greenough A (2011) Lung function prior to viral lower respiratory tract infections in prematurely born infants. Thorax 66:468–473
Hadchouel A, Decobert F, Franco-Montoya M-L, Halphen I, Jarreau PH, Boucherat O, Martin E, Benachi A, Amselem S, Bourbon J, Danan C, Delacourt C (2008) Matrix metalloproteinase gene polymorphisms and bronchopulmonary dysplasia: identification of MMP16 as a new player in lung development. PLoS One 3:e3188
Helminen M, Nuolivirta K, Virta M, Halkosalo A, Korppi M, Vesikari T, Hurme M (2008) IL-10 gene polymorphism at −1082 A/G is associated with severe rhinovirus bronchiolitis in infants. Pediatr Pulmonol 43:391–395
Lahti M, Marttila R, Hallman M (2004) Surfactant protein C gene variation in the Finnish population—association with perinatal respiratory disease. Eur J Hum Genet 12:312–320
Lemanske RF, Jackson DJ, Gangnon RE, Evans MD, Li Z, Shult PA, Kirk CJ, Reisdorf E, Roberg KA, Anderson EL, Carlson-Dakes KT, Adler KJ, Gilbertson-White S, Pappas TE, Dasilva DF, Tisler CJ, Gern JE (2005) Rhinovirus illnesses during infancy predict subsequent childhood wheezing. J Allergy Clin Immunol 116:571–577
McNally JD, Sampson M, Matheson L, McNally JD, Sampson M (2014) Vitamin D receptor (VDR) polymorphisms and severe RSV bronchiolitis: a systematic review and meta-analysis. Pediatr Pulmonol 49:790–799
Miller EK, Bugna J, Libster R, Shepherd BE, Scalzo PM, Acosta PL, Hijano D, Reynoso N, Batalle JP, Coviello S, Klein MI, Bauer G, Benitez A, Kleeberger SR, Polack FP (2012) Human rhinoviruses in severe respiratory disease in very low birth weight infants. Pediatrics 129:e60–e67
Müller L, Mack I, Tapparel C, Kaiser L, Alves MP, Kieninger E, Frey U, Regamey N, Latzin P (2015) Human rhinovirus types and association with respiratory symptoms during the first year of life. Pediatr Infect Dis J 34:907–909
Papi A, Contoli M, Adcock IM, Bellettato C, Padovani A, Casolari P, Stanciu LA, Barnes PJ, Johnston SL, Ito K, Caramori G (2013) Rhinovirus infection causes steroid resistance in airway epithelium through nuclear factor κb and c-Jun N-terminal kinase activation. J Allergy Clin Immunol 132:1075–1085 e6
Park J, Hwang S, Lee YS, Kim SC, Lee D (2007) SNP@ethnos: a database of ethnically variant single-nucleotide polymorphisms. Nucleic Acids Res 35:D711–D715
Roth DE, Jones AB, Prosser C, Robinson JL, Vohra S (2008) Vitamin D receptor polymorphisms and the risk of acute lower respiratory tract infection in early childhood. J Infect Dis 197:676–680
Roth DE, Shah R, Black RE, Baqui H (2010) Vitamin D status and acute lower respiratory infection in early childhood in Sylhet, Bangladesh. Acta Paediatr 99:389–393
Siezen CLE, Bont L, Hodemaekers HM, Ermers MJ, Doornbos G, Van’t Slot R, Wijmenga C, Houwelingen HC, Kimpen JL, Kimman TG, Hoebee B, Janssen R (2009) Genetic susceptibility to respiratory syncytial virus bronchiolitis in preterm children is associated with airway remodeling genes and innate immune genes. Pediatr Infect Dis J 28:333–335
Simpson A, Maniatis N, Jury F, Cakebread JA, Lowe LA, Holgate ST, Woodcock A, Ollier WE, Collins A, Custovic A, Holloway JW, John SL (2005) Polymorphisms in a disintegrin and metalloprotease 33 (ADAM33) predict impaired early-life lung function. Am J Respir Crit Care Med 172:55–60
Tran DN, Trinh QD, Pham NTK, Pham TM, Ha MT, Nguyen TQ, Okitsu S, Shimizu H, Hayakawa S, Mizuguchi M, Ushijima H (2015) Human rhinovirus infections in hospitalized children: clinical, epidemiological and virological features. Epidemiol Infect:1–9
van der Zalm MM, Uiterwaal CSPM, Wilbrink B, Koopman M, Verheij TJ, van der Ent CK (2011) The influence of neonatal lung function on rhinovirus-associated wheeze. Am J Respir Crit Care Med 183:262–267
van der Zalm MM, Wilbrink B, van Ewijk BE, van Ewijk BE, Overduin P, Wolfs TF, van der Ent CK (2011) Highly frequent infections with human rhinovirus in healthy young children: a longitudinal cohort study. J Clin Virol 52:317–320
van Piggelen RO, van Loon AM, Krediet TG, Verboon-Maciolek M (2010) Human rhinovirus causes severe infection in preterm infants. Pediatr Infect Dis J 29:364–365
Zomer-Kooijker K, Uiterwaal CSPM, van der Gugten AC, van der Gugten AC, Wilbrink B, Bont LJ, van der Ent CK (2014) Decreased lung function precedes severe respiratory syncytial virus infection and post-respiratory syncytial virus wheeze in term infants. Eur Respir J 44:666–674
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Funding
SBD was supported by the National Institute for Health Research (NIHR) Biomedical Research Centre at Guy’s and St Thomas’ NHS Foundation Trust/King’s College London. The research nurses (MA and TW) were supported by Abbott Laboratories. SLJ is supported by the Asthma UK Clinical Chair CH11SJ, and ERC FP7 Advanced grant 233015. SLJ and AG are MRC and Asthma UK Centre in Allergic Mechanisms of Asthma Investigators, supported by MRC Centre Grant G1000758. SLJ is an NIHR Senior Investigator.
Conflict of interest
The authors declare that they have no conflict of interest.
Informed consent
Infants whose parents gave informed written consent were recruited.
Additional information
Communicated by Patrick Van Reempts
Rights and permissions
About this article
Cite this article
Drysdale, S.B., Alcazar, M., Wilson, T. et al. Functional and genetic predisposition to rhinovirus lower respiratory tract infections in prematurely born infants. Eur J Pediatr 175, 1943–1949 (2016). https://doi.org/10.1007/s00431-016-2780-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00431-016-2780-0