Abstract
Celiac disease (CD) is a risk factor for developing small-bowel carcinoma with a 14-fold higher risk compared with general population. As small-bowel carcinomas associated with CD (CD-SBCs) are extremely rare, very few molecular data are available about their pathogenesis, and information about their transcriptomic profiling is lacking. We generated RNA-seq data on 13 CD-SBCs, taken from the largest well-characterized series published so far, collected by the Small Bowel Cancer Italian Consortium, and compared the tumor transcriptional signatures with the four Consensus Molecular Subtypes (CMS) of colorectal carcinoma by applying the “CMS classifier.” CpG Island Methylator Phenotype (CIMP) was evaluated using methylation-sensitive multiple ligation-dependent probe amplification. Up to 12 of 13 cancers fell within the two main subtypes exhibiting high immune and inflammatory signatures, i.e., subtypes 1 and 4. The first and predominant subset was commonly microsatellite unstable, and exhibited CIMP and high CD3+ and CD8+ T cell infiltration. Moreover, it showed increased expression of genes associated with T helper 1 and natural killer cell infiltration, as well as upregulation of apoptosis, cell cycle progression, and proteasome pathways. By contrast, cancers falling in subtype 4 showed prominent transforming growth factor-β activation and were characterized by complement-associated inflammation, matrix remodeling, cancer-associated stroma production, and angiogenesis. Parallel histologic and histochemical analysis confirmed such tumor subtyping. In conclusion, two molecular subtypes have been consistently identified in our series of CD-SBCs, a microsatellite instability-immune and a mesenchymal subtype, the former likely associated with an indolent and the latter with a worse tumor behavior.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Celiac disease (CD) is a chronic enteropathy induced, in genetically susceptible individuals, by gluten ingestion [1] that affects 1% of the Western population [2], and is a predisposing condition for small-bowel carcinoma (SBC). SBCs account for about 2% of gastrointestinal carcinomas and are generally associated with poor prognosis [3]. In a recent meta-analysis of eight studies, including 79,991 CD patients and 75 SBCs associated with CD (CD-SBCs), SBC risk in CD subjects has been estimated to be 14-fold higher than that of the general population [4], while in one study on 175 patients with SBC, 13% had CD [5]. Thus, CD-SBC is a rare condition, which explains why, to date, information concerning this tumor is limited to a few small series or case reports, and its molecular landscape is largely unknown [6,7,8,9,10].
In one of the largest series of CD-SBCs published so far [11, 12], we identified specific histopathological, molecular, and prognostic features of CD-SBC, distinctive from those of sporadic or Crohn’s disease–associated SBCs, and characterized by frequent microsatellite instability (MSI), associated with MLH1 methylation, and high density of tumor-infiltrating T lymphocytes (TILs). Recently, a MSI-immune subtype has been identified at gene expression level among various epithelial neoplasms, including colorectal, gastric, and pancreatic cancers [13, 14]. In particular, for colorectal cancer (CRC), the Consensus Molecular Subtype (CMS) Consortium proposed the current best description of CRC heterogeneity at the gene-expression level, after analysis of 18 different CRC gene expression datasets [15]. This transcriptomic classification enabled the categorization of most CRCs into four robust subtypes: CMS1 (MSI immune subtype, 14%), which was characterized by hypermutation, CpG Island Methylator Phenotype (CIMP) and evidence of strong immune activation (immune response factors, natural Killer (NK) cell, T helper 1 cell (TH1) and cytotoxic T cell infiltration signatures, PD1 activation), consistent with pathological descriptions of prominent tumor-infiltrating CD8+ cytotoxic T lymphocytes. Next, the remaining subtypes were CMS2 (canonical subtype 37%, with marked Wnt and MYC signaling activation); CMS3 (metabolic 13%, with evident metabolic dysregulation and KRAS mutations); and CMS4 (mesenchymal 23%, with prominent transforming growth factor (TGF)-β, stromal component, angiogenesis, and worse overall survival).
Interestingly, concerning CRCs, this transcriptomic classification showed important and clinically relevant correlations with a histologically based classification approaches [16, 17].
We aimed at investigating whether MSI CD-SBCs could correspond to the CMS1 transcriptomic subtype of CRCs, and to what extent the remaining molecular subtypes identified in CRC were also represented in CD-SBCs. For this purpose, we generated RNA-seq data on 13 CD-SBCs and compared their transcriptional signatures with those of the four CRC CMSs. Moreover, in each case, we also evaluated the presence of CIMP and of any correlation between transcriptomic and histologic, immunohistochemical, or genetic findings.
Materials and methods
Patient cohort
The study cohort included 13 CD-SBCs taken from a larger and well-characterized series previously described [11, 12], on the basis of (a) sufficient formalin-fixed paraffin-embedded (FFPE) tumor tissue still available for RNA extraction and (b) good quality of extracted RNA. Histologically normal small-bowel mucosa specimens from four pancreato-duodenectomies for pancreatic tumors in non-CD patients were used as controls.
For all tumors, MSI status, MLH1 methylation, and KRAS, NRAS, BRAF, PIK3CA, and TP53 mutations had been previously analyzed [11]. As we previously observed, two main groups of tumors among CD-SBCs, namely MSI/MLH1 hypermethylated tumors and microsatellite stable (MSS) cases exhibiting frequent TP53 mutations, both types of CD-SBCs were included in this study. The study was performed in agreement with the clinical standards laid down in the 1975. Declaration of Helsinki and its revision and was approved by the Ethics Committee of the IRCCS San Matteo Hospital Foundation, Italy (no. 20140018113, on 22nd September 2014).
RNA library preparation and sequencing
Total RNA was extracted from FFPE sections (4 normal small-bowel mucosa samples and 13 CD-SBCs) using RNeasy FFPE Kit (QIAGEN GmbH, Hilden, Germany); RNA concentration was measured using Qubit® RNA HS Assay Kit on a Qubit® 2.0 Fluorometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). Integrity was assessed using Agilent RNA 6000 Nano Kit on a 2100 Bioanalyzer instrument (Agilent Technologies, Santa Clara, CA, USA) and the percentages of fragments larger than 200 nucleotides were calculated. RNA sequencing libraries were prepared using TruSeq RNA Exome Library kit (Illumina, Inc., San Diego, CA, USA) according to the manufacturer’s protocol. In brief, RNA samples (100 ng total RNA) were fragmented at 94 °C for 8 min on a thermal cycler. First-strand cDNA syntheses were performed at 25 °C for 10 min, 42 °C for 15 min, and 70 °C for 15 min, using random hexameres and SuperScript II Reverse Transcriptase (Thermo Fisher Scientific Inc., Waltham, MA, USA). In a second-strand cDNA synthesis, the RNA templates were removed, and a second replacement strand was generated by incorporation dUTP (in place of dTTP, to keep strand information) to generate ds cDNA. The 3′ ends of the cDNA were then adenylated to facilitate adaptor ligation in the next step. In a first PCR amplification step, PCR (15 cycles of 98 °C for 10 s, 60 °C for 30 s, and 72 °C for 30 s) was used to selectively enrich those DNA fragments that have adapter molecules on both ends and to amplify the amount of DNA in the library. After validation of the libraries, using Agilent DNA 1000 kit on a 2100 Bioanalyzer instrument, the first hybridization step was performed using exome capture probes. Before hybridization, a 4-plex pool of libraries were made, by combining 200 ng of each DNA library. The hybridization was performed by 18 cycles of 1-min incubation, starting at 94 °C, and then decreasing 2 °C per cycle. Then streptavidin-coated magnetic beads were used to capture probes hybridized to the target regions. The second hybridization (18 cycles of 1-min incubation, starting at 94 °C, and then decreasing 2 °C per cycle) was required to ensure high specificity of the capture regions. A second capture with streptavidin-coated beads was performed, followed by two heated wash procedures to remove non-specific binding form the beads. The amplification step was performed by 10 cycles (98 °C for 10 s, 60 °C for 30 s, and 72 °C for 30 s). Finally, the libraries were quantitated using Qubit dsDNA HS (High Sensitivity) Assay Kit on a Qubit® 2.0 Fluorometer and validated using Agilent High Sensitivity DNA Kit on a Bioanalyzer. The size range of the DNA fragments was measured to be in the range of 200–650 bp and peaked around 270 bp. Libraries were normalized to 2 pM and sequenced on a NextSeq500 instrument 2 × 75 bp (Illumina, Inc. San Diego, CA, USA), according to the manufacturer’s instructions.
Bioinformatic analysis
For RNA sequencing, bioinformatics analysis was performed as described in Tarallo et al. [18]. In brief, sequencing reads were quality filtered and aligned to the human genome hg38 (Homo sapiens Ensembl GRCh38) using STAR v.2.5.2a [19]. HTSeq [20] was used to compute read counts across each gene, which were then used as input to R package DESeq2 [21]. DESeq2 was used to normalize read counts for library size and dispersion followed by tests for differential gene expression. Significant differentially expressed genes were determined using adjusted p value (p-adj) cutoff ≤ 0.05 and at least 1.5-fold change (FC) between conditions. Functional analyses were performed with Ingenuity Pathway Analysis suite (Ingenuity Systems).
The expression pattern of 5973 genes was used to classify CD-SBC samples into CMS subtypes exploiting a “single-sample predictor” (SSP) classifier. Indeed, SSP makes possible to predict a unique sample, and its output, considering any given sample, must remain constant whether it is predicted alone or within a series of samples. The SSP is implemented in the “CMSclassifier” R package. It is based on a similarity-to-centroid approach, with the Pearson coefficient as a similarity measure [15]. For the unclassified samples, the most similar subtype to the case has been identified using the “nearest CMS” classification.
IPA (Ingenuity Pathway Analysis, Ingenuity Systems) was used for functional enrichment and detection of significant pathways. Fisher’s Exact algorithm was used to calculate the probability of which each functional gene set was enrichment (p value < 0.05).
The gene set enrichment analysis (GSEA) was done using GSEA software version 3.0, which a collection of 70 cancer-related gene set based on Guinney et al. [15] with standard settings (with the exception that the permutation type was set to “gene_set” and gene sets < 15 genes were allowed). RNA sequencing data were deposited in the EBI ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) with Accession Number E-MTAB-7760.
CIMP phenotype analysis
Methylation study was performed using methylation-sensitive multiple ligation-dependent probe amplification (MS-MLPA) that allows the simultaneous assessment of promoter methylation of multiple genes in a single experiment. SALSA MS-MLPA ME042-C1 CIMP Kit (MRC-Holland, Amsterdam, The Netherlands) was used to perform methylation analysis on 8 gene promoters frequently methylated in CIMP tumors [22] (details in Online Resource 1). To classify a gene promoter as methylated, at least half of the probes had to show methylation (Online Resource 2). We considered CIMP positive a sample if it showed at least 3 out of 8 methylated promoters.
SALSA MS-MLPA ME001-tumor suppressor-1 Kit was used to confirm CIMP status. As previously described, we fully validated the sensitivity and specificity of this MS-MLPA assay [23]. For the CIMP status evaluation, we considered positive a sample showing at least 3 methylated promoters.
Histologic, histochemical, and genetic analysis
Histologic and histochemical patterns of the 13 CD-SBCs entering this study were carefully reinvestigated according to the procedures detailed in previous papers [11, 12]. Genetic data concerning TP53, KRAS, and PIK3CA genes or MSI status analysis, reported collectively in the previous papers [11, 12], were individually taken from our database. Finally, patient follow-up and outcome were updated.
Results
Differential gene expression in small-bowel carcinoma vs. normal intestinal mucosa
In Table 1, we summarized the main clinico-pathologic and molecular data available for all the 13 CD-SBC cases investigated. RNA-Seq was performed on FFPE samples, yielding an average of 88,654,932 ± 33,018,149 read counts per sample and ~ 73% of the reads mappable to RNA coding regions (Online Resource 3 provides a summary of RNA-sequencing statistics). After filtering lowly expressed transcripts based on the threshold of TPM (transcripts per million < 10) for all 17 tissues, 6,168 transcripts were retained. Among them, 3,142 RNAs were differentially expressed (DE) between tumor and normal tissues considering a Benjamini-Hochberg adjusted p value (p-adj) ≤ 0.05, including 1709 overexpressed genes (FC ≥ 1.5) and 1306 (FC ≤ 1.5) under expressed genes (Online Resource 4). Unsupervised hierarchical clustering analysis (HCA) of these 3,142 genes showed that normal and tumor samples clustered into distinct groups (Fig. 1a). Similar results were also revealed by a principal component analysis (PCA) (Online Resource 5), demonstrating that CD-SBCs can be distinguished from normal samples using large-scale gene expression analyses. To test whether disease-relevant pathways have been captured in our experiment, the 3142 genes identified as having significant differential expression were analyzed by Ingenuity Pathway Analyses (IPA). IPA of DE genes revealed 25 significant (p value < 0.05) biological function categories related to the catalogs Disease and Disorder, 21 resulting activated (z-score ≥ 2) and 3 inhibited (z-score ≤ − 2) (Online Resource 6). In detail, a decrease in biological functions including cell death, lipid oxidation, and transport of molecules was observed. By contrast, many differentially expressed genes were involved in activated cancer-related functions such as cellular invasion and movement, cells proliferation and survival, colony formation, checkpoint control, and progression of cell cycle. Additionally, genes involved in the inflammatory response, cellular infiltration by granulocytes, and fibrogenesis were also significantly enriched (Fig. 1b).
a Heatmap and 2-way hierarchical clustering (Kendal distance, ward.D linkage) based on 1709 overexpressed genes (FC ≥ 1.5) and 1306 (FC ≤ 1.5) underexpressed genes between CD-SBC samples vs. normal mucosa, log2(transformed RPM)-median centered expression are displayed for each RNA. Normal (blue label) and tumor (red label) samples fall in separate clusters. b IPA showing decreased and increased biological functions in CD-SBC samples vs. normal mucosa. Depicted are functions with an activation score (z-score) > 2 (increased activation, red) or ≤ 2 (decreased activation, green) with their corresponding log2 (P) (right graph)
The top overexpressed genes are cancer-related genes known to be involved in many types of cancer and included zinc-finger transcriptional regulators ZIC2 (FC 46.3) and ZIC5 (FC 48.6); a member of the paired box gene family, Pax9 (FC 44.30); the cartilage oligomeric matrix protein, COMP (FC 33.03); and the migratory indicator protein, CEMIP (FC 30.93) [24, 25]. Besides, an aberrant expression of WNT signaling components has been observed; specifically, we found 15 overexpressed and 7 underexpressed genes (underlined in Online Resource 4). In detail, in CD-SBC, CDH3 and WNT2 were the most overexpressed genes (FC > 10), and NR5A2 and SOX6 the most underexpressed (FC < 5). Remarkably, also β-catenin (CTNNB1) and the transcription factor SOX9 were upregulated, supporting at RNA level our previous findings about protein immunohistochemical overexpression [12].
Finally, we hypothesized that long non-coding RNA (lncRNA) expression may also change in CD-SBC vs. normal mucosae. Indeed, inside the DE RNAs, a total of 320 lncRNAs were identified (see Online Resource 4) including 115 overexpressed (FC ≥ 1.5) and 47 (FC ≤ 1.5) underexpressed. Above all, we found that WT1-AS and the imprinted oncofetal lncRNA H19 were significantly overexpressed in tumor tissues compared to non-tumor tissues (FC 30.85 and FC 10.12, respectively).
Interestingly, we found that 12 HOX genes, 2 lncRNA (HOXA10-AS and HOXA11-AS) encoded in HOXA cluster, and 1 miRNA gene (miRNA196A1) encoded in HOXB cluster, were strongly overexpressed in CD-SBC.
Transcriptomic subtype assessment in CD-SBC
To shed light on CD-SBC molecular features, we applied on 13 CD-SBCs the “CMS classifier” [15]. This analysis allowed to assign nine CD-SBCs (69%) into two of the four CMSs (Online Resource 7), as 6 samples fitted the CMS1 group and 3 the CMS4. The remaining four tumors (31%), probably representing either intermediate characteristics or falling out the cutoff of the algorithm, remained unclassified. The classification was also performed using the “nearest CMS” to find the most similar subtype for unclassified samples, revealing that T12 and T40 were like CMS1; T10 was closely related to CMS4 while T19 seemed to resemble CMS2 (Online Resource 7). The distribution of the CMS-like groups is shown in Fig. 2, including mixed or indeterminate samples.
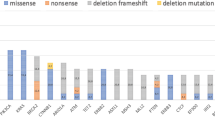
Heatmap and 2-way hierarchical clustering (Kendal distance, ward.D linkage) based on 1498 differentially expressed genes between CMS1 and CMS4 samples; log2(transformed RPM)-median centered expression are displayed for each RNA. Annotation bars on top of the heatmap visualize the membership of the different samples to the four CMS categories using “CMS classifier” (Predicted CMS) and “nearest CMS” (Nearest CMS) algorithm. The presence of MSI status (blue bars), CIMP status (yellow bars), MLH1 DNA methylation (orange bars), KRAS mutation (purple bars), TP53 mutations (green bars), and PIK3CA mutations (pink bars), for each sample, is summarized in the lower panel
Differential expression analysis between the two bona fide CD-SBC subtypes identified (6 CMS1/MSI-immune vs. 3 CMS4/mesenchymal cases) was performed, allowing the identification of 1,498 DE RNAs considering a Benjamini-Hochberg p-adj ≤ 0.05, including 537 overexpressed genes (FC ≥ 1.5) and 950 underexpressed genes (FC ≤ 1.5) (Online Resource 8). HCA analysis of these 1,498 genes showed a clear separation between the two sample groups, while the unassigned samples exhibited a mixed gene expression pattern (Fig. 2).
To gain insight into the biological characteristics of each of the subtypes, we performed the GSEA using previously described signature of pathways and well-characterized cellular processes, focusing on the two CMS groups identified in CD-SBCs and using the 9 samples confidently labeled by the CMS classifier (Fig. 3 and Fig. 4, Online Resource 9). Signatures were considered significant if the corrected FDR q-value was < 0.05. The CMS1-like/MSI-immune samples were characterized by increased expression of genes associated with diffuse immune infiltrate (51 positively correlated genes, Fig. 3 and Online Resource 9). Specially, immune activation analysis revealed in MSI-immune CD-SBCs an overrepresentation of specific gene sets related to a strong infiltration of TH1 cells, follicular helper T (TFH) cells, NK cells, and immune checkpoints, such as PD-1 (Fig. 3 and Online Resource 9), characteristics similar to what was observed in CRC of the same subtype. Worth mentioning, samples classified as CMS1-like had an elevated expression of genes involved in apoptosis (Caspase cascade), cell cycle progression and proteasome (PROTEASOME Reactome) pathways, three prominent signatures also reported in CMS1 CRCs as well as in the intestinal mucosa of untreated CD patients [24]. The MSI-immune CD-SBC group encompassed most of MSI/MLH1 methylated tumors (6 out of 7, considering the Predicted CMS, all of them referring to Nearest CMS) and of the CIMP cancers (6 out of 9 considering the Predicted CMS, 7 out of 9 referring to Nearest CMS).
Gene set enrichment analysis (GSEA) of genes highly expressed in CMS1 in comparison with CMS4. Enrichment plots and heat maps for selected gene sets are represented, complete gene set lists are available in Online Research 9. The normalized enrichment score (NES), the p value and the false discovery rate (q-value) are shown for each gene set. Heat maps shown are truncated to show only the genes identified as contributing to the enrichment score.
Gene set enrichment analysis (GSEA) of genes highly expressed in CMS4 in comparison with CMS1. Enrichment plots and heat maps for selected gene sets are represented; complete gene set lists are available in Online Research 9. The normalized enrichment score (NES), the p value, and the false discovery rate (q-value) are shown for each gene set. Heat maps shown are truncated to show only the genes identified as contributing to the enrichment score
On the other hand, the CD-SBC samples included in the CMS4 subtype exhibited gene expression profiles compatible with stromal infiltration (94 positively correlated genes, Fig. 4 and Online Resource 9), likely due to the higher density of stromal cells and/or an activated state of stromal components. Moreover, they showed expression of complement components, high activity of genes characteristic of a mesenchymal phenotype and a clear upregulation of genes implicated in epithelial mesenchymal transition, extracellular matrix remodeling, and wound response (Fig. 4 and Online Resource 9). This group of samples was also characterized by enhanced stem cell properties, like TGF-β and integrin β3 signaling activation, necessary for tumor invasion and establishment of metastases.
Correlation of transcriptomic with histologic, immunohistochemical, and genetic data
Parallel histologic and immunohistochemical analysis of the 13 CD-SBCs confirmed the presence of two distinct tumor populations, one of which characterized by glandular to medullary histology, increased intratumor T cell infiltration (> 15 CD3+ and > 9.5 CD8+ lymphocytes per high-power field), and mismatch repair protein loss, and the other one by desmoplasia-rich mixed to diffuse tumor growth, with variable T cell infiltration (Table 1 and Fig. 5), corresponding to those fitting into subtypes 1 and 4, respectively, of transcriptomic investigations. In addition, case T12, an MSI glandular case with CIMP and high TILs, closely resembled subtype 1, while case T10, a MSS case without CIMP and with a multifocal diffuse desmoplastic component, resembled subtype 4 of transcriptomic investigations, in keeping with the results of Nearest CMS analysis. The remaining two cases (T40 and T19) remained unclassified by histologic/immunohistochemical investigations.
Histologic features of small-bowel carcinomas associated with celiac disease (CD-SBC) investigated. a A medullary-type CD-SBC (corresponding to case T3 in Table 1), characterized by a trabecular-to-solid growth pattern, a pushing border, and a prominent intratumoral and peritumoral lymphocytic infiltration (H&E). b A glandular-type CD-SBC (corresponding to case T4 in Table 1), characterized by well-formed glandular structures; note the high number of CD8-positive tumor-infiltrating lymphocytes (CD8 immunohistochemistry). c A mixed-type CD-SBC (corresponding to case T2 in Table 1), with a mixture of both glandular and diffuse pattern (H&E). d A diffuse-type CD-SBC (corresponding to case T13 in Table 1), characterized by poorly cohesive cells arranged in single elements or small clusters, dispersed in a prominent, cell-rich, desmoplastic stroma (H&E). e, f The same diffuse-type carcinoma depicted in d is seen at higher magnification, to recognize cytokeratin 20 positive epithelial neoplastic cells admixed with cancer-associated fibroblasts (e H&E; f cytokeratin 20 immunohistochemistry)
TP53 mutations were mainly found in mesenchymal tumors or in unclassified tumors (Fig. 2 and Table 1). Interestingly, although most CD-SBCs investigated were characterized by a relatively indolent behavior, the only two patients who died of disease showed TP53 mutated tumors belonging to the mesenchymal group (T13) or to the unclassified CD-SBCs (T40).
MSI-immune CD-SBCs were preferentially located in the jejunum or in the ileum, frequently characterized by KRAS mutation and encompassed all the three tumors showing a medullary-type histology.
Worth of note, all the three bona fide mesenchymal CD-SBC cases presented with lymph node metastases at diagnosis in contrast to a minority (17%) of MSI-immune cases.
Discussion
In this study on CD-SBCs, two distinct types of molecular signatures were clearly identified by gene expression analysis of tumor tissue, the MSI-immune, and the mesenchymal type, resembling subtype 1 and subtype 4, respectively, of colorectal CMS [15]. In particular, the first tumor group showed increased expression of genes associated with a strong infiltration of TH1 and TFH cells, NK cells, and overexpressed immune checkpoints, such as PD-1, as well as CIMP patterns. Three of such tumors (50%) showed KRAS mutation. On the other hand, the mesenchymal subtype exhibited gene expression profiles compatible with high density and/or activation of stromal cells, including those with a frankly mesenchymal profile (possibly linked to epithelial-mesenchymal transition and matrix remodeling), and implying enhanced stem cell properties, such as a TGF-β signaling activation. As expected, mesenchymal CD-SBCs were MSS, without CIMP, and frequently showed TP53 mutations.
Parallel histological and histochemical investigation of the same CD-SBCs [11, 12] allowed to support present transcriptomic profiles, by showing the presence in 7 cases of MSI, high intratumor T cell infiltration, and glandular to medullary structure, without intratumor desmoplasia, and, in 4 MSS cases, of a desmoplasia-rich, diffuse, or mixed glandular to diffuse structure, corresponding, respectively, to type 1 MSI-immune and type 4 mesenchymal CD-SBCs of transcriptomic analysis.
Immunohistochemical analysis also showed, in most CD-SBCs [12], nuclear expression of β-catenin and SOX-9, and two crucial markers of Wnt activation [26, 27], whose overexpression was confirmed by present transcriptomic investigation. This finding would be at variance with the CRC CMS classification, where marked Wnt activation is known to be typical of the canonical CMS2 subtype [15], while MSI CRCs only rarely display immunohistochemical evidence of Wnt activation [28]. It should be recalled, however, that nuclear SOX-9 is also overexpressed in CD crypt hyperplastic cells [12, 29] as well as in related dysplastic lesions, where it is coupled with nuclear β-catenin expression and where both Wnt-related markers may precede the appearance of MSI, mostly restricted to invasive full-blown cancers [12]. Thus, the unexpectedly high Wnt activation found in CD-SBCs might reflect their retention of a special cellular and molecular background inherent to the underlying intestinal disorder, rather than the development of a specific molecular cancer subset. Indeed, in the small intestinal mucosa of untreated CD patients, nuclear NF-kB activity, known to positively regulate SOX9 expression [30, 31], has been shown to be constitutively increased [32,33,34], a change likely promoting the TH1 immune response distinctive of active CD [35] and also found to be operative in the MSI-immune subset of CD-SBCs (present study).
Interestingly, the only subtype other than the MSI-immune we consistently detected in CD-SBCs was the mesenchymal one. Several recent studies demonstrated that a peculiar feature of the mesenchymal subtype is an extensive tumor infiltration by Cancer Associated Fibroblasts (CAF) that promote inflammatory and angiogenic microenvironments [36, 37] and suggested that CMS4 together with CMS1 are the only CRC subtypes exhibiting high immune and inflammatory signatures [16].
These observations are confirmed by our transcriptomic findings and may fit in with our detection of prominent desmoplasia in such tumors, coupled with increased intratumor T cell infiltration in two out of the four mesenchymal cases. Of note, in our study, gene expression related to cytotoxic T lymphocytes did not show significant differences between MSI-immune and mesenchymal CD-SBCs, also in keeping with the increased representation of cytotoxic T signature in both CMS1 and CMS4 CRCs, as found by Becht et al [16]. In addition, our data showing that CD-SBCs fall almost exclusively into the MSI-immune and mesenchymal subtypes appear very interesting considering the role of chronic immune-inflammatory processes in CD pathogenesis. The substantial lack among CD-SBCs of tumors recalling canonical CMS2 or metabolic CMS3 subsets, which account for the majority of CRCs [15], may also suggest that more restricted immune-inflammatory pathogenetic pathways are operative among CD-SBCs. In this regard, it should be also recalled that CIMP has been found in most CD inflamed small-bowel mucosae [38], as well as in MSI-immune CD-SBCs (present study), further indicating a close molecular link between mechanisms operative in CD and SBC development.
In several cancers, including CRC [36, 37], the mesenchymal subtype has been shown to associate with worse and the MSI-immune subtype with a more favorable outcome, in keeping with our present findings in CD-SBCs, where none of patients with a MSI, high TIL, and CIMP-positive cancer showed tumor-related death or evidence of recurrence during the follow-up. Indeed, the predominance of MSI-immune subtype cases among CD-SBCs may account for the higher survival rates found in CD-SBC patients in comparison to the remaining SBC patients [8, 11, 39].
It has been shown in solid cancers of various origin, including small intestine, that MSI status, usually associated with high tumor-infiltrating T lymphocytes, may predict response to PD-1 immune checkpoint blockade therapy [40, 41]. Whether immune checkpoint inhibitors may also be effective in CD-SBC patients, especially in those with MSI-immune subtype tumors, remains to be investigated.
In conclusion, two molecular subtypes have been consistently identified in our series of CD-SBCs, a MSI-immune and a mesenchymal subtype, the former likely associated with a more indolent and the latter with a worse tumor behavior. Present clarification of CD-SBC molecular and cellular profiles, with special references to their CRC-shared immune-inflammatory pathways, may suggest novel therapeutic approaches. However, further work on larger series of this very rare tumor disease is needed before clinically relevant conclusions can be obtained.
References
Di Sabatino A, Corazza GR (2009) Coeliac disease. Lancet 373:1480–1493. https://doi.org/10.1016/S0140-6736(09)60254-3
Dube C, Rostom A, Sy R, Cranney A, Saloojee N, Garritty C, Sampson M, Zhang L, Yazdi F, Mamaladze V, Pan I, Macneil J, Mack D, Patel D, Moher D (2005) The prevalence of celiac disease in average-risk and at-risk Western European populations: a systematic review. Gastroenterology 128:S57–S67. https://doi.org/10.1053/j.gastro.2005.02.014
Aparicio T, Zaanan A, Svrcek M, Laurent-Puig P, Carrere N, Manfredi S, Locher C, Afchain P (2014) Small bowel adenocarcinoma: epidemiology, risk factors, diagnosis and treatment. Dig Liver Dis 46:97–104. https://doi.org/10.1016/j.dld.2013.04.013
Han Y, Chen W, Li P, Ye J (2015) Association between coeliac disease and risk of any malignancy and gastrointestinal malignancy: a meta-analysis. Medicine (Baltimore) 94:e1612. https://doi.org/10.1097/MD.0000000000001612
Howdle PD, Jalal PK, Holmes GK, Houlston RS (2003) Primary small-bowel malignancy in the UK and its association with coeliac disease. QJM 96:345–353. https://doi.org/10.1093/qjmed/hcg058
Bruno CJ, Batts KP, Ahlquist DA (1997) Evidence against flat dysplasia as a regional field defect in small bowel adenocarcinoma associated with celiac sprue. Mayo Clin Proc 72:320–322. https://doi.org/10.1016/S0025-6196(11)63330-2
Rampertab SD, Forde KA, Green PH (2003) Small bowel neoplasia in coeliac disease. Gut 52:1211–1214. https://doi.org/10.1136/gut.52.8.1211
Potter DD, Murray JA, Donohue JH, Burgart LJ, Nagorney DM, van Heerden JA, Plevak MF, Zinsmeister AR, Thibodeau SN (2004) The role of defective mismatch repair in small bowel adenocarcinoma in celiac disease. Cancer Res 64:7073–7077. https://doi.org/10.1158/0008-5472.CAN-04-1096
Diosdado B, Buffart TE, Watkins R, Carvalho B, Ylstra B, Tijssen M, Bolijn AS, Lewis F, Maude K, Verbeke C, Nagtegaal ID, Grabsch H, Mulder CJ, Quirke P, Howdle P, Meijer GA (2010) High-resolution array comparative genomic hybridization in sporadic and celiac disease-related small bowel adenocarcinomas. Clin Cancer Res 16:1391–1401. https://doi.org/10.1158/1078-0432.CCR-09-1773
Alvi MA, McArt DG, Kelly P, Fuchs MA, Alderdice M, McCabe CM, Bingham V, McGready C, Tripathi S, Emmert-Streib F, Loughrey MB, McQuaid S, Maxwell P, Hamilton PW, Turkington R, James JA, Wilson RH, Salto-Tellez M (2015) Comprehensive molecular pathology analysis of small bowel adenocarcinoma reveals novel targets with potential for clinical utility. Oncotarget 6:20863–20874. https://doi.org/10.18632/oncotarget.4576
Vanoli A, Di Sabatino A, Furlan D, Klersy C, Grillo F, Fiocca R, Mescoli C, Rugge M, Nesi G, Fociani P, Sampietro G, Ardizzone S, Luinetti O, Calabro A, Tonelli F, Volta U, Santini D, Caio G, Giuffrida P, Elli L, Ferrero S, Latella G, Ciardi A, Caronna R, Solina G, Rizzo A, Ciacci C, D’Armiento FP, Salemme M, Villanacci V, Cannizzaro R, Canzonieri V, Reggiani Bonetti L, Biancone L, Monteleone G, Orlandi A, Santeusanio G, Macciomei MC, D’Inca R, Perfetti V, Sandri G, Silano M, Florena AM, Giannone AG, Papi C, Coppola L, Usai P, Maccioni A, Astegiano M, Migliora P, Manca R, Martino M, Trapani D, Cerutti R, Alberizzi P, Riboni R, Sessa F, Paulli M, Solcia E, Corazza GR (2017) Small bowel carcinomas in coeliac or Crohn’s disease: clinico-pathological, molecular, and prognostic features. A Study From the Small Bowel Cancer Italian Consortium. J Crohns Colitis 11:942–953. https://doi.org/10.1093/ecco-jcc/jjx031
Vanoli A, Di Sabatino A, Martino M, Klersy C, Grillo F, Mescoli C, Nesi G, Volta U, Fornino D, Luinetti O, Fociani P, Villanacci V, D’Armiento FP, Cannizzaro R, Latella G, Ciacci C, Biancone L, Paulli M, Sessa F, Rugge M, Fiocca R, Corazza GR, Solcia E (2017) Small bowel carcinomas in celiac or Crohn’s disease: distinctive histophenotypic, molecular and histogenetic patterns. Mod Pathol 30:1453–1466. https://doi.org/10.1038/modpathol.2017.40
Bijlsma MF, Sadanandam A, Tan P, Vermeulen L (2017) Molecular subtypes in cancers of the gastrointestinal tract. Nat Rev Gastroenterol Hepatol 14:333–342. https://doi.org/10.1038/nrgastro.2017.33
Network CGAR (2014) Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513:202–209. https://doi.org/10.1038/nature13480
Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda G, Angelino P, Bot BM, Morris JS, Simon IM, Gerster S, Fessler E, De Sousa EMF, Missiaglia E, Ramay H, Barras D, Homicsko K, Maru D, Manyam GC, Broom B, Boige V, Perez-Villamil B, Laderas T, Salazar R, Gray JW, Hanahan D, Tabernero J, Bernards R, Friend SH, Laurent-Puig P, Medema JP, Sadanandam A, Wessels L, Delorenzi M, Kopetz S, Vermeulen L, Tejpar S (2015) The consensus molecular subtypes of colorectal cancer. Nat Med 21:1350–1356. https://doi.org/10.1038/nm.3967
Becht E, Giraldo NA, Lacroix L, Buttard B, Elarouci N, Petitprez F, Selves J, Laurent-Puig P, Sautes-Fridman C, Fridman WH, de Reynies A (2016) Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol 17:218. https://doi.org/10.1186/s13059-016-1070-5
Trinh A, Trumpi K, De Sousa EMF, Wang X, de Jong JH, Fessler E, Kuppen PJ, Reimers MS, Swets M, Koopman M, Nagtegaal ID, Jansen M, Hooijer GK, Offerhaus GJ, Kranenburg O, Punt CJ, Medema JP, Markowetz F, Vermeulen L (2017) Practical and robust identification of molecular subtypes in colorectal cancer by immunohistochemistry. Clin Cancer Res 23:387–398. https://doi.org/10.1158/1078-0432.CCR-16-0680
Tarallo R, Giurato G, Bruno G, Ravo M, Rizzo F, Salvati A, Ricciardi L, Marchese G, Cordella A, Rocco T, Gigantino V, Pierri B, Cimmino G, Milanesi L, Ambrosino C, Nyman TA, Nassa G, Weisz A (2017) The nuclear receptor ERbeta engages AGO2 in regulation of gene transcription, RNA splicing and RISC loading. Genome Biol 18:189. https://doi.org/10.1186/s13059-017-1321-0
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. https://doi.org/10.1093/bioinformatics/bts635
Anders S, Pyl PT, Huber W (2015) HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. https://doi.org/10.1093/bioinformatics/btu638
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. https://doi.org/10.1186/s13059-014-0550-8
Ogino S, Kawasaki T, Kirkner GJ, Kraft P, Loda M, Fuchs CS (2007) Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn 9:305–314. https://doi.org/10.2353/jmoldx.2007.060170
Furlan D, Sahnane N, Mazzoni M, Pastorino R, Carnevali I, Stefanoli M, Ferretti A, Chiaravalli AM, La Rosa S, Capella C (2013) Diagnostic utility of MS-MLPA in DNA methylation profiling of adenocarcinomas and neuroendocrine carcinomas of the colon-rectum. Virchows Arch 462:47–56. https://doi.org/10.1007/s00428-012-1348-2
Satow R, Inagaki S, Kato C, Shimozawa M, Fukami K (2017) Identification of zinc finger protein of the cerebellum 5 as a survival factor of prostate and colorectal cancer cells. Cancer Sci 108:2405–2412. https://doi.org/10.1111/cas.13419
Li L, Yan LH, Manoj S, Li Y, Lu L (2017) Central role of CEMIP in tumorigenesis and its potential as therapeutic target. J Cancer 8:2238–2246. https://doi.org/10.7150/jca.19295
Matheu A, Collado M, Wise C, Manterola L, Cekaite L, Tye AJ, Canamero M, Bujanda L, Schedl A, Cheah KS, Skotheim RI, Lothe RA, Lopez de Munain A, Briscoe J, Serrano M, Lovell-Badge R (2012) Oncogenicity of the developmental transcription factor Sox9. Cancer Res 72:1301–1315. https://doi.org/10.1158/0008-5472.CAN-11-3660
Roche KC, Gracz AD, Liu XF, Newton V, Akiyama H, Magness ST (2015) SOX9 maintains reserve stem cells and preserves radioresistance in mouse small intestine. Gastroenterology 149(1553-1563):e1510. https://doi.org/10.1053/j.gastro.2015.07.004
Panarelli NC, Vaughn CP, Samowitz WS, Yantiss RK (2015) Sporadic microsatellite instability-high colon cancers rarely display immunohistochemical evidence of Wnt signaling activation. Am J Surg Pathol 39:313–317. https://doi.org/10.1097/PAS.0000000000000380
Senger S, Sapone A, Fiorentino MR, Mazzarella G, Lauwers GY, Fasano A (2015) Celiac disease histopathology recapitulates hedgehog downregulation, consistent with wound healing processes activation. PLoS One 10:e0144634. https://doi.org/10.1371/journal.pone.0144634
Ma F, Ye H, He HH, Gerrin SJ, Chen S, Tanenbaum BA, Cai C, Sowalsky AG, He L, Wang H, Balk SP, Yuan X (2016) SOX9 drives WNT pathway activation in prostate cancer. J Clin Invest 126:1745–1758. https://doi.org/10.1172/JCI78815
Sun L, Mathews LA, Cabarcas SM, Zhang X, Yang A, Zhang Y, Young MR, Klarmann KD, Keller JR, Farrar WL (2013) Epigenetic regulation of SOX9 by the NF-kappaB signaling pathway in pancreatic cancer stem cells. Stem Cells 31:1454–1466. https://doi.org/10.1002/stem.1394
Fernandez-Jimenez N, Castellanos-Rubio A, Plaza-Izurieta L, Irastorza I, Elcoroaristizabal X, Jauregi-Miguel A, Lopez-Euba T, Tutau C, de Pancorbo MM, Vitoria JC, Bilbao JR (2014) Coregulation and modulation of NFkappaB-related genes in celiac disease: uncovered aspects of gut mucosal inflammation. Hum Mol Genet 23:1298–1310. https://doi.org/10.1093/hmg/ddt520
Maiuri MC, De Stefano D, Mele G, Fecarotta S, Greco L, Troncone R, Carnuccio R (2003) Nuclear factor kappa B is activated in small intestinal mucosa of celiac patients. J Mol Med (Berl) 81:373–379. https://doi.org/10.1007/s00109-003-0440-0
Castellanos-Rubio A, Santin I, Martin-Pagola A, Irastorza I, Castano L, Vitoria JC, Bilbao JR (2010) Long-term and acute effects of gliadin on small intestine of patients on potentially pathogenic networks in celiac disease. Autoimmunity 43:131–139. https://doi.org/10.3109/08916930903225229
Di Sabatino A, Lenti MV, Giuffrida P, Vanoli A, Corazza GR (2015) New insights into immune mechanisms underlying autoimmune diseases of the gastrointestinal tract. Autoimmun Rev 14:1161–1169. https://doi.org/10.1016/j.autrev.2015.08.004
Calon A, Lonardo E, Berenguer-Llergo A, Espinet E, Hernando-Momblona X, Iglesias M, Sevillano M, Palomo-Ponce S, Tauriello DV, Byrom D, Cortina C, Morral C, Barcelo C, Tosi S, Riera A, Attolini CS, Rossell D, Sancho E, Batlle E (2015) Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet 47:320–329. https://doi.org/10.1038/ng.3225
Isella C, Terrasi A, Bellomo SE, Petti C, Galatola G, Muratore A, Mellano A, Senetta R, Cassenti A, Sonetto C, Inghirami G, Trusolino L, Fekete Z, De Ridder M, Cassoni P, Storme G, Bertotti A, Medico E (2015) Stromal contribution to the colorectal cancer transcriptome. Nat Genet 47:312–319. https://doi.org/10.1038/ng.3224
Bergmann F, Singh S, Michel S, Kahlert C, Schirmacher P, Helmke B, Von Knebel DM, Kloor M, Blaker H (2010) Small bowel adenocarcinomas in celiac disease follow the CIM-MSI pathway. Oncol Rep 24:1535–1539. https://doi.org/10.3892/or_00001015
Giuffrida P, Vanoli A, Arpa G, Bonometti A, Luinetti O, Solcia E, Corazza GR, Paulli M, Di Sabatino A (2018) Small bowel carcinomas associated with immune-mediated intestinal disorders: the current knowledge. Cancers (Basel):11. https://doi.org/10.3390/cancers11010031
Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger SN, Kohrt HE, Horn L, Lawrence DP, Rost S, Leabman M, Xiao Y, Mokatrin A, Koeppen H, Hegde PS, Mellman I, Chen DS, Hodi FS (2014) Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 515:563–567. https://doi.org/10.1038/nature14011
Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS, Wong F, Azad NS, Rucki AA, Laheru D, Donehower R, Zaheer A, Fisher GA, Crocenzi TS, Lee JJ, Greten TF, Duffy AG, Ciombor KK, Eyring AD, Lam BH, Joe A, Kang SP, Holdhoff M, Danilova L, Cope L, Meyer C, Zhou S, Goldberg RM, Armstrong DK, Bever KM, Fader AN, Taube J, Housseau F, Spetzler D, Xiao N, Pardoll DM, Papadopoulos N, Kinzler KW, Eshleman JR, Vogelstein B, Anders RA, Diaz LA Jr (2017) Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357:409–413. https://doi.org/10.1126/science.aan6733
Acknowledgments
We thank all the Collaborators of the “Small Bowel Cancer Italian Consortium.”
Funding
This study was supported by the University of Insubria (to Prof. Furlan, FAR project 2016-2017), the Italian Association for Cancer Research (AIRC Grant: IG-17426), and Regione Campania (“La Campania lotta contro il cancro” project Rare-Plat-Net, CUP: B63D18000380007; and project GENOMAeSALUTE, CUP: B41C17000080007) and Genomix4Life to Prof. Weisz. This research was also supported by a grant of the Italian Ministry of Education, University and Research (MIUR), to the Department of Molecular Medicine of the University of Pavia under the initiative “Dipartimenti di Eccellenza” (2018–2022) to Prof. Marco Paulli and by Fondazione IRCCS Policlinico San Matteo to Dr. Ombretta Luinetti. A.S. is a PhD student of the Research Doctorate in “Sciences and Biomedical Technologies–XXXI Cycle,” University Roma 3.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Declaration of Helsinki and its revision and was approved by the Ethics Committee of the IRCCS San Matteo Hospital Foundation, Italy (no. 20140018113, on 22nd September 2014).
Conflict of interest
The authors have no conflicts of interest
Ethical responsibilities of authors section
Study design: DF, FR, AV, ES; acquisition, analysis, or interpretation of data: all authors
Drafting the work or revising it critically: all authors
Final approval of the version to be published: all authors
Agreement to be accountable for all aspects of the work: all authors
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Quality in Pathology
Rights and permissions
About this article
Cite this article
Rizzo, F., Vanoli, A., Sahnane, N. et al. Small-bowel carcinomas associated with celiac disease: transcriptomic profiling shows predominance of microsatellite instability-immune and mesenchymal subtypes. Virchows Arch 476, 711–723 (2020). https://doi.org/10.1007/s00428-019-02675-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-019-02675-w