Abstract
Primary central nervous system diffuse large B cell lymphoma (PCNS-DLBCL) is a rare and aggressive entity of diffuse large B cell lymphoma (DLBCL). Elements of the tumour microenvironment (TME) including tumour-infiltrating lymphocytes (TILs) and tumour-associated macrophages (TAMs) have been associated with survival in DLBCL but their composition and prognostic impact in PCNS-DLBCL are unknown. Programmed cell death-1 (PD1)/programmed death-ligand 1 (PD-L1) immune checkpoint may represent a therapeutic option. Here, we aimed to characterise PD1/PDL1 immune checkpoints and the composition of the TME in PCNS-DLBCL. We collected tumour tissue and clinical data from 57 PCNS-DLBCL and used immunohistochemistry to examine TAMs (CD68, CD163), TILs (CD3, CD4, CD8, PD1) and tumour B cells (PAX5/PDL1 double stains, PDL1). The PDL1 gene was evaluated by fluorescence in situ hybridization (FISH). PAX5/PDL1 identified PDL1 expression by tumour B cells in 10/57 cases (17.5%). PDL1 gene translocation was a recurrent cytogenetic alteration in PNCS-DLBCL (8/47.17%) and was correlated with PDL1 positive expression in tumour B cells. The TME consisted predominantly of CD163 (+) M2 TAMs and CD8 (+) TILs. Most TAMs expressed PDL1 and most TILs expressed PD1. The density of TAMs and TILs did not associate with outcome. We showed that expression of PD1 on TILs and PDL1 on TAMs, but not the expression of PDL1 on tumour B cells was correlated with better prognosis. These findings support a significant role of TME composition and PD1/PDL1 crosstalk in PCNS-DLBCL pathogenesis and bring new insights to the targeted therapy of this aggressive lymphoma.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Primary central nervous system diffuse large B cell lymphoma (PCNS-DLBCL) is listed in the WHO classification of tumours of haematopoietic and lymphoid tissues 2016 as a separate entity of diffuse large B cell lymphoma (DLBCL). It is characterised by a typical clinical presentation with tumour cells confined to the brain, the spinal cord, the leptomeninges or the eyes and remarkably worse outcome than patients with systemic DLBCL (s-DLBCL). The exclusively topographical restriction of PCNS-DLBCL to CNS raises the question of a particular microenvironment that could explain the aggressive behaviour of this disease. The immune checkpoints and the composition of the tumour microenvironment (TME) have emerged as the most important factors for both tumour survival and host defence. TME is mainly represented by include tumour-infiltrating lymphocytes (TILs) and tumour-associated macrophages (TAMs). TAMs in lymphomas have been extensively investigated. Noteworthy, several studies addressed outcome correlations by using both immunohistochemistry (with CD68 and CD163) and gene expression profiling. They especially showed adverse outcome in classical Hodgkin lymphoma (cHL) with increased TAMs [1,2,3]. Similar studies focusing on TILs in B cell lymphomas have demonstrated a correlation between T lymphocyte signature and a link between low amounts of TILs and a poor outcome [4]. Furthermore, high levels of CD4+ TILs have been shown to be associated with improved outcome in B cell lymphomas [5,6,7].
Programmed cell death-1 (PD1)/programmed death-ligand 1 (PD-L1) immune checkpoint provides an important escape mechanism from immune attack. The PDL1 (CD274 molecule) gene encodes an immune inhibitory receptor ligand expressed by various types of tumour cells. Interaction of the encoded protein with its receptor PD1 inhibits T cell activation and cytokine production providing an immune escape for tumour cells through cytotoxic T cell inactivation. PD1 and PDL1 expression have been shown to correlate with survival and therapeutic responses in many cancers [8,9,10]. Thus, the PD1/PDL1 pathway has emerged as an attractive target in cancer immunotherapy in order to restore immune function. Numerous clinical trials with PD1 pathway blocking agents used alone or in combination have shown significant response in lymphoid malignancies including cHL, plasma cell myeloma or DLBCL [11,12,13]. PDL1 expression in tumour cells can be induced either by extrinsic signal as an adaptive response to IFN-γ released from TILs or by intrinsic signals such as genetic aberrations involving PDL1 gene locus on chromosome 9p24.1 or latent Epstein-Barr virus (EBV) infection [14]. In cHL and primary mediastinal large B cell lymphoma (PMBL), PDL1 gene amplification is a recurrent cytogenetic alteration and strongly correlates with PDL1 protein expression [15,16,17]. PDL1 rearrangements have also been reported in lymphoid malignancies leading to aberrant expression of PDL1 favouring an immune escape and contributing to increased aggressiveness [18, 19].
The aim of this study was to evaluate the PD1/PDL1 immune checkpoints by immunohistochemistry and FISH analyses and to characterise the particular composition of the TME in PCNS-DLBCL.
Materials and methods
Patient selection
Pre-treatment biopsies from 57 immunocompetent patients with newly diagnosed PCNS-DLBCL were retrospectively retrieved from the Department of Pathology of Centre Hospitalo Universitaire (CHU) Montpellier, France. Cases with prior or concurrent low-grade B cell lymphomas and secondary CNS involvement by s-DLBCL were excluded. The study was approved by the research ethics boards of our institution (Centre des Ressources Biologiques, CRB, Montpellier) according to the Declaration of Helsinki. The written consent of patients was obtained.
Histological and immunohistochemical analysis
All cases were reviewed by five pathologists (VS, VR, VCM, LD and MP). The diagnosis of PCNS-DLBCL was made on Haematoxylin-Eosin (HE) and was based on the WHO 2016 classification of haematopoietic and lymphoid tissue. For immunohistochemical examination, 3-μm thick tissue sections from the formalin-fixed paraffin-embedded (FFPE) blocks were subjected to antigen retrieval and immunostained on a Ventana Benchmark XT autostainer (Ventana Tucson, AZ, USA). The following antibodies were used after appropriate antigen retrieval according to the manufacturer’s instructions: CD20 (clone L26, Dako, Denmark A/S, 1:300), PAX5 (clone DAK-PAX5, Dako, 1:25), CD3 (clone 2GV6,Ventana, PREP Kit Ventana), CD5 (clone 4C7, Dako, Denmark A/S, 1:100), CD4 (clone SP35, Ventana, PREP Kit Ventana), CD8 (clone SP57, Ventana, PREP Kit Ventana), CD10 (clone 56C6, Menarini, California USA, 1:10), BCL6 (clone G1191E/A8, Ventana, PREP Kit Ventana), MUM1 (clone MUM1p, Dako, 1:50), P53 (clone DO7, Ventana, PREP Kit Ventana), MYC (clone EP 121, Epitomics, Burlingame, CA, USA, 1:100), CD68 (clone KP1, Dako, 1:400), CD163 (clone MRQ-26, Ventana, PREP Kit Ventana), KI67 (clone 30-9, Ventana, PREP Kit Ventana), PD1 (clone NAT105, Abcam, Paris, France, 1:100), PDL1 (clone E1L3M, Cell Signaling, Leiden, The Netherlands, 1:200). Association with EBV was examined by in situ hybridization (ISH) using EBV-encoded early nuclear RNA (EBER). For MUM1, MYC and P53, we evaluated the percentage of positive neoplastic cells as previously described. MUM1, MYC and P53 protein expression was considered as positive if nuclear staining was observed respectively in at least 30%, 40% and 10% of the neoplastic cells [20, 21].
Evaluation of PDL1 expression by tumour cells and assessment of TME composition
For evaluation of PDL1 expression by tumour cells and for assessment of TME composition, slides were scanned at high magnification with a × 20 objective and digitised on the iScan Coreo scanner (Ventana, Roche, France) to generate an image of the whole slide. Double staining with PAX5 (nuclear B cell marker) and PDL1 was performed to evaluate PDL1 expression in the neoplastic B cells. For PDL1, both a membranous immunostaining signal on the cell surface and cytoplasmic staining within cells were recorded [22]. PD1 positive staining was evaluated in the membrane and cytoplasm of cells [23]. The percentage of positive cells was evaluated for both antibodies, and samples showing staining of any intensity in 1% or more of the respective cells were considered positive [22]. Images were obtained for PAX5/PDL1 double stains, CD3, CD4, CD8, PD1, CD68 and CD163. CD68 was used as a pan-macrophage marker staining M1 and M2 TAMs and CD163 as a M2 TAMs’ marker. Images were then compared on the same screen by five pathologists (VS, VR, VCM, MP and LD) blinded to clinical data.
PDL1 expression by tumour cells
The number of double-expressing cells was given in percentage as the rate of all PAX5 positive neoplastic B cells.
TILs
The density of TILs was estimated with CD3 as follows: (0) absent, (1) low, (2) intermediate and (3) high. The proportion of CD4 (+), CD8 (+) and PD1 (+) TILS was manually counted. Owing to their characteristic morphology, TILs could easily be distinguished from TAMs and CD4 (+) or PD1 (+) cells with apparent morphological appearance difference from TILs were excluded from the count. The CD4 (+)/CD8 (+) TIL ratio was assessed and the number of PD1 (+) TILs was given in percentage as the rate of all CD3 (+) TILs.
TAMs
The proportion of CD68 (+) cells was evaluated as follows: (0) absent, (1) low, (2) intermediate and (3) high. The CD68 (+)/CD163 (+) TAM ratio was assessed and the number of PDL1 (+) TAMs was evaluated and depicted as a proportion of all CD68 (+) cells in percentage. PAX5+/PDL1+ tumour cells were excluded from the count.
For TAM and TIL quantification, the categories low, intermediate and high were defined as follows: low, ≤ 25% of tumour area occupied by TILs or TAMs; intermediate, > 25% and ≤ 50% of tumour area occupied by TILs or TAMs; high, > 50% of tumour area occupied by TILs or TAMs.
Interphase fluorescence in situ hybridisation
Interphase FISH was performed on 3-μm thick tissue sections using split signal FISH DNA probes for BCL2/18q21 (probe Y5407; DAKO A/S), BCL6/3q27 (probe Y5408), MYC/8q24 (probe Y5410; DAKO A/S) and PDL1 (PDL1, CD274 break apart probe, 9p24.1, Empire Genomics). Digital images were captured with a Metafer slide scanning platform using a Leica Axioplan fluorescence microscope (Zeiss Axio Imager M1) equipped with a charge-coupled device (CCD) camera coupled to and driven by the ISIS software (MetaSystem, FISH Imaging System, Germany). At least 100 nuclei were evaluated independently by three scorers (VS, MA, VC). Cases were considered positive when more than 15% of the cells displayed abnormalities on the FFPE tissue sections.
Statistical analyses
Fisher’s exact test was used to calculate the P value with GraphPad Prism 6.0, GraphPad. Event-free survival (EFS) and progression-free survival (PFS) were estimated using the Kaplan-Meier method. Results were considered to be significant at P < 0.05.
Results
Patients’ characteristics and clinicopathological findings
The clinicopathological characteristics of the 57 patients are depicted in Table 1. Complete remission after first line of treatment was evaluated for patients who received treatment with curative intention whereas status at last follow-up was given for the full cohort including patients with palliative treatment.
Median of follow-up was 15 months (12–108). The majority of the patients was treated with methotrexate (MTX)-based chemotherapy and received rituximab. Most patients had poor outcome showing relapse or progression and 47.2% (25/53) died of disease. As expected, most cases had non-germinal centre (GC) phenotype. No case showed BCL2 or MYC gene breaks whereas MYC protein expression and MYC/BCL2 protein co-expression were respectively present in 32.1% (18/56) and 26.8% (15/56) of cases. BCL6 and PDL1 gene translocations were the most frequent cytogenetic aberrations occurring respectively in 22.2% (12/54) and 17% (8/47) of cases.
PDL1 protein expression in tumour cells correlates with PDL1 gene rearrangements
Using PAX5/PDL1 double stains, PDL1 protein expression by the neoplastic B cells was observed in 17.5% (10/57) of cases (Table 1, Figs. 1 and 2). When positive, at least 30% of neoplastic B cells expressed PDL1 and the mean percentage of PDL1 expressing tumour cells was 61% (30–100%).
We then investigated clinical and pathological parameters according to PDL1 protein expression in tumour cells (Table 1). We did not identify any correlation with clinical parameters although cases with PDL1 (+) tumour cells tend to present with poorer MSKCC (Memorial Sloan Kettering Cancer Center) prognostic score (p = 0.06). Of interest, PDL1 protein expression in tumour cells strongly correlated with the presence of PDL1 gene rearrangements as demonstrated by the FISH analysis (p = 0.0001) (Figs. 1 and 2). We also identified three additional cases (3/47, 6.4%) showing more than two copies of PDL1 suggesting polysomy for chromosome 9. However, these cases did not show PDL1 protein expression in tumour cells.
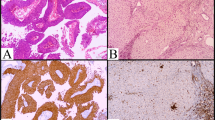
PAX5 (red)/PDL1 (brown) double stains in a case of PCNS-DLBCL with PDL1 gene translocation. a Haematoxylin and eosin stain (X20). b CD20 (X20). c PAX5 (red)/PDL1 (brown) X40. PDL1 (+) cells are PAX5 (+) and PAX5 (+) cells express PDL1. d PDL1 FISH analysis (X60). PDL1 gene translocation with one fusion signal, one red signal and one green signal (red circles)
PAX5 (red)/PDL1 (brown) double stains in a case of PCNS-DLBCL without PDL1 gene translocation. a Haematoxylin and eosin stain (X40). b CD163 (X40). The microenvironment predominately consists of CD163 (+) M2 macrophages. c PAX5 (red)/PDL1 (brown) X40. PDL1 (+) cells are PAX5 (−), and PAX5 (+) cells do not express PDL1. d PDL1 FISH analysis (X60) showing normal pattern with 2 fusion signals (red circles)
Among the ten patients with PDL1 positive expression in neoplastic B cells, two did not show PDL1 gene translocation. Both cases had normal pattern using PDL1 break apart probe with 2 fusion signals. Both patients were in complete remission at last follow-up, after anthracycline-based chemotherapy. Concerning phenotypic features, one case had GC phenotype. This case had PD1 (+) TILs and PDL1 (+) TAMs. The other case demonstrated non-GC phenotype and showed PD1 (+) TILs without PDL1 (+) TAMs. Both cases were EBER negative.
Composition of the TME
Composition of the TME is summarised in Table 2.
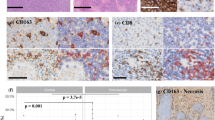
TILs identified by CD3 immunostaining were observed in all cases and usually at low abundance. TILs predominantly consisted of CD8 (+) lymphocytes whereas CD4 (+) lymphocytes were less numerous in most cases (Fig. 3b–c). Moreover, a high proportion of cases (39/56, 69.6%) had TILs PD1 (+) (Fig. 3d).
Analysis of TAMs’ abundance by CD68 immunostaining showed no macrophage in 10.9% (6/55) of cases, a low density of cells in 45.5% of cases (25/55), a moderate density in 34.5% (19/55) of cases and high density in 9.1% (5/55) of cases. CD68 (+) cells without CD163 co-expression were rare (4.1%), supporting the finding that most macrophages were M2 TAMs (Fig. 3e–f). TAMs usually expressed PDL1 (28/49, 57.1%) and rarely expressed PD1 (3/37, 6.4%) (Fig. 2b–c).
We did not identify any correlation between PD1 (+) expression in TILs and PDL1 (+) expression in tumour cells (p = 0.25). Nevertheless, PDL1 (+) expression in tumour cells was significantly associated with high density of TAMs (p = 0,015) and tended to correlate with high density of TILs (p = 0.06).
The presence of PD1 (+) TILs or PDL1 (+) TAMs is associated with better outcome
Supplementary Table 1 shows the correlation of the density of TILs or TAMs and the expression of PD1 and PDL1 in TME with clinicopathological parameters.
Density of TILs and ratio CD4 (+)/CD8 (+) did not significantly correlate with any of the clinicopathological parameters investigated although patients with ratio CD4 (+)/CD8 (+) ≥ 1 tend to present with high MSKCC prognostic score. Nevertheless, patients with TILs PD1 (+) had better outcome. PD1 positive expression in TILs was significantly correlated with complete remission at last follow-up, whereas absence of TILs PD1 (+) was significantly associated with relapse or progression and with death of disease or progressive pathology (Table 2). Furthermore, presence of TILs PD1 (+) was significantly associated with favourable impact on event-free and progression-free survival (Fig. 4a–b).
Prognostic impact of PD1 protein expression in TILs and PDL1 protein expression in TAMs. a PD1 expression in TILs is significantly associated with a favourable impact on event-free survival. b PD1 expression in TILs is significantly associated with a favourable impact on progression-free survival. c PDL1 expression in TAMs is significantly associated with a favourable impact on event-free survival. d PDL1 expression in TAMs is significantly associated with a favourable impact on progression-free survival
Density of TAMs did not correlate with the clinical outcome. Although patients with high density of TAMs were more frequently in complete remission after first line of treatment, we did not identify any impact on clinical presentation at diagnosis (both with MSKCC prognostic class and IPCG prognostic score) and on status at last follow-up. We observed a significant positive relationship between high density of TAMs and PDL1 positive expression in tumour cells. Moreover, the presence of PDL1 (+) TAMs significantly correlated with better survival. Patients with TAMs PDL1 (+) underwent less frequently at least 2 lines of treatment, usually achieved complete remission and showed fewer relapses/progression. Furthermore, presence of TAMs PDL1 (+) was significantly associated with favourable impact on event-free and progression-free survival (Fig. 4c–d).
Discussion
In order to develop within immunocompetent hosts, tumour cells are able to evolve several immune escape strategies. Mechanisms leading to immune evasion that have been identified in lymphomas especially include recruitment of immunosuppressive cells such as TILs or TAMs, and dysregulation of immune checkpoints involved in the interaction of PD1 at the surface of T lymphocytes with its ligand PDL1 found at the surface of macrophages and some tumour cells [24, 25]. In this study, we aimed to characterise the TME and the immune checkpoint profiles and their association with the outcome in patients with PCNS-DLBCL.
In s-DLBCL, PDL1 may be expressed by both tumour B cells and TAMs. PDL1 expression in DLBCL has been reported around 20–30% of cases depending on the cell compartment analysed [8, 26] and on the antibody used for PDL1 identification. Of interest, using double staining of PAX5 and PDL1, some identified PDL1 positive expression in 10.5% of tumour B cells and in 15.3% of TAMs [8]. In the present work, the antibody used for PDL1 (E1L3N, Cell Signaling) detection has been previously used in several series investigating s-DLBCL and has shown a better signal-to-noise result than SP142 clone [27]. Using double staining of PAX5 and PDL1, we identified PDL1 positive expression in 17.5% (10/57) of tumour B cells. In a previous report of PCNS-DLBCL, we observed PDL1 protein expression in 37% of tumour B cells. Nevertheless, the PDL1 clone used was SP142, and we did not perform double staining to discriminate PDL1 (+) TAMs from PDL1 (+) tumour B cells.
We did not identify any correlation between PDL1 positive expression in tumour B cell and prognosis. In the largest series of s-DLBCL, patients with PDL1 (+) tumour cells had poorer prognosis [8, 28]. Of interest, as observed in our work, Pollari et al. showed, in their series of primary testicular lymphoma, that the presence of PDL1 (+) lymphoma cells was not associated with the outcome. Of interest, primary testicular and primary central nervous system lymphomas are both immune-privileged site-associated B cell lymphomas and share similar genetic and molecular features [19] rarely observed in s-DLBCL. Such distinctive biology could explain our data concerning the absence of clinical impact of PDL1 positive expression in lymphoma cells.
The structural anomalies on the chromosome 9p24.1 including gains, amplifications or translocations of the PDL1 locus have been significantly correlated with PDL1 expression in B lymphomas [19, 29]. In the present series, expression of PDL1 in tumour B cells was observed in all samples with translocations. However, in 2 cases, we identified PDL1 expression in lymphoma B cells without any cytogenetic aberrations (including polysomy for chromosome 9), indicating that there are other underlying mechanisms leading to the expression of the protein in PCNS-DLBCL. EBV infection can explain the expression of PDL1 in some cases of lymphomas [30]; nevertheless, all cases in the present series were EBER negative. Active JAK/STAT3 signalling has been associated with stimulation of PDL1 expression in lymphomas [29]. Of interest, oncogenic mutations of the MYD88 gene lead to the activation of the JAK kinase [31] and these mutations are common in PCNS-DLBCL [32]. We thus hypothesised that MYD88 mutations in PCNS-DLBCL are associated with the JAK kinase reactivation of STAT3 that in turn stimulates the expression PDL1.
We identified PDL1 gene rearrangements in 17% (8/47) of cases. Translocations involving the PDL1 locus have already been reported in PCNS-DLBCL and other immune-privileged site-associated B cell lymphomas. In their series, Chapuy et al. identified PDL1 or PDL2 translocations in 6% (4/66) of PCNS-DLBCL and in 4% (2/50) of primary testicular lymphomas [19]. PDL1 gene translocations have been reported at higher frequency in PMBL and primary cutaneous diffuse large B cell lymphoma, leg type, occurring respectively in 20% (25/125) [33] and 40% (4/10) [34] of cases. Using the FISH analysis, PDL1/PDL2 locus translocations have been reported in s-DLBCL at low frequency occurring in 3% (6/179) of cases [35]. Taken together, these results suggest that recurrent genomic rearrangements of PDL1 underlie immune-privileged site-associated B cell lymphomas.
TILs are composed of a mixture of lymphocytes with different functions and phenotypes. The activation of both CD4 (+) and CD8 (+) TILs is needed for an efficient immune response [36]. Of interest, the ratio CD4 (+)/CD8 (+) T cells may be different for different types of cancers and DLBCL have been reported to have several fold more CD8 (+) TILs than some other cancers [37] In classical Hodgkin lymphoma, a predominance of CD4 (+) TILs is observed [38] and a high ratio CD4 (+)/CD8 (+) is associated with treatment failure [39]. On the contrary, s-DLBCL patients with ratio CD4 (+)/CD8 (+) ≥ 2 have better overall survival [6]. Here, we reported a predominance of CD8 (+) TILs in the background of tumour cells but we did not observe an association between prognostic and ratio CD4 (+)/CD8 (+). Presence of TILs PD1 (+) in DLBCL has been associated either with favourable, unfavourable or no prognostic effect in different studies [24]. These differences may result from methodologic and technical disparities among studies [40]. In their series of 74 primary testicular lymphoma patients, Pollari et al. showed that presence of PD1 (+) TILs predicted favourable survival [41]. In this series, we also identified a positive correlation between the outcome and the presence of PD1 (+) TILs.
Two main macrophage phenotypes have been described. They include the classically activated (or M1) macrophages or the alternatively activated (or M2) macrophages [42]. M1 macrophages are involved in inflammatory response and antitumoral defence whereas M2 macrophages reduce inflammation and have tumour-promoting effect [43]. M2 macrophages are able to suppress the adaptive immune response through mechanisms including inhibition of T cell proliferation [42]. Part of their immunosuppressive activity is exerted by their release of chemokines (CCL17, CCL18 and CCL22) that preferentially attract T cell subsets devoid of cytotoxic function [44]. Nevertheless, in this series, although TAMs had M2 phenotype, TILs preferentially showed CD8+ cytotoxic phenotype.
Some have demonstrated that tumour cells are able to recruit and shift TAMs toward M2 function [45, 46]. In most studies, TAM density is associated with poor prognosis [47], and the presence of M2 TAMs has been shown to correlate with poor prognosis in DLBCL [48]. In the present work, we identified a polarisation to M2 macrophages in nearly all cases but we did not show any correlation between TAM density and adverse prognosis. Of interest, we identified an association between the presence of TAMs PDL1 (+) and better outcome. Such association has also been reported in primary testicular lymphoma [41], another aggressive large B cell lymphoma arising in immune-privileged site. Although PDL1 expression has been largely investigated in B cell lymphomas, its expression in TME has not been well defined and probably not separated from tumour cell expression in most studies. This could explain some discrepancies. In their series of 29 primary cutaneous DLBCL, Menguy et al. found a predominant M2 macrophages’ infiltrate and demonstrated that M2 TAMs expressed PDL1 [49]. Nevertheless, they did not study any correlation with the outcome. In their series of primary intestinal DLBCL, Ishikawa et al. identified PDL1 negativity on microenvironment immune cells as a poor independent prognostic factor for overall survival. Thus, although our cases were associated with M2 macrophages in TME, usually reported as a pejorative biomarker in DLBCL, the presence of PDL1 (+) macrophage should counteract this adverse effect.
In conclusion, the TME in PNCS-DLBCL consists predominantly of M2 PDL1 (+) TAMs and CD8 (+), PD1 (+) TILs. PDL1 gene translocation is a recurrent cytogenetic alteration in PNCS-DLBCL and correlates with PDL1 positive expression in tumour B cells. Expression of PD1 on TILs and PDL1 on TAMs, but not expression of PDL1 on tumour B cells, correlates with better prognosis. PD1 antibodies are currently approved by the US Food and Drug Administration (FDA) and tested in multiple clinical trials in B cell lymphomas with promising results. A retrospective analysis of PNCS-DLBCL and primary testicular lymphoma, which usually have PDL1 genetic alterations, concluded that both these immune-privileged site-associated B cell lymphomas could be suitable for anti-PD1 immunotherapy [50], and a preclinical murine model concluded that anti-PD1 antibody had significant therapeutic activity against PNCS-DLBCL. Therefore, immune checkpoint inhibition are promising therapeutic options in PNCS-DLBCL and the results of underway clinical trials to test these drugs are eagerly awaited (CheckMate 647).
References
Steidl C, Lee T, Shah SP, Farinha P, Han G, Nayar T, Delaney A, Jones SJ, Iqbal J, Weisenburger DD, Bast MA, Rosenwald A, Muller-Hermelink HK, Rimsza LM, Campo E, Delabie J, Braziel RM, Cook JR, Tubbs RR, Jaffe ES, Lenz G, Connors JM, Staudt LM, Chan WC, Gascoyne RD (2010) Tumor-associated macrophages and survival in classic Hodgkin’s lymphoma. N Engl J Med 362:875–885. https://doi.org/10.1056/NEJMoa0905680
Tan KL, Scott DW, Hong F, Kahl BS, Fisher RI, Bartlett NL, Advani RH, Buckstein R, Rimsza LM, Connors JM, Steidl C, Gordon LI, Horning SJ, Gascoyne RD (2012) Tumor-associated macrophages predict inferior outcomes in classic Hodgkin lymphoma: a correlative study from the E2496 intergroup trial. Blood 120:3280–3287. https://doi.org/10.1182/blood-2012-04-421057
Jakovic LR, Mihaljevic BS, Perunicic Jovanovic MD, Bogdanovic AD, Andjelic BM, Bumbasirevic VZ (2011) The prognostic relevance of tumor associated macrophages in advanced stage classical Hodgkin lymphoma. Leuk Lymphoma 52:1913–1919. https://doi.org/10.3109/10428194.2011.580026
Leivonen S-K, Pollari M, Brück O, Pellinen T, Autio M, Karjalainen-Lindsberg M-L et al (2018) T-cell inflamed tumor microenvironment predicts favorable prognosis in primary testicular lymphoma. Haematologica. https://doi.org/10.3324/haematol.2018.200105
Keane C, Gill D, Vari F, Cross D, Griffiths L, Gandhi M (2013) CD4(+) tumor infiltrating lymphocytes are prognostic and independent of R-IPI in patients with DLBCL receiving R-CHOP chemo-immunotherapy. Am J Hematol 88:273–276. https://doi.org/10.1002/ajh.23398
Xu Y, Kroft SH, McKenna RW, Aquino DB (2001) Prognostic significance of tumour-infiltrating T lymphocytes and T-cell subsets in de novo diffuse large B-cell lymphoma: a multiparameter flow cytometry study. Br J Haematol 112:945–949
Ansell SM, Stenson M, Habermann TM, Jelinek DF, Witzig TE (2001) Cd4+ T-cell immune response to large B-cell non-Hodgkin’s lymphoma predicts patient outcome. J Clin Oncol Off J Am Soc Clin Oncol 19:720–726. https://doi.org/10.1200/JCO.2001.19.3.720
Kiyasu J, Miyoshi H, Hirata A, Arakawa F, Ichikawa A, Niino D, Sugita Y, Yufu Y, Choi I, Abe Y, Uike N, Nagafuji K, Okamura T, Akashi K, Takayanagi R, Shiratsuchi M, Ohshima K (2015) Expression of programmed cell death ligand 1 is associated with poor overall survival in patients with diffuse large B-cell lymphoma. Blood 126:2193–2201. https://doi.org/10.1182/blood-2015-02-629600
Wang Y, Wu L, Tian C, Zhang Y (2017) PD-1-PD-L1 immune-checkpoint blockade in malignant lymphomas. Ann Hematol 97:229–237. https://doi.org/10.1007/s00277-017-3176-6
Taube JM, Klein A, Brahmer JR, Xu H, Pan X, Kim JH, Chen L, Pardoll DM, Topalian SL, Anders RA (2014) Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res Off J Am Assoc Cancer Res 20:5064–5074. https://doi.org/10.1158/1078-0432.CCR-13-3271
Ansell SM, Minnema MC, Johnson P, Timmerman JM, Armand P, Shipp MA et al (2019) Nivolumab for relapsed/refractory diffuse large B-cell lymphoma in patients ineligible for or having failed autologous transplantation: a single-arm, phase II study. J Clin Oncol Off J Am Soc Clin Oncol:JCO1800766. https://doi.org/10.1200/JCO.18.00766
von Keudell G, Younes A (2019) Novel therapeutic agents for relapsed classical Hodgkin lymphoma. Br J Haematol 184:105–112. https://doi.org/10.1111/bjh.15695
Oliva S, Troia R, D’Agostino M, Boccadoro M, Gay F (2018) Promises and pitfalls in the use of PD-1/PD-L1 inhibitors in multiple myeloma. Front Immunol 9:2749. https://doi.org/10.3389/fimmu.2018.02749
Dong Z-Y, Wu S-P, Liao R-Q, Huang S-M, Wu Y-L (2016) Potential biomarker for checkpoint blockade immunotherapy and treatment strategy. Tumour Biol J Int Soc Oncodevelopmental Biol Med 37:4251–4261. https://doi.org/10.1007/s13277-016-4812-9
Rosenwald A, Wright G, Leroy K, Yu X, Gaulard P, Gascoyne RD, Chan WC, Zhao T, Haioun C, Greiner TC, Weisenburger DD, Lynch JC, Vose J, Armitage JO, Smeland EB, Kvaloy S, Holte H, Delabie J, Campo E, Montserrat E, Lopez-Guillermo A, Ott G, Muller-Hermelink HK, Connors JM, Braziel R, Grogan TM, Fisher RI, Miller TP, LeBlanc M, Chiorazzi M, Zhao H, Yang L, Powell J, Wilson WH, Jaffe ES, Simon R, Klausner RD, Staudt LM (2003) Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. J Exp Med 198:851–862. https://doi.org/10.1084/jem.20031074
Joos S, Otaño-Joos MI, Ziegler S, Brüderlein S, du Manoir S, Bentz M et al (1996) Primary mediastinal (thymic) B-cell lymphoma is characterized by gains of chromosomal material including 9p and amplification of the REL gene. Blood 87:1571–1578
Green MR, Monti S, Rodig SJ, Juszczynski P, Currie T, O’Donnell E et al (2010) Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood 116:3268–3277. https://doi.org/10.1182/blood-2010-05-282780
Van Roosbroeck K, Ferreiro JF, Tousseyn T, van der Krogt J-A, Michaux L, Pienkowska-Grela B et al (2016) Genomic alterations of the JAK2 and PDL loci occur in a broad spectrum of lymphoid malignancies. Genes Chromosomes Cancer 55:428–441. https://doi.org/10.1002/gcc.22345
Chapuy B, Roemer MGM, Stewart C, Tan Y, Abo RP, Zhang L et al (2016) Targetable genetic features of primary testicular and primary central nervous system lymphomas. Blood 127:869–881. https://doi.org/10.1182/blood-2015-10-673236
Johnson NA, Slack GW, Savage KJ, Connors JM, Ben-Neriah S, Rogic S et al (2012) Concurrent expression of MYC and BCL2 in diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol Off J Am Soc Clin Oncol. https://doi.org/10.1200/JCO.2011.41.0985
Chatzitolios A, Venizelos I, Tripsiannis G, Anastassopoulos G, Papadopoulos N (2010) Prognostic significance of CD95, P53, and BCL2 expression in extranodal non-Hodgkin’s lymphoma. Ann Hematol 89:889–896. https://doi.org/10.1007/s00277-010-0945-x
Shukuya T, Carbone DP (2016) Predictive markers for the efficacy of anti-PD-1/PD-L1 antibodies in lung cancer. J Thorac Oncol Off Publ Int Assoc Study Lung Cancer. https://doi.org/10.1016/j.jtho.2016.02.015
Krishnan C, Warnke RA, Arber DA, Natkunam Y (2010) PD-1 expression in T-cell lymphomas and reactive lymphoid entities: potential overlap in staining patterns between lymphoma and viral lymphadenitis. Am J Surg Pathol 34:178–189. https://doi.org/10.1097/PAS.0b013e3181cc7e79
Kwon D, Kim S, Kim P-J, Go H, Nam SJ, Paik JH, Kim YA, Kim TM, Heo DS, Kim CW, Jeon YK (2016) Clinicopathological analysis of programmed cell death 1 and programmed cell death ligand 1 expression in the tumour microenvironments of diffuse large B cell lymphomas. Histopathology 68:1079–1089. https://doi.org/10.1111/his.12882
Carey CD, Gusenleitner D, Lipschitz M, Roemer MGM, Stack EC, Gjini E, Hu X, Redd R, Freeman GJ, Neuberg D, Hodi FS, Liu XS, Shipp MA, Rodig SJ (2017) Topological analysis reveals a PD-L1-associated microenvironmental niche for Reed-Sternberg cells in Hodgkin lymphoma. Blood 130:2420–2430. https://doi.org/10.1182/blood-2017-03-770719
Laurent C, Charmpi K, Gravelle P, Tosolini M, Franchet C, Ysebaert L, Brousset P, Bidaut A, Ycart B, Fournié JJ (2015) Several immune escape patterns in non-Hodgkin’s lymphomas. Oncoimmunology 4:e1026530. https://doi.org/10.1080/2162402X.2015.1026530
Menter T, Bodmer-Haecki A, Dirnhofer S, Tzankov A (2016) Evaluation of the diagnostic and prognostic value of PDL1-expression in Hodgkin and B-cell lymphomas. Hum Pathol. https://doi.org/10.1016/j.humpath.2016.03.005
Qiu L, Zheng H, Zhao X (2019) The prognostic and clinicopathological significance of PD-L1 expression in patients with diffuse large B-cell lymphoma: a meta-analysis. BMC Cancer 19:273. https://doi.org/10.1186/s12885-019-5466-y
Green MR, Monti S, Rodig SJ, Juszczynski P, Currie T, O’Donnell E et al (2010) Integrative analysis reveals selective 9p24.1 amplification, increased PD-1 ligand expression, and further induction via JAK2 in nodular sclerosing Hodgkin lymphoma and primary mediastinal large B-cell lymphoma. Blood 116:3268–3277. https://doi.org/10.1182/blood-2010-05-282780
Chen BJ, Chapuy B, Ouyang J, Sun HH, Roemer MGM, Xu ML, Yu H, Fletcher CD, Freeman GJ, Shipp MA, Rodig SJ (2013) PD-L1 expression is characteristic of a subset of aggressive B-cell lymphomas and virus-associated malignancies. Clin Cancer Res Off J Am Assoc Cancer Res 19:3462–3473. https://doi.org/10.1158/1078-0432.CCR-13-0855
Ngo VN, Young RM, Schmitz R, Jhavar S, Xiao W, Lim K-H, Kohlhammer H, Xu W, Yang Y, Zhao H, Shaffer AL, Romesser P, Wright G, Powell J, Rosenwald A, Muller-Hermelink HK, Ott G, Gascoyne RD, Connors JM, Rimsza LM, Campo E, Jaffe ES, Delabie J, Smeland EB, Fisher RI, Braziel RM, Tubbs RR, Cook JR, Weisenburger DD, Chan WC, Staudt LM (2011) Oncogenically active MYD88 mutations in human lymphoma. Nature 470:115–119. https://doi.org/10.1038/nature09671
Montesinos-Rongen M, Van Roost D, Schaller C, Wiestler OD, Deckert M (2004) Primary diffuse large B-cell lymphomas of the central nervous system are targeted by aberrant somatic hypermutation. Blood 103:1869–1875. https://doi.org/10.1182/blood-2003-05-1465
Twa DDW, Chan FC, Ben-Neriah S, Woolcock BW, Mottok A, Tan KL, Slack GW, Gunawardana J, Lim RS, McPherson A, Kridel R, Telenius A, Scott DW, Savage KJ, Shah SP, Gascoyne RD, Steidl C (2014) Genomic rearrangements involving programmed death ligands are recurrent in primary mediastinal large B-cell lymphoma. Blood 123:2062–2065. https://doi.org/10.1182/blood-2013-10-535443
Zhou XA, Louissaint A, Wenzel A, Yang J, Martinez-Escala ME, Moy AP et al (2018) Genomic analyses identify recurrent alterations in immune evasion genes in diffuse large B-cell lymphoma, leg type. J Invest Dermatol 138:2365–2376. https://doi.org/10.1016/j.jid.2018.04.038
Georgiou K, Chen L, Berglund M, Ren W, de Miranda NFCC, Lisboa S, Fangazio M, Zhu S, Hou Y, Wu K, Fang W, Wang X, Meng B, Zhang L, Zeng Y, Bhagat G, Nordenskjöld M, Sundström C, Enblad G, Dalla-Favera R, Zhang H, Teixeira MR, Pasqualucci L, Peng R, Pan-Hammarström Q (2016) Genetic basis of PD-L1 overexpression in diffuse large B-cell lymphomas. Blood 127:3026–3034. https://doi.org/10.1182/blood-2015-12-686550
Pardoll DM, Topalian SL (1998) The role of CD4+ T cell responses in antitumor immunity. Curr Opin Immunol 10:588–594
Steele KE, Tan TH, Korn R, Dacosta K, Brown C, Kuziora M, Zimmermann J, Laffin B, Widmaier M, Rognoni L, Cardenes R, Schneider K, Boutrin A, Martin P, Zha J, Wiestler T (2018) Measuring multiple parameters of CD8+ tumor-infiltrating lymphocytes in human cancers by image analysis. J Immunother Cancer 6:20–14. https://doi.org/10.1186/s40425-018-0326-x
Alvaro-Naranjo T, Lejeune M, Salvadó-Usach MT, Bosch-Príncep R, Reverter-Branchat G, Jaén-Martínez J, Pons-Ferré LE (2005) Tumor-infiltrating cells as a prognostic factor in Hodgkin’s lymphoma: a quantitative tissue microarray study in a large retrospective cohort of 267 patients. Leuk Lymphoma 46:1581–1591. https://doi.org/10.1080/10428190500220654
Alonso-Álvarez S, Vidriales MB, Caballero MD, Blanco O, Puig N, Martin A, Peñarrubia MJ, Zato E, Galende J, Bárez A, Alcoceba M, Orfão A, González M, García-Sanz R (2017) The number of tumor infiltrating T-cell subsets in lymph nodes from patients with Hodgkin lymphoma is associated with the outcome after first line ABVD therapy. Leuk Lymphoma 58:1144–1152. https://doi.org/10.1080/10428194.2016.1239263
McClanahan F, Sharp TG, Gribben JG (2016) Catching up with solid tumor oncology: what is the evidence for a prognostic role of programmed cell death-ligand 1/programmed cell death-1 expression in B-cell lymphomas? Haematologica 101:1144–1158. https://doi.org/10.3324/haematol.2016.145904
Pollari M, Brück O, Pellinen T, Vähämurto P, Karjalainen-Lindsberg M-L, Mannisto S, Kallioniemi O, Kellokumpu-Lehtinen PL, Mustjoki S, Leivonen SK, Leppä S (2018) PD-L1+ tumor-associated macrophages and PD-1+ tumor-infiltrating lymphocytes predict survival in primary testicular lymphoma. Haematologica 103:1908–1914. https://doi.org/10.3324/haematol.2018.197194
Gordon S, Martinez FO (2010) Alternative activation of macrophages: mechanism and functions. Immunity 32:593–604. https://doi.org/10.1016/j.immuni.2010.05.007
Biswas SK, Chittezhath M, Shalova IN, Lim J-Y (2012) Macrophage polarization and plasticity in health and disease. Immunol Res 53:11–24. https://doi.org/10.1007/s12026-012-8291-9
Adema GJ, Hartgers F, Verstraten R, de Vries E, Marland G, Menon S, Foster J, Xu Y, Nooyen P, McClanahan T, Bacon KB, Figdor CG (1997) A dendritic-cell-derived C-C chemokine that preferentially attracts naive T cells. Nature 387:713–717. https://doi.org/10.1038/42716
Hu H, Nie Y, Huang H, Guo M, Chen J, Wu W et al (2019) Breast phyllodes tumors recruit and repolarize tumor-associated macrophages via secreting CCL5 to promote malignant progression which can be inhibited by CCR5 inhibition therapy. Clin Cancer Res Off J Am Assoc Cancer Res. https://doi.org/10.1158/1078-0432.CCR-18-3421
Mu X, Shi W, Xu Y, Xu C, Zhao T, Geng B et al (2018) Tumor-derived lactate induces M2 macrophage polarization via the activation of the ERK/STAT3 signaling pathway in breast cancer. Cell Cycle Georget Tex 17:428–438. https://doi.org/10.1080/15384101.2018.1444305
Bingle L, Brown NJ, Lewis CE (2002) The role of tumour-associated macrophages in tumour progression: implications for new anticancer therapies. J Pathol 196:254–265. https://doi.org/10.1002/path.1027
Nam SJ, Go H, Paik JH, Kim TM, Heo DS, Kim CW, Jeon YK (2014) An increase of M2 macrophages predicts poor prognosis in patients with diffuse large B-cell lymphoma treated with rituximab, cyclophosphamide, doxorubicin, vincristine and prednisone. Leuk Lymphoma 55:2466–2476. https://doi.org/10.3109/10428194.2013.879713
Menguy S, Prochazkova-Carlotti M, Beylot-Barry M, Saltel F, Vergier B, Merlio J-P et al (2017) PD-L1 and PD-L2 are differentially expressed by macrophages or tumor cells in primary cutaneous diffuse large B-cell lymphoma. Leg Type Am J Surg Pathol. https://doi.org/10.1097/PAS.0000000000000983
Nayak L, Iwamoto FM, LaCasce A, Mukundan S, Roemer MGM, Chapuy B, Armand P, Rodig SJ, Shipp MA (2017) PD-1 blockade with nivolumab in relapsed/refractory primary central nervous system and testicular lymphoma. Blood 129:3071–3073. https://doi.org/10.1182/blood-2017-01-764209
Author information
Authors and Affiliations
Contributions
Vanessa Szablewski, Melissa Alame, Valérie Costes-Martineau, Valérie Rigau and Valère Cacheux designed the research project. Vanessa Szablewski, Marion Pirel, Valérie Rigau, Luc Durand and Valérie Costes-Martineau evaluated the histological and immunohistochemical findings. Vanessa Szablewski, Melissa Alame, Laura De Oliveira, Alicia Tourneret and Valère Cacheux evaluated the cytogenetic findings. Vanessa Szablewski and Melissa Alame obtained data and wrote the main part of the manuscript. Marion Pirel, Valérie Costes-Martineau, Luc Bauchet, Michel Fabbro, Pascal Roger, Samia Gonzalez, Luc Durand, Laura De Oliveira, Alicia Tourneret Tempier, Valérie Rigau and Valère Cacheux reviewed the draft with critical comments.
Corresponding author
Ethics declarations
This study was carried out in agreements with the Declaration of Helsinki and was approved by the Centre des Ressources Biologiques (CRB) of the Centre Hospitalo Universitaire (CHU) of Montpellier, France.
Conflict of interest
The authors declare that they have no conflict of interest.
Informed consent
Written informed consent for the study was obtained from the patient.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Quality in Phatology
Electronic supplementary material
ESM 1
(DOCX 134 kb)
Rights and permissions
About this article
Cite this article
Alame, M., Pirel, M., Costes-Martineau, V. et al. Characterisation of tumour microenvironment and immune checkpoints in primary central nervous system diffuse large B cell lymphomas. Virchows Arch 476, 891–902 (2020). https://doi.org/10.1007/s00428-019-02695-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-019-02695-6