Abstract
Primary gastrointestinal neuroendocrine carcinoma (GI-NEC) cannot be distinguished morphologically from pulmonary neuroendocrine carcinoma (P-NEC). This can present a significant diagnostic challenge in cases where site of origin cannot be readily determined. To identify immunohistochemical (IHC) markers that can be used to reliably distinguish between GI-NECs and P-NECs, we constructed 3-mm tissue microarrays, one containing 13 GI-NECs and one containing 20 P-NECs. IHC was performed on both microarrays using 21 stains: AE1/AE3, CK7, CK20, synaptophysin, chromogranin, CD56, INSM1, SSTR2A, CDX2, SATB2, TTF1, Napsin A, PR, GATA3, PAX8, ISL1, beta-catenin, AFP, SMAD4, Rb, and p53. For GI-NEC, the most strongly expressed marker was synaptophysin (mean H-score 248), while AE1/AE3 was the most strongly expressed in P-NEC (mean H-score 230), which was stronger than in GI-NEC (p = 0.011). Other markers that were stronger overall in P-NEC than in GI-NEC included CK7 (p < 0.0001) and TTF1 (p < 0.0001). Markers that were stronger overall in GI-NEC than in P-NEC included SSTR2A (p = 0.0021), SATB2 (p = 0.018), CDX2 (p = 0.019), and beta-catenin (nuclear; p = 0.029). SMAD4, Rb, and p53 showed similar rates of abnormal protein expression. Based on these results, a stepwise algorithmic approach utilizing CK7, TTF1, beta-catenin, CDX2, and SSTR2A had a 91% overall accuracy in distinguishing these GI-NEC from P-NEC. This was tested on a second cohort of 10 metastatic GI-NEC and 10 metastatic P-NEC, with an accuracy in this cohort of 85% and an overall accuracy of 89% for the 53 cases tested. Our algorithm reasonably discriminates GI-NEC from P-NEC using currently available IHC stains.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Neuroendocrine neoplasms can broadly be divided into well-differentiated neuroendocrine tumors (NETs) and poorly differentiated neuroendocrine carcinomas (NECs), with the latter made up of small cell neuroendocrine carcinoma and large cell neuroendocrine carcinoma (LCNEC). The two most common organ systems for development of neuroendocrine neoplasms are the digestive system and the respiratory system [1]. In the former, well-differentiated tumors are more common in most organs, though NEC can occur. In the latter, well-differentiated tumors (carcinoids and atypical carcinoids) are less common than NEC due to the high prevalence of small cell lung carcinoma (SCLC); LCNEC can also occur in the lung but is far less common.
Gastrointestinal NECs (GI-NECs), though rare, are very aggressive; 60–80% patients with GI-NEC have distant metastatic disease at the time of presentation [2,3,4,5]. Median survival is only 1 month without chemotherapy, compared to 11–14 months with palliative chemotherapy [3, 5,6,7].
SCLC, which is seen almost solely in smokers, is the preferred term for pulmonary small cell NEC and currently makes up 15–25% of invasive lung cancer worldwide; approximately 45,000 new cases are diagnosed annually in the USA alone [8, 9]. It has a rapid doubling time [10], with a more aggressive clinical course than non-SCLC [11, 12]. Like GI-NEC, it is often metastatic at presentation [13, 14]. LCNEC, in contrast, accounts for approximately 3% of lung malignancies in the USA [15]; it also has a higher incidence in smokers [16, 17]. Overall, the lungs are the most common sites for primary NEC to occur, with the digestive system coming in second [18].
Histologically, NEC from the gastrointestinal tract and the lung appear essentially identical, with sheets of high-grade neuroendocrine cells demonstrating abundant mitoses and areas of necrosis. Furthermore, as noted, both GI-NECs and pulmonary NECs (P-NEC) metastasize early. As a result, some patients may present with the metastatic foci representing the first-encountered or most obvious site of disease. In situations where these foci are sampled for histologic evaluation, pathologists may initiate workup to attempt to identify site of origin. This is becoming increasingly important, as site of origin is emerging as a relevant factor in selecting oncologic treatment regimens for NECs. However, relatively few immunohistochemical (IHC) markers have been interrogated for their ability to reliably distinguish between GI-NECs and P-NECs. Indeed, it has been stated that “immunohistochemistry has a more limited role in assigning NEC site of origin” [19]. Prior reports have indicated that TTF1 is a reliable, though variable, marker for P-NEC [20], while CDX2 and SATB2 can reportedly be useful to diagnose GI-NEC [17, 21]. Our study sought to confirm and expand these findings by performing and comparing a large number of IHC stains on a series of GI-NEC and P-NEC.
Material and methods
Following approval by the Institutional Review Board (Dana Farber Cancer Institute, Office for Human Research Studies, 19-085, 27/2/2019), primary GI-NECs and P-NECs were identified from the surgical pathology archives of the Beth Israel Deaconess Medical Center. Mixed neuroendocrine–non-neuroendocrine neoplasms were excluded from this search. Original glass slides were reviewed, the diagnoses were confirmed (based on histologic appearance, IHC expression profile, and lack of other potential primary site), and the most representative tumor block was identified from each case. Cases excluded after review included multiple grade 3 pancreatic NETs, two pancreas NEC biopsies with insufficient remaining tissue, one SCLC metastatic to the pancreas, and one colonic medullary carcinoma with spurious synaptophysin positivity (negative upon repeat staining).
Two tissue microarrays (TMAs) were constructed with the EZ-TMA Manual Tissue Microarray Kit 3—3 mm × 24 Core (IHC World, LLC, Ellicott City, MD), one containing 13 confirmed GI-NEC and the other containing 20 confirmed P-NEC. As NEC cases are often sampled via biopsy, resulting in a small amount of diagnostic tissue (as were many cases in this study), we included one 3-mm core from each case in the TMAs; however, in 4 GI-NECs and 1 P-NEC with relatively scant available tissue, two cores were included in order to ensure lesional tissue remained present in the resultant TMA sections.
In order to identify potential IHC markers that could help distinguish GI-NEC from P-NEC, the following 21 IHC stains were performed on 4-micron sections cut from both TMAs: AE1/AE3 cocktail (expressed in most carcinomas [22]); CK7 and CK20 (useful in carcinoma localization [22]); synaptophysin, chromogranin, CD56, and INSM1 (neuroendocrine markers [22]); SSTR2A (variably expressed in NETs [22]); CDX2, SATB2, TTF1, Napsin A, PR, GATA3, PAX8, AFP, and ISL1 (often considered site- or system-specific markers [22]); and beta-catenin, SMAD4, Rb, and p53 (often correlating to genetic mutations [22, 23]). Antibody details are provided in Table 1. Staining protocol was as follows:
-
Staining for AE1/AE3, CK7, CK20, synaptophysin, chromogranin, CD56, CDX2 (initial stain), TTF1, PR, GATA3, Rb, and p53 was performed using the Leica Bond III automated staining platform with the Leica Refine detection kit (Leica Biosystems, Wetzlar, Germany).
-
Staining for INSM1, SSTR2A, SATB2, PAX8, ISL1, and SMAD4 was performed using the Dako Envision Plus detection system (Agilent Technologies, Santa Clara, CA, USA).
-
Staining for CDX2 (repeat stain), Napsin A, beta-catenin, and AFP was performed using the Dako Envision Flex HRP detection system (Agilent).
AE1/AE3, CK7, CK20, synaptophysin, chromogranin, CD56, INSM1, SSTR2A, CDX2, SATB2, TTF1, Napsin A, PR, GATA3, PAX8, ISL1, beta-catenin (nuclear only), and AFP expression were evaluated for intensity (0–3+) and percent of corresponding intensity in each TMA core, and an overall H-score was calculated for each: 3*(percent 3+) + 2*(percent 2+) + 1*(percent 1+). While there is no generally accepted universal H-score cutoffs for positivity and negativity, due to variation among stains and among labs (Dr. Andrew M. Bellizzi, personal communication), for the purposes of this study, a cutoff of 50 was used (namely, 0–49 was interpreted as negative). SMAD4 and Rb were assessed for complete loss of nuclear staining, whereas p53 was assessed for mutant-pattern (strong and diffuse, or null) staining. H-scores were compared between GI-NEC and P-NEC using the t-test, and rates of normal versus abnormal staining for SMAD4, Rb, and p53 were compared using Fisher’s exact test on a 2 × 2 contingency table, with statistical significance set at p < 0.05. All statistical tests were performed using GraphPad online (http://graphpad.com/quickcalcs, GraphPad Software, San Diego, CA, USA, last accessed 3/9/2021).
Once potentially useful markers were identified in the cohort above, 20 additional cases of metastatic NEC foci were identified to create a second, validation cohort. These included 10 metastatic GI-NEC and 10 metastatic P-NEC (based on clinical history and imaging). Four-micron whole-section unstained tissue slides from these cases were stained for CK7, TTF1, beta-catenin, CDX2 (Agilent), and SSTR2A as above, and an H-score was determined for each slide. Statistical analysis was performed on this cohort in the same manner as for the first cohort.
Results
The GI-NECs included 6 small cell and 7 large cell NECs, from the esophagus (n = 2), stomach (n = 1), small bowel (n = 4), and colorectum (n = 6). The P-NECs included 7 small cell carcinomas and 13 large cell NECs. All 33 cases expressed at least one neuroendocrine marker (synaptophysin, chromogranin, CD56, INSM1); among these, synaptophysin had the strongest staining and INSM1 the weakest staining overall. For the five cases with two cores in the TMA, the higher H-score expression was included, but the two numbers were almost always very similar within a case. Only one GI case yielded a situation where the two core H-scores were < 50 and > 50 (for AE1/AE3 and CD56; the “negative” core showed very little lesional tissue).
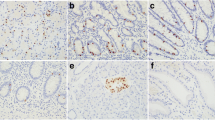
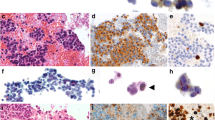
IHC expression for the 13 primary GI-NEC and 20 primary P-NEC is summarized in Table 2 (leftmost columns). The most strongly expressed IHC markers among the 13 GI-NECs (Fig. 1a) included synaptophysin (Fig. 1b), chromogranin, AE1/AE3, CD56, SSTR2A (Fig. 1c), SATB2 (Fig. 1d), and nuclear beta-catenin (Fig. 1e). CDX2 expression was low in this cohort (mean H-score 43; Fig. 1f), on both on the original CDX2 stain and on a confirmatory CDX2 stain using a different antibody. No cases were positive for TTF1, PR, Napsin A, or AFP. Only one case showed abnormal loss of SMAD4 expression, compared to 6 with abnormal Rb expression and 10 with abnormal p53 expression. The most strongly expressed IHC markers among the 20 P-NECs (Fig. 2a) included AE1/AE3 (Fig. 2b), CK7 (Fig. 2c), TTF1 (Fig. 2d), synaptophysin, chromogranin, and CD56. Napsin A was only convincingly positive in one case. No cases were positive for PR, CK20, CDX2, or AFP. Only one case showed abnormal loss of SMAD4 expression, compared to 11 with abnormal Rb expression and 18 with abnormal p53 expression.
a This colonic neoplasm showed H&E features compatible with a large cell neuroendocrine carcinoma. b All GI-NEC were positive for at least one neuroendocrine marker, confirming the diagnosis. Synaptophysin is shown here. c SSTR2A was positive in most GI-NEC in our study. d Similarly, SATB2 was positive in most GI-NEC. e Nuclear beta-catenin staining was often seen in GI-NEC. f CDX2 was positive in this GI-NEC, but overall was negative in most
a This lung neoplasm was readily diagnosable as a small cell neuroendocrine carcinoma on H&E. Neuroendocrine markers (e.g., synaptophysin) were positive to confirm. b AE1/AE3 cocktail was strikingly positive in this and most P-NEC. GI-NEC was on average less strikingly positive. c CK7 was positive in this and most P-NEC. d Similarly, TTF1 was positive in this and most P-NEC
Markers with significantly different mean H-scores between primary GI-NEC and primary P-NEC included AE1/AE3 (149 vs. 230, p = 0.011), CK7 (30 vs. 209, p < 0.0001), TTF1 (1 vs. 177, p < 0.0001), SSTR2A (123 vs. 14, p = 0.0021), SATB2 (106 vs. 35, p = 0.018), CDX2 (43 vs. 0, p = 0.019), and nuclear beta-catenin (106 vs. 23, p = 0.029).
SSTR2A expression was highest in foregut GI-NEC (n=3), with a mean H-score of 257, which was significantly higher than that in midgut (n = 3; mean H-score of 13, p = 0.002). SSTR2A levels were comparable between foregut and hindgut (n = 7), and between midgut and hindgut. There were no significant differences in SATB2 expression levels across foregut, midgut, and hindgut. We also compared H-score expression of all the markers in small cell vs. large cell GI-NEC and in small cell vs. large cell P-NEC. SATB2 demonstrated higher expression in large cell carcinoma in both the GI tract (median H-score 176, vs. 8 for small cell, p = 0.0017) and the lung (median H-score 54, vs. 0 for small cell, p = 0.0498); other markers showed no significant difference (all p > 0.05).
Based on the above findings and on relative antibody availability (i.e., most labs offer CK7, TTF1, CDX2, and beta-catenin, while SSTR2A is offered in fewer labs), we created a three-step algorithmic approach to IHC workup of NEC (Fig. 3). In step 1, a case is stained for CK7 and TTF1. Positive staining for both suggested P-NEC, with 65% sensitivity and 100% specificity in our initial cohort of primary cases. If either stain is negative, then in step 2, a case is stained for beta-catenin and CDX2. Positive staining for either suggested GI-NEC, with 62% sensitivity and 86% specificity in our initial cohort of primary cases. If both stains are negative, then in step 3, a case is stained for SSTR2A. A positive result suggested GI-NEC, with 60% specificity and 100% sensitivity in our initial cohort of primary cases; leftover cases are most likely P-NEC. This algorithm correctly localized 30 of the 33 primary cases (accuracy of 91%). The 3 mis-localized cases included one small cell GI-NEC negative for all 5 stains, one large cell GI-NEC negative for all 5 stains, and one large cell P-NEC negative for everything but nuclear beta-catenin. While AE1/3 and SATB2 showed significant differences in staining between GI-NEC and P-NEC, the former was not included in the algorithm because the mean H-score was high (i.e., positive) in both malignancy types, and the latter was not included because it is not widely available and because pairing it with SSTR2A in the final step did not improve overall accuracy of the algorithm.
An algorithmic approach for differentiating between P-NECs and GI-NECs using IHC markers based on study findings. Sensitivities and specificities provided apply to the entire study cohort of 53 cases, not the smaller cohorts of only primary or only metastatic cases. * indicates only nuclear staining for beta-catenin should be considered a positive result
The second cohort of 20 metastatic cases (10 GI-NEC and 10 P-NEC) was assessed for CK7, TTF1, CDX2, nuclear beta-catenin, and SSTR2A. Not all five markers showed significantly different expression in GI-NEC vs. P-NEC in this cohort (Table 2, middle columns), though when the H-score results of the primary and metastatic lesions were combined, all five markers retained statistical significance across the entire cohort of 53 cases (Table 2, rightmost columns). The above algorithm correctly localized 17 of the 20 metastatic cases (accuracy of 85%). The 3 mis-localized cases included a large cell GI-NEC that was positive for CK7 and TTF1, a small cell GI-NEC positive for CK7 and negative for everything else, and a small cell GI-NEC negative for all 5 stains.
When considering the entire cohort of 53 cases, our algorithm properly localized 47 (accuracy of 89%). As provided in Fig. 3, the initial step of CK7 and TTF1 staining had a sensitivity of 67% and specificity of 96% for P-NEC overall, the second step of beta-catenin and CDX2 staining had a sensitivity of 54% and specificity of 90% for GI-NEC overall, and the third step of SSTR2A staining had a sensitivity of 56% and specificity of 100% for GI-NEC overall. Of note, three of the six cases mis-localized by this algorithm were GI-NEC negative for all 5 markers. Therefore, if the stepwise algorithm indicates a case is likely P-NEC at the third step, a GI-NEC should still be considered if both CK7 and TTF1 were negative.
Discussion
Several prior studies have investigated the utility of IHC in determining site of origin for NEC [20]. Reported positivity rates have varied widely for most proteins, likely due to different cutoff thresholds and different laboratory conditions (including different antibodies). As NEC is rare in most organ systems, many studies have also been relatively small in size (as is ours), which may also contribute to variation in reported positivity rates. For example, reported TTF1 positivity rates in studies examining at least 20 SCLC have ranged from 57 [24] to 100% [25]; our rate in this study was 71% (5 of 7). TTF1 has also been reported as positive in small cell NEC from other sites, including prostate and cervix [26, 27]. Reported rates are generally lower for pulmonary LCNEC, ranging from 24 [25] to 48% [28]. La Rosa et al. [29] and Masai et al. [30] reported different TTF1 positivity rates in LCNEC with different antibody clones (SPT24 giving higher rates than 8G7G3/1; our study used the latter). Napsin A is rarely positive in P-NEC, as observed in our study and others [31, 32].
CDX2 is usually used to confirm gastrointestinal origin in malignancies, and some authors have reported high rates of positivity in GI-NEC. This includes Barbareschi et al. [33], who reported CDX2 positivity in 13 of 16 (81%) GI-NECs. However, La Rosa et al. [34] reported weak CDX2 staining in 3 of 27 (11%) GI-NECs, along with one of 14 (7%) P-NECs. Similarly, only one of our GI-NEC expressed strong CDX2 IHC; the stain was repeated with a different antibody, yielding similar results. Our metastatic GI-NEC similarly showed minimal CDX2 expression. CDX2 is reportedly expressed in half of NECs of the uterine cervix as well [35].
Along these lines, Lee et al. [36] performed TTF1, CDX2, and ISL1 IHC on 38 gastroenteropancreatic NECs and 36 P-NECs. They found that the former were more likely to express CDX2, and the latter were more likely to express TTF1 and ISL1. (We had analogous results for TTF1 and CDX2, though we found no significant difference in ISL1 expression between GI-NEC and P-NEC.) Cheuk et al. also reported that 43 of 52 (83%) P-NECs in their study were TTF1 positive, though 8 of 15 (53%) GI-NECs were as well [37].
SATB2 has relatively recently come into use as an IHC marker indicating gastrointestinal origin [38, 39]. Li et al. [40] reported positive SATB2 staining in 17% of foregut, 12% of midgut, and 90% of hindgut NETs; our study did not find significant SATB2 staining differences among NECs in these sites. Bellizzi recently reported that SATB2 was expressed in 60% of extrapulmonary visceral NECs, compared to 33% of P-NECs, though highest expression was seen in Merkel cell carcinoma (79%) [19, 21]. Similarly, we found SATB2 positivity in 54% of our GI-NECs and only 25% of our P-NECs (p = 0.018). It was therefore more reliable than CDX2 in our study to indicate a diagnosis of GI-NEC, though the stain is less widely available. We also observed that SATB2 was the only marker we tested that stained significantly stronger in one form of NEC than the other (namely, more common in large cell than small cell, for both GI-NEC and P-NEC).
TTF1, CDX2, and SATB2 are members of the superfamily of homeobox genes. Other examples utilized in our study include ISL1 and PAX8. ISL1 was originally described as a marker for pancreatic NETs but was subsequently demonstrated in NETs and NECs of various sites, including Merkel cell carcinoma, SCLC, and head and neck NECs [41]. As above, Lee et al. showed that ISL1 was more frequently expressed in P-NECs than in GI-NECs [36]. The data on PAX8 expression in NECs appear limited and inconsistent.
CK7 was preferentially expressed in P-NEC in our study, though this has undergone little investigation in the literature. Only two of our GI-NECs were positive for CK7; one was an ampullary LCNEC that also expressed CK20 weakly (H-score of 40). Accordingly, Nassar et al. [42] reported that 87% of ampullary NECs express CK7, and 38% express CK20. To our knowledge, these keratin markers have not been systematically evaluated in GI-NECs of other sites. While most of our GI-NECs were from the colorectum (including the other CK7-positive case) or the small bowel, both esophagus cases and the gastric case were CK7-negative. Nearly all NECs expressed AE1/AE3 in our study, though mean H-score was higher among P-NECs. AE1/AE3 expression has not been well studied in GI-NEC, though it has been reported as positive in 88% of SCLCs [43]. It is unclear why P-NECs expressed AE1/AE3 more strongly than GI-NECs in our study, as well as why some cases demonstrated weak or no staining.
It has been suggested that poorly differentiated GI-NECs exhibit reduced SSTR2A expression [44]. However, we found IHC positivity for SSTR2A in nearly half (6/13, 46%) of our GI-NECs. One other study has evaluated SSTR2A in GI-NEC, finding positivity in 2 of 9 (22%) colorectal NECs [45]. Only one of our P-NECs (5%) was positive for SSTR2A, a significantly lower rate than for our GI-NEC (p = 0.0021). Other authors have reported higher rates (36% [46] and 68% [47]) of SSTR2A positivity in P-NECs.
A few other markers we investigated have not been thoroughly studied in NEC in the literature. These include INSM1, a somewhat recently publicized neuroendocrine marker [22] that was moderately positive in both GI-NECs and P-NECs in our study, and beta-catenin, which often shows nuclear positivity in colorectal adenocarcinomas [48] and accordingly was a useful marker in our study to distinguish GI-NEC (sometimes positive) from P-NEC (rarely positive). We are not aware of other studies evaluating beta-catenin for NEC localization.
Finally, mutations in TP53 and RB1 [49,50,51] have frequently been reported in P-NECs, and accordingly, p53 and Rb IHC expression was frequently abnormal in the P-NECs in our study and in others [52]. This aberrancy was observed at a similar rate in our GI-NECs.
It should be noted that some prior studies were performed when a different definition of NEC was generally accepted outside the respiratory system; prior to the 4th Edition of the WHO Classification of Tumours of Endocrine Organs, published in 2017 [53], any digestive tract neuroendocrine neoplasm that was grade 3 by mitotic rate or Ki67 index was considered a poorly differentiated NEC, even when resembling a well-differentiated NET histologically [54]. Such tumors are now considered grade 3 NET. Some earlier studies may have included both in their cohorts, introducing a small but potentially confounding factor into their results. All cases in our study were carefully confirmed as NEC using current criteria.
Based on our findings, CK7 and TTF1 combined have a high specificity for P-NECs, and CDX2, nuclear beta-catenin, and SSTR2A staining are decently specific for GI-NECs. Using this information, we designed an algorithm for differentiation between P-NECs and GI-NECs. This had a high accuracy both in the original cohort of 33 primary lesions and in a confirmatory cohort of 20 metastatic lesions. While there were some outliers in both cohorts, this approach remains practical, as 4 of the 5 markers are readily available in most IHC labs.
Traditional therapy for NECs of any site (most notably SCLC) consists of platinum/etoposide [55]. However, site of origin is becoming increasingly relevant in treatment of NECs (Dr. Andrew M. Bellizzi, personal communication). This is most notable for Merkel cell carcinomas, where checkpoint inhibitor monotherapy is considered the preferred first-line treatment [56, 57]. However, both the National Comprehensive Cancer Network and the European Neuroendocrine Tumor Society recommend therapies such as FOLFOX and FOLFIRI (typically employed in colorectal adenocarcinoma) and capecitabine/temozolomide (typically employed in pancreatic NETs) as potential treatment options for extrapulmonary NEC but not for pulmonary NEC [58, 59]. Such approaches are bolstered by recent molecular discoveries and classifications, which indicate that NECs are often genetically similar to adenocarcinomas of the same site [60]. These recommendations and categorizations are evolving, and a handful of pertinent clinical trials are ongoing, but determining NEC site of origin is necessary in order to select patients appropriately for such clinical trials in order to further explore this frontier of treatment for NEC patients.
Our study was designed to be narrow and focused in scope, which resulted in several self-imposed limitations. First, we excluded NECs at other, less common, sites (e.g., endometrium) in order to perform a clean, direct comparison between GI-NEC and P-NEC. We also did not include NEC of the skin (namely, Merkel cell carcinoma) or well-differentiated NETs of any site. Including such cases would likely have yielded more convoluted results, and the additional subgroups would have been small as well, since NECs in general are rare.
Second, we used a TMA to evaluate the primary NEC cases. As a result, lesions with significant immunohistochemical heterogeneity could have been misrepresented. This approach was undertaken to identify key potential markers, which were then validated on a cohort of metastatic NEC; we used whole tissue sections to evaluate this second cohort, and our proposed algorithm performed similarly in those cases. Therefore, the use of a TMA does not appear to have significantly skewed our findings.
Third, our study utilized the H-score, which has been used to semi-quantitate expression of protein biomarkers and which offers a broad dynamic range against which clinical outcomes might be measured [61]. H-scores are admittedly utilized more in research settings than in daily diagnostic practice, where markers with clear and striking differences in positivity and negativity are the most practical for use. Fortunately, such differences were found in three of the five markers in our algorithm (TTF1, beta-catenin, SSTR2A), wherein NECs from one location were usually strikingly positive and NECs from the other location totally negative. CDX2 was not often positive in GI-NEC but was often striking in positive cases, and it was totally negative in all P-NECs. CK7 was usually negative in GI-NEC but was strongly positive in six of them, and for this reason it was paired with TTF1 in our algorithm. Therefore, the H-score helped in devising our algorithm but is not required when employing it.
Finally, we included small cell and large cell NECs from both the GI tract and the lung in our cohorts, as GI-NECs are sufficiently rare that we could not have otherwise compiled a cohort of reasonable size. As shown above, the H-scores for the IHC markers used in our algorithm did not significantly differ between small cell and large cell GI-NEC or P-NEC.
While most of the IHC information provided in this manuscript is not novel in isolation, we feel our algorithm has utility given the tools currently available to practicing pathologists, and hopefully it lays the groundwork for additional investigation into an increasingly clinically relevant matter. Improved localization of metastatic NECs will likely require additional markers that are currently unknown or uninvestigated in this setting.
In conclusion, we found that IHC for AE1/AE3, CK7, TTF1, CDX2, beta-catenin, SSTR2A, and SATB2 can be utilized to help distinguish GI-NEC from P-NEC, which may be helpful in the workup of metastatic lesions. Given the varied rates of positivity reported in the literature for these markers in GI-NEC and P-NEC, it appears generally advisable to use a panel containing multiple markers rather than rely on one result for diagnosis, as demonstrated in our algorithm utilizing most of these markers. If possible, individual labs may also wish to test available in-house cases to determine how these markers perform under local conditions.
References
Dasari A, Shen C, Halperin D, Zhao B, Zhou S, Xu Y, Shih T, Yao JC (2017) Trends in the incidence, prevalence, and survival outcomes in patients with neuroendocrine tumors in the United States. JAMA Oncol 3:1335–1342
Milione M, Maisonneuve P, Spada F, Pellegrinelli A, Spaggiari P, Albarello L, Pisa E, Barberis M, Vanoli A, Buzzoni R, Pusceddu S, Concas L, Sessa F, Solcia E, Capella C, Fazio N, la Rosa S (2017) The clinicopathologic heterogeneity of grade 3 gastroenteropancreatic neuroendocrine neoplasms: morphological differentiation and proliferation identify different prognostic categories. Neuroendocrinology 104:85–93
Heetfeld M, Chougnet CN, Olsen IH, Rinke A, Borbath I, Crespo G, Barriuso J, Pavel M, O'Toole D, Walter T, other Knowledge Network members (2015) Characteristics and treatment of patients with G3 gastroenteropancreatic neuroendocrine neoplasms. Endocr Relat Cancer 22:657–664
Basturk O, Saka B, Balci S et al (2015) Substaging of lymph node status in resected pancreatic ductal adenocarcinoma has strong prognostic correlations: proposal for a revised N classification for TNM staging. Ann Surg Oncol 22(Suppl 3):S1187–S1195
Walter T, Tougeron D, Baudin E, le Malicot K, Lecomte T, Malka D, Hentic O, Manfredi S, Bonnet I, Guimbaud R, Coriat R, Lepère C, Desauw C, Thirot-Bidault A, Dahan L, Roquin G, Aparicio T, Legoux JL, Lombard-Bohas C, Scoazec JY, Lepage C, Cadiot G, Stephanie L, Borbath I, Castex, Petorin C, Terrebonne E, Bouhier-Leporrier K, Suc E, Hautefeuille V, Bourgeois V, Cany L, Dewaele F, Niccoli P, Seitz JF, Lecaille C, Rebischung C, Rossi V, Baconnier M, Dubreuil O, Ferec M, Deplanque G, Geslin G, Wanicki Caron I, Lavau Denes S, Bedenne L, Ligeza C, Maringe E, Ran-Royo AL, Guigay J, Rougier P (2017) Poorly differentiated gastro-entero-pancreatic neuroendocrine carcinomas: are they really heterogeneous? Insights from the FFCD-GTE national cohort. Eur J Cancer 79:158–165
Sorbye H, Welin S, Langer SW, Vestermark LW, Holt N, Osterlund P, Dueland S, Hofsli E, Guren MG, Ohrling K, Birkemeyer E, Thiis-Evensen E, Biagini M, Gronbaek H, Soveri LM, Olsen IH, Federspiel B, Assmus J, Janson ET, Knigge U (2013) Predictive and prognostic factors for treatment and survival in 305 patients with advanced gastrointestinal neuroendocrine carcinoma (WHO G3): the NORDIC NEC study. Ann Oncol 24:152–160
Yamaguchi T, Machida N, Morizane C, Kasuga A, Takahashi H, Sudo K, Nishina T, Tobimatsu K, Ishido K, Furuse J, Boku N, Okusaka T (2014) Multicenter retrospective analysis of systemic chemotherapy for advanced neuroendocrine carcinoma of the digestive system. Cancer Sci 105:1176–1181
Howe HL, Wingo PA, Thun MJ, Ries LAG, Rosenberg HM, Feigal EG, Edwards BK (2001) Annual report to the nation on the status of cancer (1973 through 1998), featuring cancers with recent increasing trends. J Natl Cancer Inst 93:824–842
Mulshine JL, Treston AM, Brown PH, Birrer MJ, Shaw GL (1993) Initiators and promoters of lung cancer. Chest 103:4s–11s
Usuda K, Saito Y, Sagawa M, Sato M, Kanma K, Takahashi S, Endo C, Chen Y, Sakurada A, Fujimura S (1994) Tumor doubling time and prognostic assessment of patients with primary lung cancer. Cancer 74:2239–2244
Tamura T (2001) New state of the art in small-cell lung cancer. Oncology (Williston Park) 15:8–10
Zochbauer-Muller S, Pirker R, Huber H (1999) Treatment of small cell lung cancer patients. Ann Oncol 10:83–91
Adjei AA, Marks RS, Bonner JA (1999) Current guidelines for the management of small cell lung cancer. Mayo Clin Proc 74:809–816
Nicholson SA, Beasley MB, Brambilla E, Hasleton PS, Colby TV, Sheppard MN, Falk R, Travis WD (2002) Small cell lung carcinoma (SCLC): a clinicopathologic study of 100 cases with surgical specimens. Am J Surg Pathol 26:1184–1197
Fasano M, Della Corte CM, Papaccio F, Ciardiello F, Morgillo F (2015) Pulmonary large-cell neuroendocrine carcinoma: from epidemiology to therapy. J Thorac Oncol 10:1133–1141
Kinslow CJ, May MS, Saqi A, Shu CA, Chaudhary KR, Wang TJC, Cheng SK (2020) Large-Cell Neuroendocrine Carcinoma of the Lung: A Population-Based Study. Clin Lung Cancer 21:e99–e113
Cao L, Li ZW, Wang M, Zhang TT, Bao B, Liu YP (2019) Clinicopathological characteristics, treatment and survival of pulmonary large cell neuroendocrine carcinoma: a SEER population-based study. PeerJ 7:e6539
Dasari A, Mehta K, Byers LA, Sorbye H, Yao JC (2018) Comparative study of lung and extrapulmonary poorly differentiated neuroendocrine carcinomas: a SEER database analysis of 162,983 cases. Cancer 124:807–815
Bellizzi AM (2020) Immunohistochemistry in the diagnosis and classification of neuroendocrine neoplasms: what can brown do for you? Hum Pathol 96:8–33
Bellizzi AM (2013) Assigning site of origin in metastatic neuroendocrine neoplasms: a clinically significant application of diagnostic immunohistochemistry. Adv Anat Pathol 20:285–314
Bellizzi AM (2020) SATB2 in neuroendocrine neoplasms: strong expression is restricted to well-differentiated tumours of lower gastrointestinal tract origin and is most frequent in Merkel cell carcinoma among poorly differentiated carcinomas. Histopathology 76:251–264
Bellizzi AM (2020) An algorithmic immunohistochemical approach to define tumor type and assign site of origin. Adv Anat Pathol 27:114–163
Schatoff EM, Leach BI, Dow LE (2017) Wnt signaling and colorectal cancer. Curr Colorectal Cancer Rep 13:101–110
Hiroshima K, Iyoda A, Shida T, Shibuya K, Iizasa T, Kishi H, Tanizawa T, Fujisawa T, Nakatani Y (2006) Distinction of pulmonary large cell neuroendocrine carcinoma from small cell lung carcinoma: a morphological, immunohistochemical, and molecular analysis. Mod Pathol 19:1358–1368
Kargi A, Gurel D, Tuna B (2007) The diagnostic value of TTF-1, CK 5/6, and p63 immunostaining in classification of lung carcinomas. Appl Immunohistochem Mol Morphol 15:415–420
Hu J, Han B, Huang J (2020) Morphologic spectrum of neuroendocrine tumors of the prostate: an updated review. Arch Pathol Lab Med 144:320–325
McCluggage WG, Kennedy K, Busam KJ (2010) An immunohistochemical study of cervical neuroendocrine carcinomas: neoplasms that are commonly TTF1 positive and which may express CK20 and P63. Am J Surg Pathol 34:525–532
Sturm N, Rossi G, Lantuejoul S, Papotti M, Frachon S, Claraz C, Brichon PY, Brambilla C, Brambilla E (2002) Expression of thyroid transcription factor-1 in the spectrum of neuroendocrine cell lung proliferations with special interest in carcinoids. Hum Pathol 33:175–182
La Rosa S, Chiaravalli AM, Placidi C, Papanikolaou N, Cerati M, Capella C (2010) TTF1 expression in normal lung neuroendocrine cells and related tumors: immunohistochemical study comparing two different monoclonal antibodies. Virchows Arch 457:497–507
Masai K, Tsuta K, Kawago M, Tatsumori T, Kinno T, Taniyama T, Yoshida A, Asamura H, Tsuda H (2013) Expression of squamous cell carcinoma markers and adenocarcinoma markers in primary pulmonary neuroendocrine carcinomas. Appl Immunohistochem Mol Morphol 21:292–297
Rekhtman N, Pietanza CM, Sabari J, Montecalvo J, Wang H, Habeeb O, Kadota K, Adusumilli P, Rudin CM, Ladanyi M, Travis WD, Joubert P (2018) Pulmonary large cell neuroendocrine carcinoma with adenocarcinoma-like features: napsin A expression and genomic alterations. Mod Pathol 31:111–121
Zhang C, Schmidt LA, Hatanaka K, Thomas D, Lagstein A, Myers JL (2014) Evaluation of napsin A, TTF-1, p63, p40, and CK5/6 immunohistochemical stains in pulmonary neuroendocrine tumors. Am J Clin Pathol 142:320–324
Barbareschi M, Roldo C, Zamboni G, Capelli P, Cavazza A, Macri E, Cangi MG, Chilosi M, Doglioni C (2004) CDX-2 homeobox gene product expression in neuroendocrine tumors: its role as a marker of intestinal neuroendocrine tumors. Am J Surg Pathol 28:1169–1176
La Rosa S, Rigoli E, Uccella S, Chiaravalli AM, Capella C (2004) CDX2 as a marker of intestinal EC-cells and related well-differentiated endocrine tumors. Virchows Arch 445:248–254
Inzani F, Santoro A, Angelico G, Feraco A, Spadola S, Arciuolo D, Valente M, Carlino A, Piermattei A, Scaglione G, Scambia G, Rindi G, Zannoni GF (2020) Neuroendocrine carcinoma of the uterine cervix: a clinicopathologic and immunohistochemical study with focus on novel markers (Sst2-Sst5). Cancers 12:1211
Lee H, Fu Z, Koo BH, Sheehan CE, Young GQ, Lin J, Patil DT, Yang Z (2018) The expression of TTF1, CDX2 and ISL1 in 74 poorly differentiated neuroendocrine carcinomas. Ann Diagn Pathol 37:30–34
Cheuk W, Kwan MY, Suster S, Chan JK (2001) Immunostaining for thyroid transcription factor 1 and cytokeratin 20 aids the distinction of small cell carcinoma from Merkel cell carcinoma, but not pulmonary from extrapulmonary small cell carcinomas. Arch Pathol Lab Med 125:228–231
Magnusson K, de Wit M, Brennan DJ, Johnson LB, McGee SF, Lundberg E, Naicker K, Klinger R, Kampf C, Asplund A, Wester K, Gry M, Bjartell A, Gallagher WM, Rexhepaj E, Kilpinen S, Kallioniemi OP, Belt E, Goos J, Meijer G, Birgisson H, Glimelius B, Borrebaeck CAK, Navani S, Uhlén M, O'Connor DP, Jirström K, Pontén F (2011) SATB2 in combination with cytokeratin 20 identifies over 95% of all colorectal carcinomas. Am J Surg Pathol 35:937–948
Dragomir A, de Wit M, Johansson C, Uhlen M, Ponten F (2014) The role of SATB2 as a diagnostic marker for tumors of colorectal origin: results of a pathology-based clinical prospective study. Am J Clin Pathol 141:630–638
Li Z, Yuan J, Wei L, Zhou L, Mei K, Yue J, Gao H, Zhang M, Jia L, Kang Q, Huang X, Cao D (2015) SATB2 is a sensitive marker for lower gastrointestinal well-differentiated neuroendocrine tumors. Int J Clin Exp Pathol 8:7072–7082
Agaimy A, Erlenbach-Wunsch K, Konukiewitz B et al (2013) ISL1 expression is not restricted to pancreatic well-differentiated neuroendocrine neoplasms, but is also commonly found in well and poorly differentiated neuroendocrine neoplasms of extrapancreatic origin. Mod Pathol 26:995–1003
Nassar H, Albores-Saavedra J, Klimstra DS (2005) High-grade neuroendocrine carcinoma of the ampulla of vater: a clinicopathologic and immunohistochemical analysis of 14 cases. Am J Surg Pathol 29:588–594
Ordonez NG (2013) Broad-spectrum immunohistochemical epithelial markers: a review. Hum Pathol 44:1195–1215
Charoenpitakchai M, Liu E, Zhao Z, Koyama T, Huh WJ, Berlin J, Hande K, Walker R, Shi C (2017) In liver metastases from small intestinal neuroendocrine tumors, SSTR2A expression is heterogeneous. Virchows Arch 470:545–552
Konukiewitz B, Schlitter AM, Jesinghaus M, Pfister D, Steiger K, Segler A, Agaimy A, Sipos B, Zamboni G, Weichert W, Esposito I, Pfarr N, Klöppel G (2017) Somatostatin receptor expression related to TP53 and RB1 alterations in pancreatic and extrapancreatic neuroendocrine neoplasms with a Ki67-index above 20. Mod Pathol 30:587–598
Righi L, Volante M, Tavaglione V, Billè A, Daniele L, Angusti T, Inzani F, Pelosi G, Rindi G, Papotti M (2010) Somatostatin receptor tissue distribution in lung neuroendocrine tumours: a clinicopathologic and immunohistochemical study of 218 ‘clinically aggressive’ cases. Ann Oncol 21:548–555
Tsuta K, Wistuba II, Moran CA (2012) Differential expression of somatostatin receptors 1–5 in neuroendocrine carcinoma of the lung. Pathol Res Pract 208:470–474
White BD, Chien AJ, Dawson DW (2012) Dysregulation of Wnt/β-catenin signaling in gastrointestinal cancers. Gastroenterology 142:219–232
Vijayvergia N, Boland PM, Handorf E, Gustafson KS, Gong Y, Cooper HS, Sheriff F, Astsaturov I, Cohen SJ, Engstrom PF (2016) Molecular profiling of neuroendocrine malignancies to identify prognostic and therapeutic markers: a Fox Chase Cancer Center Pilot Study. Br J Cancer 115:564–570
Rekhtman N, Pietanza MC, Hellmann MD, Naidoo J, Arora A, Won H, Halpenny DF, Wang H, Tian SK, Litvak AM, Paik PK, Drilon AE, Socci N, Poirier JT, Shen R, Berger MF, Moreira AL, Travis WD, Rudin CM, Ladanyi M (2016) Next-Generation sequencing of pulmonary large cell neuroendocrine carcinoma reveals small cell carcinoma-like and non-small cell carcinoma-like subsets. Clin Cancer Res 22:3618–3629
Derks JL, Leblay N, Thunnissen E, van Suylen R, den Bakker M, Groen HJM, Smit EF, Damhuis R, van den Broek E, Charbrier A, Foll M, McKay J, Fernandez-Cuesta L, Speel EM, Dingemans AC, PALGA-Group (2018) Molecular subtypes of pulmonary large-cell neuroendocrine carcinoma predict chemotherapy treatment outcome. Clin Cancer Res 24:33–42
Liu H, Zhang Y, Chang J, Liu Z, Tang N (2018) Differential expression of neuroendocrine markers, TTF-1, p53, and Ki-67 in cervical and pulmonary small cell carcinoma. Medicine (Baltimore) 97:e11604
Lloyd RV, Osamura RY, Klöppel G, Rosai J (eds) (2017) WHO classification of tumours of endocrine organs. 4th ed. IARC Press, Lyon
Bosman FT, Carneiro F, Hruban RH, Theise ND (eds) (2010) WHO classification of tumours of the digestive system. 4th ed. IARC Press, Lyon
National Comprehensive Cancer Network. NCCN clinical practice guidelines in oncology: small cell lung cancer. https://www.nccn.org/professionals/physician_gls/pdf/sclc.pdf (Accessed 3/9/2021).
Kaufman HL, Russell J, Hamid O, Bhatia S, Terheyden P, D'Angelo SP, Shih KC, Lebbé C, Linette GP, Milella M, Brownell I, Lewis KD, Lorch JH, Chin K, Mahnke L, von Heydebreck A, Cuillerot JM, Nghiem P (2016) Avelumab in patients with chemotherapy-refractory metastatic Merkel cell carcinoma: a multicentre, single-group, open-label, phase 2 trial. Lancet Oncol 17:1374–1385
National Comprehensive Cancer Network. NCCN clinical practice guidelines in oncology: merkel cell carcinoma. https://www.nccn.org/professionals/physician_gls/pdf/mcc.pdf (Accessed 3/9/2021).
National Comprehensive Cancer Network. NCCN clinical practice guidelines in oncology: neuroendocrine and adrenal tumors. https://www.nccn.org/professionals/physician_gls/pdf/neuroendocrine.pdf (Accessed 3/9/2021).
Garcia-Carbonero R, Sorbye H, Baudin E, Raymond E, Wiedenmann B, Niederle B, Sedlackova E, Toumpanakis C, Anlauf M, Cwikla JB, Caplin M, O''Toole D, Perren A, all other Vienna Consensus Conference participants (2016) ENETS consensus guidelines for high-grade gastroenteropancreatic neuroendocrine tumors and neuroendocrine carcinomas. Neuroendocrinology 103:186–194
Uccella S, La Rosa S, Metovic J et al (2021) Genomics of high-grade neuroendocrine neoplasms: well-differentiated neuroendocrine tumor with high-grade features (G3 NET) and neuroendocrine carcinomas (NEC) of various anatomic sites. Endocr Pathol 32:192–210
Pierceall WE, Wolfe M, Suschak J, Chang H, Chen Y, Sprott KM, Kutok JL, Quan S, Weaver DT, Ward BE (2011) Strategies for H-score normalization of preanalytical technical variables with potential utility to immunohistochemical-based biomarker quantitation in therapeutic response diagnostics. Anal Cell Pathol (Amst) 34:159–168
Acknowledgements
Teri Bowman, HT, ASCP assisted with immunohistochemical staining for this project.
Author information
Authors and Affiliations
Contributions
SY analyzed stains and data, devised the algorithm, and wrote the manuscript. JLH assisted in case selection, performed some tissue staining, and offered expert advice. RSG conceived the study, assisted in case selection, arranged tissue staining, and edited the manuscript.
Corresponding author
Ethics declarations
Compliance with ethical standards
The authors adhered to institutional ethical standards.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yu, S., Hornick, J.L. & Gonzalez, R.S. An algorithmic approach utilizing CK7, TTF1, beta-catenin, CDX2, and SSTR2A can help differentiate between gastrointestinal and pulmonary neuroendocrine carcinomas. Virchows Arch 479, 481–491 (2021). https://doi.org/10.1007/s00428-021-03085-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-021-03085-7