Abstract
Main conclusion
The investigation is the first report on genome-wide identification and characterization of NBLRR genes in pearl millet. We have shown the role of gene loss and purifying selection in the divergence of NBLRRs in Poaceae lineage and candidate CaNBLRR genes for resistance to Magnaporthe grisea infection.
Abstract
Plants have evolved multiple integral mechanisms to counteract the pathogens' infection, among which plant immunity through NBLRR (nucleotide-binding site, leucine-rich repeat) genes is at the forefront. The genome-wide mining in pearl millet (Cenchrus americanus (L.) Morrone) revealed 146 CaNBLRRs. The variation in the branch length of NBLRRs showed the dynamic nature of NBLRRs in response to evolving pathogen races. The orthology of NBLRRs showed a predominance of many-to-one orthologs, indicating the divergence of NBLRRs in the pearl millet lineage mainly through gene loss events followed by gene gain through single-copy duplications. Further, the purifying selection (Ka/Ks < 1) shaped the expansion of NBLRRs within the lineage of pear millet and other members of Poaceae. Presence of cis-acting elements, viz. TCA element, G-box, MYB, SARE, ABRE and conserved motifs annotated with P-loop, kinase 2, RNBS-A, RNBS-D, GLPL, MHD, Rx-CC and LRR suggests their putative role in disease resistance and stress regulation. The qRT-PCR analysis in pearl millet lines showing contrasting responses to Magnaporthe grisea infection identified CaNBLRR20, CaNBLRR33, CaNBLRR46 CaNBLRR51, CaNBLRR78 and CaNBLRR146 as putative candidates. Molecular docking showed the involvement of three and two amino acid residues of LRR domains forming hydrogen bonds (histidine, arginine and threonine) and salt bridges (arginine and lysine) with effectors. Whereas 14 and 20 amino acid residues of CaNBLRR78 and CaNBLRR20 showed hydrophobic interactions with 11 and 9 amino acid residues of effectors, Mg.00g064570.m01 and Mg.00g006570.m01, respectively. The present investigation gives a comprehensive overview of CaNBLRRs and paves the foundation for their utility in pearl millet resistance breeding through understanding of host–pathogen interactions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pearl millet is one of the important millets for climate-resilient agriculture owing to its withstanding ability to harsh environmental conditions such as drought, low rainfall, high pH, poor quality soil and considerable nutritional and economic significance, especially in areas with arduous cultivation conditions (Kumar et al. 2021). Pearl millet is cultivated in approximately 27 million hectares in Asia and Africa. India leads the pearl millet production, with a production of 8.61 million tons, a productivity of 1243 kg ha−1, and occupies an area of 6.93 million ha (http://dmd.dacnet.nic.in/). It is highly nutritious and can cater to economically weaker sections with 80–90% energy demands (Govindaraj et al. 2019). However, pearl millet production has declined due to many biotic and abiotic factors. The blast disease caused by Magnaporthe grisea has emerged as one of the major production constraints during the last decade in pearl millet cultivation, causing severe loss in grain yield and quality (Singh et al. 2021). The chemical-based management of pearl millet demands high-cost inputs. Further, it poses the risk of environmental contamination and the development of fungicide-resistant pathogen strains, thereby necessitating the identification of alternate means. Thus, the genetic management of pearl millet blast by exploiting host–pathogen interaction mechanisms is the most effective, alternative and sustainable approach, especially in low-input agriculture systems (Nelson et al. 2018; Jeevan et al. 2020).
During pathogen infection, the plant exhibits two levels of immunity, viz. pathogen-associated molecular patterns (PAMPs)-triggered immunity (PTI) and effector-triggered-immunity (ETI) (Jones and Dangl 2006). As the pathogen attacks the plant, the cell wall acts as the primary defence barrier that triggers PTI response by recognizing the PAMPs through pattern recognition receptors (PRRs) on the cell membrane. Subsequently, the PRRs send signals inside the plant cell through the plasma membrane to activate defence-related pathways. The pathogens escape the PTI by secreting effectors, which are perceived by plants' resistance (R) genes, which activate a more intense and fast defence mechanism termed ETI (Yuan et al. 2021). Despite different defence mechanisms, PTI and ETI work co-ordinately (Zipfel 2014). Resistance against pathogens is a function of interaction between the avirulence (Avr) gene or pathogen effectors and the R gene of a plant (Ameline-Torregrosa et al. 2008). The availability of advanced molecular technologies and the need to develop robust resistance mechanisms led to the isolation and characterization of several R genes in various crop species. The largest class of functionally characterized R genes belong to nucleotide binding (NB) and leucine-rich repeat (LRR) types (NBLRRs) (Meyers et al. 2003). According to the zig-zag model, the R gene meant only those receptors that possessed nucleotide binding (NB) and leucine-rich repeat (LRR) domains, i.e. NBLRRs, which activates ETI responsible for pathogen resistance and induce hypersensitive response (HR) (Yuan et al. 2021). NBLRR proteins primarily consist of two domains; the NBS domain is an acronym of the NB-ARC domain, which is part of a signalling cascade that serves as a nucleotide-binding pocket that hydrolyses ATP to bring about conformational change in R proteins (Tameling et al. 2002). On the other hand, LRR domains act as ON/OFF switches for plant defence by recognizing pathogen effector molecules via protein–protein interactions (Michelmore et al. 2013). The NBS domain plays an essential role in the defence signalling mechanism and triggers a downstream hypersensitive response in plants, whereas the LRR domain is responsible for R-Avr interactions (Zhang et al. 2022).
NBLRR proteins may directly or indirectly recognize the pathogen. Several R genes may be activated to recognise various infections precisely. According to the guard theory, plant targets for pathogen effector proteins are protected by NBLRR proteins; in such a case, pathogen effector molecules serve as virulence factors to increase the sensitivity of the host plant in the absence of recognition. The indirect recognition theory states that the limited number of R proteins can recognise a wide range of pathogens (Meyers et al. 2003; McHale et al. 2006). The NBLRR genes PmTR1 and PmTR3 from rye have conferred resistance to powdery mildews from the three-leaf stage and all-stage resistance in wheat-rye introgressed lines (Han et al. 2023). In rice, 33 NBLRR genes were reported to assign resistance to various blast stages (Kalia and Rathour 2019; Li et al. 2019b, a). For instance, Pb1 (Fujii et al. 2000), Pb2 (Yu et al. 2022), and Pb3 (Ma et al. 2022) for panicle blast, and Pia (Okuyama et al. 2011), Pik-e (Chen et al. 2015), Pik-p (Yuan et al. 2011) for leaf blast, etc. Considering the importance of NBLRR genes in assigning disease resistance, the genome-wide identification and their evolutionary and functional characterization were undertaken in various plant species, such as Arabidopsis thaliana (Meyers et al. 1999), Oryza sp. (Rawal et al. 2018), Triticum aestivum (Andersen et al. 2020), Vitis vinifera (Goyal et al. 2020), etc. Further, several gene families, viz. MYB (Lin et al. 2023; Lv et al. 2023), RWP-RK (Jin et al. 2023), WRKY (Chanwala et al. 2020), etc., were characterized in the genus pearl millet and related species and voluminous information was generated on NBLRRs in various crop species. However, there is no information on detailed evolution and genome-wide analysis of NBLRR genes in pearl millet. Deciphering NBLRRs in pearl millet and their characterization is crucial for paving the foundation for molecular breeding of disease resistance in pearl millet. Therefore, we framed the current investigation firstly to mine the NBLRR genes in pearl millet using HMMscan and homology-based approaches and secondly to comprehensively study the evolution and functional response of NBLRRs to pearl millet blast caused by Magnaporthe grisea.
Materials and methods
Identification of putative NBLRR proteins in pearl millet proteome
A total of 88 and 126 NBLRR query proteins of Arabidopsis thaliana and Oryza sativa, respectively, were retrieved from TAIR (http://www.arabidopsis.org/) and the rice genome annotation project (http://rice.uga.edu) using the search terms 'NBLRR' and 'NBSLRR'. The query sequences were BLAST (BLAST + 2.10.1; https://www.ncbi.nlm.nih.gov/) searched with an e-value cut-off of 1e-5 against pearl millet proteome (PM.genechr.trans.pep) retrieved from ICRISAT- Centre of Excellence in Genomics & Systems Biology server (https://cegresources.icrisat.org/data_public/PearlMillet_Genome/v1.1/). Subsequently, the HMM models generated for Arabidopsis thaliana and Oryza sativa NBLRR query protein sequences with hmmbuild (HMMER v3.3.2) function and the NB-ARC Pfam domain (PF00931; http://pfam.xfam.org) were used to execute the hmmsearch (e-value < 0.01; HMMER 3.3.2) among NBLRR blast hits from pearl millet proteome (PM.genechr.trans.pep). The non-redundant hits from the BLAST and HMMSearch were used for HMMScan (HMMER 3.3.2) with the Pfam database (http://pfam.xfam.org). The protein sequences showing the NBS domain (PF00931) towards the amino (N) terminal and at least one LRR domain (PF00560, PF07725, PF12799, PF13306, PF13516, PF13855 and PF14580) towards the carboxyl (C) terminal were considered for subsequent analysis. Further, the non-redundant gene models were arranged in ascending order based on chromosomal positions and named CaNBLRRs (CaNBLRR1-146). The CaNBLRRs were plotted on seven chromosomes of pearl millet using TBtools v2.052 (https://bio.tools/tbtools) (Chen et al. 2023). The physicochemical properties and subcellular localization of CaNBLRRs were predicted using the ProtParam tool (https://web.expasy.org/protparam) (Gasteiger et al. 2005) and WoLFPSORT (https://wolfpsort.hgc.jp) (Horton 2006) servers.
Structural attributes of CaNBLRR sequences
The gene structure and intronic phases of CaNBLRR genes were visualized in TBtools v2.052 (https://bio.tools/tbtools) (Chen et al. 2023) using the GFF file of pearl millet (PM.genechr.trans.gff) retrieved from ICRISAT- Centre of Excellence in Genomics & Systems Biology server (https://cegresources.icrisat.org/data_public/PearlMillet_Genome/v1.1/). The best ten conserved motifs of CaNBLRRs were discovered using MEME suit (https://meme-suite.org/meme) (Bailey et al. 2015) with default parameters.
Multiple sequence alignment and phylogenetic analysis
For evolutionary analysis, the NBLRR proteins were mined from five related species, viz. purple false brome (Brachypodium distachyon (L.) P.Beauv.), finger millet (Eleusine coracana), rice (Oryza sativa L.), sorghum (Sorghum bicolor L. (Moench)) and foxtail millet (Setaria italica (L.) P.Beauv.) using the similar approach used for mining CaNBLRRs (Balamurugan et al. 2024). The phylogenetic analysis was performed to determine the evolutionary association among NBLRR sequences of pearl millet with related Poaceae members. The multiple sequence alignment of NBLRRs was carried out using Clustal Omega (Sievers et al. 2011) with default parameters. Subsequently, the multiple alignments were trimmed using the trimAL v1.2 tool (http://trimal.cgenomics.org) (Capella-Gutiérrez et al. 2009). Then, the aligned and trimmed sequences were used for phylogenetic tree construction based on the maximum likelihood method using the IQ TREE v16.12 (http://www.iqtree.org) (Minh et al. 2020) with 1000 bootstraps.
Duplication, orthology and selection pressure analyses
The self-BLASTp search (e-value < 1e-5) within the pearl millet proteome was performed with standalone BLAST + 2.10.1 (https://www.ncbi.nlm.nih.gov/) for duplication analysis, and the resulting BLAST output was subjected to DupGen_finder software to harvest the various duplications (Qiao et al. 2019). The NBLRRs of C. americanus, B. distachyon, E. coracana, O. sativa, S. bicolor and S. italica were used to predict orthology using OrthoFinder software (https://github.com/davidemms/OrthoFinder) (Emms and Kelly 2019). Each paralog and ortholog pair of NBLRRs were aligned with the ParaAT2.0 program (https://ngdc.cncb.ac.cn/tools/paraat) (Zhang et al. 2012). Subsequently, the aligned homolog pair were used to calculate the nonsynonymous rate (Ka), synonymous rate (Ks) and evolutionary constraint (Ka/Ks) using the KaKs calculator v.3.0 (https://ngdc.cncb.ac.cn/tools/kaks) (Zhang 2022).
Synteny and collinearity of NBLRRs among pearl millet and other target species
The whole proteomes of B. distachyon, E. coracana, O. sativa, S. bicolor and S. italica were aligned with C. americanus using the BLASTp program with the parameters of e-value < 1e-5 and best five hits. The BLASTp outputs were used to decipher the collinear blocks using MCScanX program (Wang et al. 2012). Later, the syntenic and collinear blocks of target NBLRRs were fetched and visualised with TBtools v2.052 (https://bio.tools/tbtools) (Chen et al. 2023).
Promoter analysis of CaNBLRR genes
The 1.5 kb upstream promoter region from the start codon of each CaNBLRR gene was fetched from the pearl millet genome (Pearl_millet_v1_1.fa) downloaded from the ICRISAT- Center of Excellence in Genomics & Systems Biology server (https://cegresources.icrisat.org/data_public/PearlMillet_Genome/v1.1/). Further, all the promoter sequences were analysed for cis-acting elements with the PlantCare server (http://bioinformatics.psb.ugent.be/webtools/plantcare/html) (Lescot et al. 2002). The predicted cis-acting elements were classified into subcategories, and the distribution was visualized within the 1.5 kb upstream regions of CaNBLRRs.
Plant materials and pathogen inoculation
The highly virulent strain of Magnaporthe grisea (PMg_Dl) was used to infect two pearl millet genotypes; viz. ICMB 95444 (susceptible) and IP 11353 (resistant) showed contrasting responses to blast infection. Pathogen revival and culture inoculation were done as per our previous protocol (Bansal et al. 2024), and the control plants were sprayed with sterile water. In order to facilitate the infection, all the plants were kept in a growth chamber with 26 ± 1 ℃ and 90% relative humidity. The experiment was plotted in CRD design with three replications for each treatment and control, and each replication carried ten plants. The blast severity was recorded on pearl millet based on a 0–9 disease scale grade on seven days post-inoculation (dpi) (Ganesan et al. 2016) (Table S1). The mean disease scale scores were considered for statistical analysis using the web-agri-stat software (WASP 2.0) package (https://ccari.icar.gov.in/wasp2.0/index.php). The plant samples were collected at 0, 24 and 72 h (h) post-inoculation (hpi) for further analysis.
RNA isolation, cDNA synthesis and qRT-PCR analysis
Total RNA was isolated using a Trizol reagent (Invitrogen, Carlsbad, CA, USA). RNA was converted to cDNA using a cDNA synthesis kit (Thermo Fisher Scientific, Waltham, MA, USA) as per the manufacturer’s protocol. The cDNA was checked using endogenous control provided with the cDNA synthesis kit and subsequently diluted to be used as a template for qRT-PCR. Gene-specific primer pairs were designed using IDT (https://www.idtdna.com/pages/tools/primerquest) and Primer 3 (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) servers (Table S2). The actin gene was taken as an endogenous control for qRT-PCR expression analysis. The qRT-PCR reaction mixture comprised of 50 ng cDNA, 5 pmol of each forward and reverse primer, 5 μl of SYBR Green PCR master mix 2X (Promega, Madison, WI, USA) and the final volume was maintained with nuclease-free water to 10 μl. All the reactions were carried out in triplicates. The reactions were carried out on LightCycler® 96 System (Roche Life Science, Penzberg, Germany) with the following reaction conditions: 95 ℃ for 2 min for one cycle, 95 ℃ for 30 s and 55 ℃ for 1 min for 40 cycles and melting curve analysis with 95 ℃ for 15 s, 60 ℃ for 1 min and 95 ℃ for 15 s. The relative gene expression levels were calculated using 2–∆∆Ct or Livak method (Livak and Schmittgen 2001).
Prediction and evaluation of protein 3D models
The amino acid sequences of ten CaNBLRRs used for expression analysis were selected for constructing the 3D models using the Robetta server (https://robetta.bakerlab.org/) (Baek et al. 2021) with a confidence score of > 0.75. The predicted protein structures were evaluated using UCLA-DOE LAB—SAVES v6.0 (https://saves.mbi.ucla.edu/) server for ERRAT (Colovos and Yeates 1993), VERIFY 3D (Bowie et al. 1991; Lüthy et al. 1992; Eisenberg et al. 1997) and PROCHECK (Laskowski et al. 1993) scores.
NBLRR-effector interaction through molecular docking analysis
The cloned effector proteins of M. oryzae, viz. CCN97897.1 (Avr-Pi54), AIS23643.1 (Avr-Pi9), BAH59485.1 (Avr-Pii) and AAK00131.1 (Avr-Pita) retrieved from NCBI database (https://www.ncbi.nlm.nih.gov/) were BLASTp (e-value < 1e-5) aligned against the proteome of M. grisea (Prakash et al. 2019). The five effector hits from M.grisea based on BLASTp hits, viz. Mg.00g020410.m01, Mg.00g064570.m01, Mg.00g013260.m01, Mg.00g012780.m01 and Mg.00g006570.m01 with BLAST + 2.10.1 were used to construct the 3D models using Robetta server (https://robetta.bakerlab.org/). Further, for docking analysis, CaNBLRR20 and CaNBLRR78 host proteins were selected based on qRT-PCR expression results. The LZerD web server (https://lzerd.kiharalab.org/about/) (Christoffer et al. 2021b, a) was employed for docking of CaNBLRR20 and CaNBLRR78 with the above five effector proteins using default parameters. The best docking models were selected based on the lowest rank sum score. The docking interactions between LRR domains of CaNBLRRs and effector protein were considered for subsequent analysis and visualization. The docking models were visualized and analysed using the LigPlot+ v.2.2 software (Laskowski and Swindells 2011).
Results
Mining, chromosomal distribution and characterization of NBLRR of pearl millet
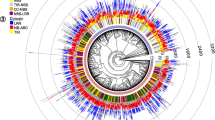
The genome-wide mining of NBLRR genes in pearl millet using homology-based BLASTp search, HMMsearch and HMMScan resulted in 146 CaNBLRRs, showing characteristic features of NB-ARC domain towards N-terminal and LRR domain(s) towards the C-terminal (Fig. 1a; Table S3). The mined NBLRRs in pearl millet were named based on physical location in ascending order from CaNBLRR1 to CaNBLRR146. The CaNBLRRs showed an uneven distribution among the seven chromosomes of Cenchrus americanus (Fig. 1b and c). Chromosome 4 showed the highest number of CaNBLRRs (N = 37, CaNBLRR63-CaNBLRR99), followed by chromosome 1 (N = 35, CaNBLRR1-CaNBLRR35). Where the chromosome 6 contains the least CaNBLRRs (N = 9; CaNBLRR114-CaNBLRR122).
The pearl millet NBLRR genes. a The protein domain and features considered for mining NBLRR proteins and genes from the pearl millet genome. b The chromosome-wise distribution of NBLRR numbers in pearl millet. c The distribution of NBLRR numbers in the pearl millet chromosome. Note: The NBLRR genes are named in ascending order according to physical location
The subcellular localization showed 43% of CaNBLRRs in the cytoplasm, followed by the nucleus, chloroplast, plasma membrane, etc. (Fig. S1). The physicochemical properties of CaNBLRRs were studied to decipher the protein length, molecular weight, isoelectric point and GRAVY indices. The protein length of CaNBLRRs ranged from 461 (CaNBLRR32) to 1584 (CaNBLRR62) amino acids with the corresponding molecular weight (MWs) of 52.56 and 177.66 kDa. The 76 CaNBLRR proteins showed an isoelectric point of pI < 7 (5.2–6.88) with acidic properties, and 65 showed pI > 7 (7.02–9.24) with basic properties. Interestingly, CaNBLRR141 was found to have a neutral pH, i.e. 7.00. The GRAVY index measures hydropathy, and all the CaNBLRR proteins showed negative GRAVY values, indicating the hydrophilic nature of CaNBLRR proteins (Table S3).
Gene structure and motif analysis of CaNBLRRs
The domains are the essential features of any gene/protein family. The NB and LRR motifs were visualized on target CaNBLRR proteins to know their distribution (Fig. S2a). The motifs are short, recurring patterns in protein sequences that mediate the regulatory functions of the sequence. The MEME server identified the ten conserved motifs distributed among the 146 CaNBLRR proteins. The conserved motifs sequences showed the general order motif 6, 2, 10, 8, 3, 1, 7, 4, 9 and 5. Among the ten motifs, motif 5 was present in all the genes in multiple copies except for CaNBLRR34, which has only one copy. The position of motif 5 remained conserved in 91% of CaNBLRRs. The 56% CaNBLRRs harboured all ten motifs, where motif 6 was missing in 25.3% CaNBLRRs, followed by motif 2 (19.1%) and motif 10 (17.1%) (Fig. S2b, Fig. S3).
Deciphering the number of introns and exons Introns is crucial to knowing the splice variants or protein repertoire by alternative splicing mechanisms. The CaNBLRR coding sequences ranged in size from 1.0 kb to 5.0 kb, with the majority (54%) being between 2 and 3 kb, followed by 3 to 4 kb (31%), > 4 kb (10%), and 5% < 2 kb. The gene structure analysis showed that the number of coding sequences present in the gene sequences of pearl millet ranges from one (28%) to seven (CaNBLRR43) (Table S3; Fig. S2c). The majority of the intronic sequences were found in phase 0 (69.01%), followed by phase 1 (20.08%) and phase 2 (10.12%) (Fig. S2c).
Evolutionary analysis of NBLRR genes in pearl millet and target species
Duplication and selection pressure analysis
Gene duplications mediate the expansion of gene families and generate new members that can add to functional variation and the evolution of new functions. Gene duplication analysis was performed to decipher the kind of duplications and selection shaping the expansion of NBLRRs within the pearl millet genome. The duplication analysis showed the contribution of single gene duplications in the expansion of CaNBLRRs (Fig. 2a; Table S4). A total of 143 CaNBLRRs showed duplication encompassing dispersed (86), proximal (34) and tandem (23) duplications (Fig. 2b; Table S3). Further, the 143 duplicated CaNBLRRs formed 158 duplication pairs encompassing 19 proximal 12 tandem and 127 dispersed duplication pairs (Fig. 2c). Among the proximal duplication, CaNBLRR3, CaNBLRR22, CaNBLRR92, and CaNBLRR104 showed one-to-two duplication (Fig. 2a; Table S3). Similarly, CaNBLRR132 exhibited one-to-two tandem duplication. In the case of dispersed duplication, 127 pairs have harboured all kinds of duplications, i.e. one-to-one, one-to-many and many-to-many. The selection pressure analysis was performed by deciphering the non-synonymous (Ka) and synonymous substitutions (Ks) ratio, which is used to infer the direction and magnitude of natural selection acting on protein-coding genes. The CaNBLRR duplication pairs showed the mean Ka/Ks ratio of 0.34 with the range of 0.14 (CaNBLRR56-CaNBLRR57) to 0.74 (CaNBLRR49-CaNBLRR50), suggesting the expansion of CaNBLRRs under strong purifying selection (Fig. 2d; Table S4).
The expansion of NBLRRs in the pearl millet genome. a The chromosomal location and kinds of duplication of CaNBLRR genes in the pearl millet genome. The orange, blue and green coloured linked lines in a duplication circos represent dispersed, proximal and tandem duplications, respectively. b The Venn diagram shows the number and percentages of CaNBLRR genes showing different kinds of duplications. c The Venn diagram shows the number and percentages of different kinds of duplication pairs among CaNBLRRs. d The box-plot shows non-synonymous (Ka), synonymous (Ks) and the ratio of non-synonymous to synonymous (Ka/Ks) substitutions among CaNBLRR duplication pairs
Phylogenetic analysis of NBLRR genes
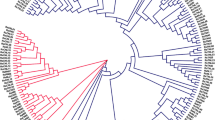
Phylogenetics of target protein sequences of a gene family among different species gives an in-depth insight into how the protein sequences are related and evolved through evolutionary changes. The phylogenetics of NBLRRs was worked out to decipher the evolutionary relationship among 1103 NBLRR sequences from B. distachyon, C. americanus, E. coracana, O. sativa, S. bicolor and S. italica. The phylogenetic tree was constructed using the maximum likelihood method with the best-fit model JTT + F + R7 (Fig. 3; Fig. S4). The 1103 NBLRRs were clustered into eight different clades, viz. I to VIII (Fig. 3a). The cluster-VIII encompasses the maximum number of NBLRRs (284) followed by cluster-IV (187), cluster-III (179), cluster-I (131), cluster-V (114), cluster-VII (89), cluster-II (68) and cluster-VI (51) (Fig. 3b). Further, every cluster showed representation from all the six target species, indicating the mixed grouping patterns of NBLRRs in Poaceae lineage. The branch length in phylogenetics indicates the extent of genetic changes in terms of the average number of nucleotide or protein substitutions per site. Thus, the branch length of entities typically depicts the genetic changes and divergence of the entities. The phylogenetics of NBLRRs showed broad variations in branch length ranging from 2.021 × 10–6 (SbNBLRR75) to 2.52 (OsNBLRR175) (Fig. 3a). The significant variation in the branch length indicates the active divergence of NBLRRs in response to pathogen pressure during the process of evolution. Further, the mixed grouping of NBLRRs indicates the evolution of founder NBLRRs prior to the divergence of target species in the Poaceae lineage.
The phylogenetics of NBLRRs of Poaceae members. a The phylogenetic tree based on the maximum likelihood method with 1000 bootstraps shows an evolutionary association among 1103 NBLRRs from C. americanus, B. distachyon, E. coracana, O. sativa, S. bicolor and S. itlaica. b The bar graph shows the clade and species-wise distribution of NBLRRs in the phylogenetic tree. The numerals in different coloured boxes below the bar graphs represent the total number of NBLRRs in the respective coloured clade
Ortholog and synteny of pearl millet NBLRRs with other target species
The ortholog analysis was performed to decipher the orthology and evolutionary forces shaping the divergence of NBLRRs in the Poaceae lineage. The CaNBLRRs showed maximum orthology with S. italica (73.97%) followed by S. bicolor (69.18%), O. sativa (65.75%), B. distachyon (46.58%) and E. coracana (45.21%). At the same time, the NBLRRs from S. italica, S. bicolor, E. coracana, B. distachyon and O. sativa showed 61.73, 58.59, 55.17, 52.83 and 43.98% of orthology with CaNBLRRs, respectively. The orthology analysis showed a higher representation of many-to-one orthologs compared to other categories with a range of 19–53%, indication the loss of NBLRRs in the lineage of target species. Further, a higher proportion of one-to-one orthology was observed among the closely related lineages, for instance CaNBLRRs- SbNBLRRs (51%), CaNBLRRs- SiNBLRRs (44%), SbNBLRRs- SiNBLRRs (48%), SbNBLRRs- CaNBLRRs (45%), whereas the inverse association of one-to-many and many-to-many orthologs were observed with genetic relatedness of the species (Fig. 4a). The reconciliation of species tree with the phylogenetic tree of NBLRRs also showed the predominance of gene loss events than gene gains in the ancestral lineage of target species (Fig. 4b). The ratio of mean non-synonymous (mean = 0.33; range: 0.01–0.82) and synonymous (mean = 1.07; range = 0.06–2.96) showed the predominance of purifying or negative selection pressure (mean = 0.34; range = 0.04–0.76) in the expansion of NBLRRs in Poaceae lineage (Table S5).
The orthology among the NBLRRs of Poaceae members. a The Venn diagrams show the different kinds of orthologous analysis among the NBLRR members of the Poaceae species. The blue, orange, grey and yellow coloured slices represent percent one-to-one, one-to-many, many-to-one and many-to-many orthologs. The values in the diagonal boxes represent the total number of NBLRRs in each species. b The summary of NBLRRs' loss and gain events across the phylogenetic tree shows NBLRRs' divergence among the target Poaceae members
The syntenic analysis of pearl millet NBLRRs with NBLRRs from S. italica, S. bicolor, E. coracana, B. distachyon and O. sativa was performed to decipher the synteny and collinearity association (Fig. 5a–e; Table S6). The CaNBLRRs showed maximum syntenic blocks with E. coracana (37) followed by S. italica (32), S. bicolor (29), O. sativa (25) and B. distachyon (16). In comparison, the maximum syntenic gene pairs were detected with S. italica (48) followed by S. bicolor (47), E. coracana (42), O. sativa (32) and B. distachyon (17) (Fig. 5f).
The syntenic association of CaNBLRRs with NBLRRs of other Poaceae members. a Pearl millet (orange bars) with E. coracana (dark green bars). b Pearl millet (orange bars) with B. distachyon (green bars). c Pearl millet (orange bars) with O. sativa (red bars). d Pearl millet (orange bars) with S. bicolor (blue bars). e Pearl millet (orange bars) with S. itlaica (yellow bars). f The bar graphs show the syntenic alignment blocks and collinear pairs between CaNBLRRs and NBLRRs of other Poaceae members
Functional analysis of the NBLRR genes in pearl millet
Promotor analysis of NBLRR genes
The cis-acting elements are known to modulate the gene expression. The PlantCARE database generated a list of 96 different kinds of cis-acting elements in the 1.5 kb upstream sequence of 146 CaNBLRR genes. The 96 types of cis-elements are categorized under 5 categories, viz. core promotor (CPE), growth and development (GDE), light responsive (LRE), hormone-responsive (HRE) and stress-responsive (SRE) elements (Fig. S5-S10). The core promotor sequence comprised of mainly four elements: TATA Box, CAAT Box, AT ~ TATA and TATA present in all 146 genes. TATA Box was also present in all genes except in CaNBLRR5, CaNBLRR32 and CaNBLRR125, while AT ~ TATA was found only in 41.7% of genes (Fig. S6). A total of 452 growth and development (GDE)-related elements belonging to 21 different types were identified in all the genes, except CaNBLRR32, CaNBLRR47 CaNBLRR60, CaNBLRR64, CaNBLRR70, CaNBLRR78, CaNBLRR89, CaNBLRR94 and CaNBLRR96 (Fig. S7). Elements associated with meristem expression (CAT-Box), growth-responsive element (AAGAA-motif), meristem specific activation (CCGTCC motif), and zein metabolism regulation (O2-site) were found in maximum frequency in nearly 50.6%, 45.2%, 40.41% and 31.5% of the genes (Fig. S7). In response to light, plants undergo a wide array of physiological activities, some of which include gravitropism, seed germination, circadian rhythms, etc. Several cis-elements further regulate the light-controlled gene expression. The 145 promoters showed 1098 cis-elements of 35 distinct categories, except CaNBLRR122, which was devoid of any light-sensitive elements. G-Box, Box4 and Sp1 were predominant light-responsive elements of CaNBLRR genes with percent frequency of 77.39, 41.78 and 41.78%, respectively (Fig. S8).
Plants encounter a variety of abiotic and biotic stresses, which causes the plant to undergo a reprogramming in gene expression to withstand them. This adaptation is accomplished by cis-elements-transcription factors (TFs) interactions (Sheshadri et al. 2016). In total, 3825 stress-responsive elements (SRE) belonging to 29 categories were identified in CaNBLRR promoter sequences. Among the SREs, the maximum representations are observed from MYB (14.24%) followed by MYC (11.11%), STRE (9.43%), ABRE (7.47%), CGTCA-motif, TGACG-motif, as-1 (6.35%), and ARE (5.12%). The distribution of these specific SRE varied among different genes. The MYB elements are present in 97.94% of CaNBLLRs. Similarly, MYC, ABRE, STRE, CGTCA-motif, TGACG-motif and as-1 were present in 73–88% of CaNBLLRs. Interestingly, the CaNBLRR32 did not show any SRE (Fig. S9). The hormone-responsive elements (HRE) constituted seven types of cis-acting distributed in 108 CaNBLRR elements with minimum representation (183) among all the five functional categories. The TGA-element (auxin-responsive element) was found in 52 CaNBLRRs, followed by the two gibberellin-responsive elements, viz. P-box (42), GARE-motif (31), (Fig. S10).
Blast severity and disease incidence
Two pearl millet genotypes, namely ICMB 95444 (susceptible) and IP 11353 (resistant), challenged with M. grisea virulent inoculum, showed a contrasting disease reaction under artificial epiphytotic conditions. The initial symptoms start with tiny specks or lesions that broaden and turn necrotic, resulting in widespread chlorosis and, ultimately, drying of young leaves at seven days post inoculation (Fig. 6). The blast severity was calculated based on the percent disease incidence revealed that susceptible line (ICMB 95444) showed up to 86.66% disease severity and resistant line (IP 11353) showed 12.22% blast severity. No blast symptoms were observed in the absolute control plants, which were remains healthy during the study period (Table 1).
The phenotypic response of pearl millet lines ICMB 95444 (susceptible) and IP 11353 (resistant) to M.grisea infection. a The phenotypic response of ICMB 95444 (susceptible) line at 0 h of inoculation. b The phenotypic response of ICMB 95444 (susceptible) line at seventh-day post-inoculation. c Close-up view of blast symptoms on ICMB 95444 (susceptible) genotype. d The phenotypic response of IP 11353 (resistant) line at 0 h of inoculation. e The phenotypic response of IP 11353 (resistant) line at the seventh-day post-inoculation. f Close-up view of blast symptoms on IP 11353 (resistant) genotype. g The comparative blast incidence on control and inoculated leaves of ICMB 95444 (susceptible) on the seventh-day of post-inoculation. h The comparative blast incidence on control and inoculated leaves of IP 11353 (resistant) on the seventh-day of post-inoculation
Relative expression analysis of candidate CaNBLRR genes against M. grisea infection
The qRT-PCR analysis expression was performed to study the CaNBLRRs response to M. grisea infection analysis. The ten CaNBLRRs are selected based on chromosomal distributions. The upregulated expression of CaNBLRR20, CaNBLRR33 and CaNBLRR146 up to 8, 2.5 and 2.5 folds, respectively, at 24 hpi, followed by declined expressions at 72 h was observed in the resistant line (IP 11353) compared to the susceptible line (ICMB 95444) (Fig. 7). The similar trend of expression was also observed for CaNBLRR113; however, the expression fold change was very low (< 1). The CaNBLRR46, CaNBLRR51 and CaNBLRR78 showed consistent enhanced expression in the resistant genotype (IP 11353) compared to susceptible genotype ICMB 95444 at both 24 and 72 hpi (Fig. 7). On the contrary, the significant downregulation of CaNBLRR52 and CaNBLRR120 was observed in resistant genotype (IP 11353) compared to susceptible genotype ICMB 95444 at both 24 and 72 hpi (Fig. 7). However, CaNBLRR92 showed significant downregulation at 24 hpi and non-significant expression at 72 hpi in resistant line IP 11353 as compared to susceptible genotype ICMB 95444 (Fig. 7). Overall, it can be summarized that out of 10 CaNBLRR candidate genes, three genes (CaNBLRR20, CaNBLRR33, CaNBLRR146) were positively regulated by infection process at 24 hpi indicating these could be involved in early responsiveness during the infection process. Whereas CaNBLRR51, CaNBLRR78 and CaNBLRR46 were positively regulated by the infection process at 24 and 72 hpi suggesting their role in early and late infection processes (Fig. 7). Overall, the qRT-PCR results indicate that CaNBLRR20, CaNBLRR33, CaNBLRR46, CaNBLRR51, CaNBLRR78 and CaNBLRR146 the idea that these genes may have a role in plant defence against M. grisea but up to what extent that needs to be confirmed by the more detailed study. Also, the variations between susceptible and resistant lines may be attributed to the genetic makeup of both lines.
The expression profiles of ten CaNBLRR genes in the susceptible (ICMB 95444) and resistant (IP11353) genotyping showing contrasting responses to M. grisea infection at 24 h and 72 h of post-inoculation periods. The main bars represent mean fold change values over the control (0 h), and the error bars represent standard error values. The two-way ANOVA analysis was carried out to determine the statistical significance at *P ≤ 0.05, **P ≤ 0.001, and ***P ≤ 0.0001
Modelling and validation of 3D protein structure
The amino acid sequences of ten genes used for expression analysis were used to model the 3D structures. The modelled 3D structures showing a confidence score of > 0.75 were selected from the Robetta server. The CaNBLRR 3D structures analysis through the Ramachandran plot revealed the distribution of 90–93.6% residues in the most favoured region. Further, only three proteins, viz. CaNBLRR33 (0.5%), CaNBLRR52 (0.5%) and CaNBLRR92 (0.2%) had their residues in the disallowed region. The remaining residues for all proteins were mainly concentrated in the additionally allowed region (6.1–10%). Further, CaNBLRR33, CaNBLRR46, CaNBLRR52, CaNBLRR78, CaNBLRR92, CaNBLRR113 and CaNBLRR120 showed 0.2 to 0.8% residues in the generously allowed regions (Fig. S11). The ERRAT score of the ten CaNBLRRs was in the range of 92 (CaNBLRR46) – 99 (CaNBLRR78, CaNBLRR92), while the verify-3D score was in the range of 80.76–97.81 (Fig. 8a; Table 2).
The 3D models and docking analysis of CaNBLRR proteins. a The 3D structure of CaNBLRRs. The indigo and red colours represent the NB-ARC and LRR domains, respectively. b The molecular docking pose of CaNBLRR20 with Mg.00g006570.m01 (Avr-Pita). The red portion shows the LRR domain of CaNBLRR20, and the cyan portion represents the effector protein Mg.00g006570.m01 (Avr-Pita). c The amino acid residues of CaNBLRR20 and Mg.00g006570.m01 (Avr-Pita) showing various molecular interactions. c The molecular docking of CaNBLRR78 with Mg.00g064570.m01 (Avr-Pi9). The red portion shows the LRR domain of CaNBLRR78, and the green portion represents the effector protein Mg.00g064570.m01 (Avr-Pi9). The amino acids of CaNBLRR78 and Mg.00g064570.m01 (Avr-Pi9) showing various molecular interactions. Note: The green and red dotted lines in sections b and c represent the hydrogen bonds and salt bridges, respectively, and the amino acid residues with ray lines indicate hydrophobic interactions
Molecular docking of NBLRRs and effector interaction
The molecular interactions between the R proteins of the host and the effectors of the pathogen molecular basis for host–pathogen interactions. Molecular docking of proteins allows the investigation of the various putative binding patterns and interactions with target receptors. The online LzerD web server (https://lzerd.kiharalab.org/about/) was employed for docking CaNBLRRs with effector proteins using default parameters. Since CaNBLRR20 and CaNBLRR78 genes showed the highest expressions, their protein models were considered as receptors. Where the homologs of Avr-Pi54 (Mg.00g020410.m01), Avr-Pi9 (Mg.00g064570.m01), Avr-Pii (Mg.00go13260.m01) and Avr-Pita (Mg.00g006570.m01 and Mg.00g012780.m01) effectors from M. grisea were considered for docking and 3D models were predicted with Robetta server. Among all the docking interactions, the LRR domains of CaNBLRR20 and CaNBLRR78, showing docking interaction with effectors Mg.00g006570.m01 and Mg.00g064570.m01, respectively, were selected for docking analysis (Fig. S12; Fig. 8b, c). The molecular interaction between CaNBLRR20 and effectors Mg.00g006570.m01 three hydrogen bonds between His594 (CaNBLRR20)-Tyr390 (Mg.00g006570.m01) in the LRR, and Gly180 (CaNBLRR20)—Arg471 (Mg.00g006570.m01) and Arg111 (CaNBLRR20)—Glu470 & Leu467 (Mg.00g006570.m01) in non-LRR regions of CaNBLRR20 (Fig. 8b). Similarly, CaNBLRR78 and effectors Mg.00g064570.m01 showed all three hydrogen bonds in the LRR region, viz. Arg932 (CaNBLRR78)-Asn33 (Mg.00g064570.m01) and Thr931 (CaNBLRR78)-Arg112 (Mg.00g064570.m01) (Fig. 8c). Besides the amino acids showing hydrogen bonds, 20 (Ser757, Thr758, Lys779, Phe803, Ile805, Ser806, Lys807, Ala808, His829, Ile831, Cys832, Ser834, Arg853, Pro855, Asn857, Ile880, Ser882, Asp915, His930, Gly933), 11 (Met1, Leu3, Phe6, Val7, Ile9, Leu10, Thr11, Val13, Ile14, Cys15, Gln71), 14 (Thr79, Ala82, Ser88, Asn89, Phe97, Phe100, Val112, His115, Gln119, Asp122, Leu568, Ser569, Lys591, Phe593) and 9 (Pro386, Ser387, Phe388, Asp389, Gly391, Val394, Val395, Thr510, Gln646) amino acid residues in CaNBLRR78, Mg.00g064570.m01, CaNBLRR20 and Mg.00g006570.m01, respectively showed exclusively hydrophobic interactions (Fig. 8b, c).
Discussion
Pearl millet is an important member of the millet family and has been immensely valued for its nutritional richness and ability to withstand considerable drought and high-temperature stresses. However, pearl millet's potential cannot be exploited to its fullest owing to various biotic constraints, including pests and diseases. In order to minimize the damage of pathogens, the precise development of resistant lines against pathogens and identify the genes involved in plant defence mechanisms are most crucial. Unfortunately, despite the benefits pearl millet offers, it has been less explored for host–pathogen interactions at the molecular level. Plant–pathogen interaction involves the interaction of virulence (vir/effector) factors with plant resistance (R) genes. It has been found that most of the R genes in plants are comprised of NBS and LRR domains, which impart resistance to plants. The widespread existence of NBLRRs across the plant kingdom, viz. angiosperms, gymnosperms and non-vascular plants, indicates their importance in plants (McHale et al. 2006).
The present investigation mined 146 CaNBLRRs based on homology-based BLAST followed by HMMSearch and HMMScan with the Pfam database. In order to investigate the evolutionary patterns of pearl millet NBLRRs, we have included the 159, 116, 241, 198 and 243 NBLRR homologs from B. distachyon, E. coracana, O. sativa, S. bicolor and S. italica, respectively, using the similar approach used for mining CaNBLRRs (Balamurugan et al. 2024). The protein length of the mined NBLRR genes ranges from 461 to 1584 amino acids with the corresponding molecular weight (MWs) of 52.56 and 177.66 kDa, which is on par with previous reports in various crops, viz. 663 to 1613 amino acids having molecular weights of 75.57 and 182.73 kDa in Chinese cabbage (Liu et al. 2021), 352–2101 amino acids with 40.2 kDa and 240.4 kDa in grapes (Goyal et al. 2020) and 111–1632 amino acids with 12.72–182.40 kDa in Raphanus sativus (Ma et al. 2021). The pI value is the nature of the predicted protein, which may be either acidic or basic. In our study, the pI value ranges from 5.22 to 9.24. A similar range was also found in NBLRR members of Raphanus sativus (4.77–9.60) (Ma et al. 2021), cucumber (5.57–9.15) (Zhang et al. 2022) and Chinese cabbage (5.42–8.98) (Liu et al. 2021).
The significance of R proteins in pathogen resistance is substantially impacted by their subcellular location. Numerous R proteins have been found to localize in the nucleus and cytoplasm; even most R proteins lacked nuclear localization signals (Meier and Somers 2011). In the case of maize, RppM was known to target both the nucleus and cytoplasm, which happens to be essential for regulating the resistance against southern corn rust (Wang et al. 2022). The NBLRR protein Pi64 in rice provides resistance against M. oryzae and also shows subcellular localization in the nucleus and cytoplasm (Ma et al. 2015). Accordingly, the subcellular localization of CaNBLRRs in the current investigation showed major distribution in the cytoplasm and nucleus (Fig. S1). Besides, the Trifolium alexandrinum showed the distribution of NBLRRs in 12 different cellular compartments in addition to cytoplasm and nucleus (Nasir et al. 2021). In the present study, genes were unevenly distributed across the seven chromosomes. The uneven distribution of NBLRRs in plant genomes is common, viz. Actinidia chinensis (Wang et al. 2020), Medicago truncatula (Ameline-Torregrosa et al. 2008), and soybean (Kang et al. 2012). The maximum number of genes were located in chromosome 4 (37), followed by chromosome 1 (35). Accordingly, the recent studies on QTL mapping for blast resistance in pearl millet showed five major QTL with phenotypic variations ranging from 11.21 to 18.45% on chromosome 4 and three major QTL with 10.08 to 13.44% on chromosome 1 (Pujar et al. 2023). Furthermore, chromosome 4 has major QTLs with phenotypic variations (16.70 to 78.00%) for Downey mildew resistance in pearl millet (Chelpuri et al. 2019). The co-occurrence of NBLRRs with disease resistance associated with QTL on the same chromosome has also been highlighted in various crop species (Goyal et al. 2020).
Structural analysis of CaNBLRR genes showed varying numbers of exons across the genes ranging from 1 to 7, while introns in the range of 0–6. Except for CaNBLRR17, CaNBLRR18, CaNBLRR40, and CaNBLRR41, all the genes carry introns of different phases. The majority of introns are in phase 0 (64%), which implies they are located between the two codons. Few introns are in phase 1, implying that these introns disrupted a codon between the first and second nucleotides. Phase 2 has the fewest introns, which obstruct the codon at bases 2 and 3. The NBLRRs showed similar kinds of exon-intronic distributions in grass pea (up to 7 exons and 6 introns) (Alsamman et al. 2023) and sweet orange (1–12 exons and 0–11 introns) (Yin et al. 2023). The exon/intronic diversity in NBLRR genes is pivotal to understanding the diverged evolution of gene families, which is further indicative of gain or loss or insertion or deletion events during evolution, leading to structural and functional variability (Roy and Gilbert 2005; Xu et al. 2012).
NBLRR protein is composed of NB-ARC, which is constituted of multiple motifs and leucine-rich repeats. Each of these component domains is crucial for the proper functioning of NBLRRs. In pearl millet NBLRRs, the best ten motifs were studied for their presence and distribution. Motifs 2, 3, 7–10 were annotated to the NB-ARC domain, motif 5 to the LRR domain, and motif 6 to the Rx-CC domain. Nearly similar motif distribution patterns were observed in chickpeas (Sagi et al. 2017) and Vitis vinifera (Goyal et al. 2020). The repeated occurrence of motif 5 corresponding to leucine-rich repeats in CaNBLRRs is an indication of the important role it plays in pathogenesis. The LRR region is known for mediating protein (R)-protein (Avr) interactions, besides cell adhesion and transport (Wang et al. 2023). Despite the conserved nature, the LRR regions undergo diversifying selection, which is attributed to their ability to recognize specific effector molecules to impart specific resistance against a wide array of pathogens and their races (Marone et al. 2013). Further, motif 6 with conserved sequence EDVID facilitates intramolecular interaction with the NB domain, as was seen in Rx (CC-NB-LRR protein), which provides resistance to Potato virus X (Rairdan et al. 2008). The motif 2 corresponding to the P-loop harbouring GXXXXGK (T/S) is linked to nucleotide binding through phosphate and Mg2+, and any mutation in the P-loop leads to impaired gene function (Williams et al. 2011). Motif 9 is present in 94.5% of CaNBLRR codes for methionine, histidine and aspartate (MHD) motifs. MHD histidine binds to the beta phosphate of ADP, and mutation in it could disrupt the inert ADP-bound protein complex, permitting the exchange of nucleotides and protein activation. Aspartate mutations may cause the histidine to displace from its location, which would reduce its ability to suppress the R protein. It could also have a negative impact on the interaction between the NB and ARC2 subdomains, disrupting the closed, inactive protein conformation (van Ooijen et al. 2008). Kinase 2, corresponding to motif 8, channelizes phosphate transfer reaction and is comprised of four hydrophobic amino acid moieties followed by aspartic acid moieties important for co-ordinating metal ions (Mg2+) (Wang et al. 2023). Motif 3 represents the GLPL motif and mainly acts as a binding site, while motif 7 corresponds to the RNBS-D motif and is involved in forming the NBLRR interface (Martin et al. 2022). The signal transduction mechanism of the NBS domain is modulated by the CC and LRR domains, both of which function in a recognition-dependent manner. This benefits the development of the NBLRR function model recognition specificities.
The gene duplications and losses are crucial for evolution and structural and functional diversifications in nature. The gene duplications and losses contributed to the expansion and diversification of NBLRRs in the Poaceae lineage (Fig. 4b). The single gene duplication events shaped the CaNBLRRs expansion under purifying selection pressure in Poaceae lineage (Fig. 4b). The prominence of purifying selection on the expansion of NBLRRs within the pearl millet and Poaceae lineage showed the elimination of deleterious mutations during evolutionary lineages. The major portion of many-to-one NBLRRs orthologs in the Poaceae lineage also confirms the role of gene loss events in diversifying NBLRRs in the lineage of pearl millet and Poaceae members. The same results were also observed among the Poaceae NBLRRs, excluding CaNBLRRs (Balamurugan et al. 2024). The differential gains and losses lead to considerable variation in the number of genes in a gene family. Further, some of the families show high turnover rates over other gene families having similar sizes across species (Wang et al. 2018; Fernández and Gabaldón 2020). The lineage-specific losses and gain of NBLRRs in Poacease reflect the rapid changes in response to the evolution of new races or pathotypes in the environment during the divergence process (Wang et al. 2018).
Cis-acting elements recruit the transcriptional factors near the transcription site and regulate corresponding gene expression. The promoter sequence analysis of NBLRRs resulted in various cis-acting elements falling under core promoter, stress response, growth and development, light response and hormone response. The four hormone-responsive elements (methyl jasmonate, salicylic acid, abscisic acid, ethylene-responsive) observed in CaNBLRRs in our current investigation were also reported in grass pea NBLRRs (Alsamman et al. 2023). The second highest contribution of stress-responsive elements to the pool of cis-acting elements indicates the involvement of CaNBLRR genes in various biotic and abiotic stress responses. The MYB binding elements in model plant Arabidopsis are known to provide pathogen resistance by modulating the salicylic acid (SA) biosynthetic pathway and are also known to counteract the drought, salt and oxidative stresses simultaneously (Ambawat et al. 2013; Kim et al. 2020), and insect infestations, viz. also Helicoverpa armigera (Shen et al. 2018). The conserved elements, viz. W box, TCA element, S box, and GCC box have been visualized in pathogen-induced gene promoters (Muthusamy et al. 2017). Pathogen infection triggers the accumulation of SA, which activates natriuretic peptide receptor 1 protein, followed by its transport to the nucleus to bind to TGA class elements. As a result, SA-responsive elements (TCA-element, SARE) present in pathogenesis-related genes interact with TGA factors, marking the onset of systemic acquired resistance. Two elements found in CaNBLRR gene As-1 (243) and G-box (323) have been reported in rice, and these elements are associated with themselves and other defence-related elements, viz. GSTs, TGA family, and H-box to induce pathogen resistance (Kong et al. 2018).
The qRT-PCR expression analysis of 10 CaNBLRRs in response to M. grisea infection exhibited a characteristic genotype and time intervals specific expression pattern in pearl millet. The CaNBLRR20, CaNBLRR33, CaNBLRR46, CaNBLRR51, CaNBLRR78 and CaNBLRR146 upregulated in pathogen challenged resistant genotype (IP11353) when compared to pathogen challenged susceptible genotype (ICMB 95444). The CaNBLRR20, CaNBLRR33 and CaNBLRR146 showed upregulation at 24 h, followed by a drastic decline at 72 h. Meanwhile, CaNBLRR78 and CaNBLRR46 showed enhanced expression at 24 and 72 h. Variations in expression of some NBLRR genes at 24 h and some at 72 h were also evident in sugarcane. T difference could be attributed to the difference in response mechanism of NBLRRs to pathogenesis (Jiang et al. 2023). CaNBLRR92 and CaNBLRR120 showed significant down-regulation or non-significant expressions.
NBLRR stimulate a strong ETI immune response through directly or indirectly interacting with pathogen effectors and results in an allergic reaction characterized by programmed cell death to resist the invasion of the pathogen (Dangl and Jones 2001). Thus, the CaNBLRR20 and CaNBLRR78 were selected for R-Avr molecular docking studies owing to their maximum contrasting expression patterns among susceptible and resistant cultivars. In the present study, the effectors Avr-Pi9 (Mg.00g064570.m01) and Avr-Pita (Mg.00g006570.m01) interacted with LRR domains of CaNBLRR78 than CaNBLRR20, respectively, through hydrogen bonds, which contributes to the protein complex stability. The ligand binding process to proteins is facilitated through various inter-atomic interactions, predominantly comprising electrostatic and van der Waals interactions (Böhm 2003; Dill and MacCallum 2012). The hydrogen bonding, salt bridges, and metal interactions are crucial for electrostatic complementarity between the proteins at the binding interface. The hydrogen bonds are primary forms of electrostatic interactions that contribute to this complementarity and play a crucial role in the directional interactions that form the basis of protein folding, protein structure, and molecular recognition. The specific and directional nature of the interaction between a protein and its interacting partner through hydrogen bonding is a fundamental aspect of molecular recognition (Hubbard and Kamran Haider 2010). The hydrogen bonds formed in CaNBLRR20-Mg.00g006570.m01 interaction by Lys93, Gly108, Arg111 and His594 of CaNBLRR20, and Arg471, Leu467, Tyr390, Glu383 of Mg.00g006570.m01. Similarly, CaNBLRR78-Mg.00g064570.m01 showed Thr931 and Arg 932 from CaNBLRR78 and Asn93 and Arg112 from Mg.00g064570.m01 forming hydrogen bonds. The Arg residues contribute a maximum (4) for the hydrogen-bond formation in CaNBLRR-effector interactions. Arginine plays a significant role in the energetic stabilization of protein–protein complexes and is frequently in the "hot spot" protein interfaces (Bogan and Thorn 1998). Furthermore, Arg111 of CaNBLRR20 formed the salt bridge with Glu470 of Mg.00g006570.m01. Salt bridges influence protein stability positively through electrostatic interactions (Pylaeva et al. 2018).
Besides H-bonds, hydrophobic bonds are the additional driving force required to maintain the conformational stability of proteins (Pace 1995). Diverse amino acids were involved in hydrophobic interactions in variable numbers for both proteins (Fig. 8b, c). Hydrophobic interactions entail the interaction of non-polar segments of a molecule. In protein–ligand complexes, the non-polar segments located at the interacting surfaces become buried upon binding. This results in the displacement of water molecules, leading to an increase in entropy. The hydrophobic interactions are driven by entropy and have been demonstrated to play a critical role in protein–ligand/protein interactions (Böhm 2003; Bissantz et al. 2010).
Conclusions
The current investigation showed 146 CaNBLRRs in a pearl millet genome. The motif analysis revealed highly conserved signature sequences annotated with NB-ARC and LRR domains. The significant association of cis-acting elements with hormonal and stress responsiveness showed the relevance of CaNBLRRs in regulating biotic stress tolerance. Evolutionary analysis revealed that the NBLRR in the Poaceae lineage showed the expansion of CaNBLRRs through single gene duplication events under purifying selection and divergence through gene loss and gain events in the lineage of pearl millet. The qRT-PCR expression analysis identified the CaNBLRR33, CaNBLRR46, CaNBLRR51, CaNBLRR78 and CaNBLRR146 genes associated with M.grisea infection and pathogenesis. The molecular docking of CaNBLRR20 and CaNBLRR78 with Avr-Pita (Mg.00g006570.m01) and Avr-Pi9 (Mg.00g064570.m01), respectively, showed the interacting residues and kind of interactions associated with pearl millet and M.grisea interactions. The current investigation is the first report on genome-wide identification and structural and evolutionary analysis of NBLRRs in pearl millet. The results paved the foundation for molecular resistance breeding by identifying the set of CaNBLRRs, which could be further validated through in-depth functional analysis using transgenics and genome editing approaches.
Data availability
All the raw data sets used in this study were downloaded from publicly available databases. The necessary supporting data is provided as a supplementary file.
Abbreviations
- ETI:
-
Effector-triggered immunity
- hpi:
-
Hours post inoculation
- NBLRR:
-
Nucleotide-binding site leucine-rich repeats
- R:
-
Resistance
- SRE:
-
Stress-responsive element
References
Alsamman AM, Mousa KH, Nassar AE et al (2023) Identification, characterization, and validation of NBS-encoding genes in grass pea. Front Genet 14:1187597. https://doi.org/10.3389/fgene.2023.1187597
Ambawat S, Sharma P, Yadav NR, Yadav RC (2013) MYB transcription factor genes as regulators for plant responses: an overview. Physiol Mol Biol Plants 19:307–321. https://doi.org/10.1007/s12298-013-0179-1
Ameline-Torregrosa C, Wang B-B, O’Bleness MS et al (2008) Identification and characterization of nucleotide-binding site-leucine-rich repeat genes in the model plant Medicago truncatula. Plant Physiol 146:5–21. https://doi.org/10.1104/pp.107.104588
Andersen EJ, Nepal MP, Purintun JM et al (2020) Wheat disease resistance genes and their diversification through integrated domain fusions. Front Genet 11:898. https://doi.org/10.3389/fgene.2020.00898
Baek M, DiMaio F, Anishchenko I et al (2021) Accurate prediction of protein structures and interactions using a three-track neural network. Science 373:871–876. https://doi.org/10.1126/science.abj8754
Bailey TL, Johnson J, Grant CE, Noble WS (2015) The MEME Suite. Nucleic Acids Res 43:W39–W49. https://doi.org/10.1093/nar/gkv416
Balamurugan A, Mallikarjuna MG, Bansal S et al (2024) Genome-wide identification and characterization of NBLRR genes in finger millet (Eleusine coracana L.) and their expression in response to Magnaporthe grisea infection. BMC Plant Biol. 24(1):75. https://doi.org/10.1186/s12870-024-04743-z
Bansal S, Mallikarjuna MG, Reddy B et al (2024) Characterization and validation of hypothetical virulence factors in recently sequenced genomes of Magnaporthe species. Physiol Mol Plant Pathol 124:101969. https://doi.org/10.1016/j.pmpp.2023.101969
Bissantz C, Kuhn B, Stahl M (2010) A medicinal chemist’s guide to molecular interactions. J Med Chem 53:5061–5084. https://doi.org/10.1021/jm100112j
Bogan AA, Thorn KS (1998) Anatomy of hot spots in protein interfaces. J Mol Biol 280:1–9. https://doi.org/10.1006/jmbi.1998.1843
Böhm H-J (2003) Prediction of non-bonded interactions in drug design. In: Mannhold R, Kubinyi H, Folkers G (eds) Methods and principles in medicinal chemistry. Wiley, pp 3–20
Bowie JU, Lüthy R, Eisenberg D (1991) a method to identify protein sequences that fold into a known three-dimensional structure. Science. 253:164–170. https://doi.org/10.1126/science.1853201
Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. https://doi.org/10.1093/bioinformatics/btp348
Chanwala J, Satpati S, Dixit A et al (2020) Genome-wide identification and expression analysis of WRKY transcription factors in pearl millet (Pennisetum glaucum) under dehydration and salinity stress. BMC Genomics 21:231
Chelpuri D, Sharma R, Durga KK et al (2019) Mapping quantitative trait loci (QTLs) associated with resistance to major pathotype-isolates of pearl millet downy mildew pathogen. Eur J Plant Pathol 154:983–994. https://doi.org/10.1007/s10658-019-01718-x
Chen J, Peng P, Tian J et al (2015) Pike, a rice blast resistance allele consisting of two adjacent NBS–LRR genes, was identified as a novel allele at the Pik locus. Mol Breeding 35:117. https://doi.org/10.1007/s11032-015-0305-6
Chen C, Wu Y, Li J et al (2023) TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol Plant 16(11):1733–1742. https://doi.org/10.1016/j.molp.2023.09.010
Christoffer C, Bharadwaj V, Luu R, Kihara D (2021a) LZerD protein-protein docking webserver enhanced with de novo structure prediction. Front Mol Biosci 8:724947. https://doi.org/10.3389/fmolb.2021.724947
Christoffer C, Chen S, Bharadwaj V et al (2021b) LZerD webserver for pairwise and multiple protein–protein docking. Nucleic Acids Res 49:W359–W365. https://doi.org/10.1093/nar/gkab336
Colovos C, Yeates TO (1993) Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci 2:1511–1519. https://doi.org/10.1002/pro.5560020916
Dangl JL, Jones JDG (2001) Plant pathogens and integrated defence responses to infection. Nature 411:826–833. https://doi.org/10.1038/35081161
Dill KA, MacCallum JL (2012) The protein-folding problem, 50 years on. Science. 338:1042–1046. https://doi.org/10.1126/science.1219021
Eisenberg D, Lüthy R, Bowie JU (1997) VERIFY3D: Assessment of protein models with three-dimensional profiles. Methods Enzymol 277:396–404
Emms DM, Kelly S (2019) OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol 20:1–14. https://doi.org/10.1186/S13059-019-1832-Y
Fernández R, Gabaldón T (2020) Gene gain and loss across the metazoan tree of life. Nat Ecol Evol 4:524–533. https://doi.org/10.1038/s41559-019-1069-x
Fujii K, Hayano-Saito Y, Saito K et al (2000) Identification of a RFLP marker tightly linked to the panicle blast resistance gene, Pb1, in rice. Breed Sci 50:183–188. https://doi.org/10.1270/jsbbs.50.183
Ganesan P, Srinivasa N, Shankar M et al (2016) Standardization of pearl millet blast (Magnaporthe grisea) phenotyping under artificial conditions. Ann Agric Res 37:200–205
Gasteiger E, Hoogland C, Gattiker A et al (2005) Protein identification and analysis tools on the ExPASy server. The proteomics protocols handbook. Humana Press, Totowa, pp 571–607. https://doi.org/10.1385/1-59259-890-0:571
Govindaraj M, Rai KN, Cherian B et al (2019) Breeding biofortified pearl millet varieties and hybrids to enhance millet markets for human nutrition. Agriculture 9:106. https://doi.org/10.3390/agriculture9050106
Goyal N, Bhatia G, Sharma S et al (2020) Genome-wide characterization revealed role of NBS-LRR genes during powdery mildew infection in Vitis vinifera. Genomics 112:312–322. https://doi.org/10.1016/j.ygeno.2019.02.011
Han G, Liu H, Zhu S et al (2023) Two functional CC-NBS-LRR proteins from rye chromosome 6RS confer differential age-related powdery mildew resistance to wheat. Plant Biotechnol J 22:66–81. https://doi.org/10.1111/pbi.14165
Horton S (2006) The economics of food fortification. J Nutr 136:1068–1071. https://doi.org/10.1093/jn/136.4.1068
Hubbard RE, Kamran Haider M (2010) Hydrogen bonds in proteins: role and strength. Encyclopedia of life sciences. Wiley, New York. https://doi.org/10.1002/9780470015902.a0003011.pub2
Jeevan B, Gogoi R, Sharma D et al (2020) Genetic analysis of maydis leaf blight resistance in subtropical maize (Zea mays L.) germplasm. J Genet. 99:89. https://doi.org/10.1007/s12041-020-01245-3
Jiang Z, Zhao M, Qin H et al (2023) Genome-wide analysis of NBS-LRR genes revealed contribution of disease resistance from Saccharum spontaneum to modern sugarcane cultivar. Front Plant Sci 14:1091567. https://doi.org/10.3389/fpls.2023.1091567
Jin Y, Luo J, Yang Y et al (2023) The evolution and expansion of RWP-RK gene family improve the heat adaptability of elephant grass (Pennisetum purpureum Schum.). BMC Genomics 24(1):510. https://doi.org/10.1186/s12864-023-09550-8
Jones JDG, Dangl JL (2006) The plant immune system. Nature 444:323–329. https://doi.org/10.1038/nature05286
Kalia S, Rathour R (2019) Current status on mapping of genes for resistance to leaf- and neck-blast disease in rice. 3 Biotech. 9:209. https://doi.org/10.1007/s13205-019-1738-0
Kang YJ, Kim KH, Shim S et al (2012) Genome-wide mapping of NBS-LRR genes and their association with disease resistance in soybean. BMC Plant Biol 12:139. https://doi.org/10.1186/1471-2229-12-139
Kim SH, Lam PY, Lee M-H et al (2020) The Arabidopsis R2R3 MYB transcription factor MYB15 is a key regulator of lignin biosynthesis in effector-triggered immunity. Front Plant Sci 11:583153. https://doi.org/10.3389/fpls.2020.583153
Kong W, Ding L, Cheng J, Wang B (2018) Identification and expression analysis of genes with pathogen-inducible cis-regulatory elements in the promoter regions in Oryza sativa. Rice 11:52. https://doi.org/10.1186/s12284-018-0243-0
Kumar RR, Bhargava DV, Pandit K et al (2021) Lipase—The fascinating dynamics of enzyme in seed storage and germination—a real challenge to pearl millet. Food Chem 361:130031. https://doi.org/10.1016/j.foodchem.2021.130031
Laskowski RA, Swindells MB (2011) LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model 51:2778–2786. https://doi.org/10.1021/ci200227u
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291. https://doi.org/10.1107/S0021889892009944
Lescot M, Déhais P, Thijs G et al (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327. https://doi.org/10.1093/nar/30.1.325
Li C, Wang D, Peng S et al (2019a) Genome-wide association mapping of resistance against rice blast strains in South China and identification of a new Pik allele. Rice 12:1–9. https://doi.org/10.1186/S12284-019-0309-7
Li W, Chern M, Yin J et al (2019b) Recent advances in broad-spectrum resistance to the rice blast disease. Curr Opin Plant Biol 50:114–120. https://doi.org/10.1016/j.pbi.2019.03.015
Lin M, Dong Z, Zhou H et al (2023) Genome-wide identification and transcriptional analysis of the MYB gene family in pearl millet (Pennisetum glaucum). Int J Mol Sci 24:2484. https://doi.org/10.3390/ijms24032484
Liu Y, Li D, Yang N et al (2021) Genome-wide identification and analysis of CC-NBS-LRR family in response to downy mildew and black rot in Chinese cabbage. Int J Mol Sci 22:4266. https://doi.org/10.3390/ijms22084266
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Lüthy R, Bowie JU, Eisenberg D (1992) Assessment of protein models with three-dimensional profiles. Nature 356:83–85. https://doi.org/10.1038/356083a0
Lv J, Xu Y, Dan X et al (2023) Genomic survey of MYB gene family in six pearl millet (Pennisetum glaucum) varieties and their response to abiotic stresses. Genetica 15:251–265. https://doi.org/10.1007/s10709-023-00188-8
Ma J, Lei C, Xu X et al (2015) Pi64, encoding a novel CC-NBS-LRR protein, confers resistance to leaf and neck blast in rice. Mol Plant Microbe Interact 28:558–568. https://doi.org/10.1094/MPMI-11-14-0367-R
Ma Y, Chhapekar SS, Lu L et al (2021) Genome-wide identification and characterization of NBS-encoding genes in Raphanus sativus L. and their roles related to Fusarium oxysporum resistance. BMC Plant Biol. 21:47. https://doi.org/10.1186/s12870-020-02803-8
Ma L, Yu Y, Li C et al (2022) Genome-wide association study identifies a rice panicle blast resistance gene Pb3 encoding NLR protein. Int J Mol Sci 23:14032. https://doi.org/10.3390/ijms232214032
Marone D, Russo M, Laidò G et al (2013) Plant nucleotide binding site–leucine-rich repeat (NBS-LRR) genes: active guardians in host defense responses. Int J Mol Sci 14:7302–7326. https://doi.org/10.3390/ijms14047302
Martin EC, Spiridon L, Goverse A, Petrescu A-J (2022) NLRexpress—a bundle of machine learning motif predictors—reveals motif stability underlying plant Nod-like receptors diversity. Front Plant Sci 13:975888. https://doi.org/10.3389/fpls.2022.975888
McHale L, Tan X, Koehl P, Michelmore RW (2006) Plant NBS-LRR proteins: adaptable guards. Genome Biol 7:212. https://doi.org/10.1186/gb-2006-7-4-212
Meier I, Somers DE (2011) Regulation of nucleocytoplasmic trafficking in plants. Curr Opin Plant Biol 14:538–546. https://doi.org/10.1016/j.pbi.2011.06.005
Meyers BC, Dickerman AW, Michelmore RW et al (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J 20:317–332. https://doi.org/10.1046/j.1365-313X.1999.t01-1-00606.x
Meyers BC, Kozik A, Griego A et al (2003) Genome-wide analysis of NBS-LRR–encoding genes in Arabidopsis. Plant Cell 15:809–834. https://doi.org/10.1105/tpc.009308
Michelmore RW, Christopoulou M, Caldwell KS (2013) Impacts of resistance gene genetics, function, and evolution on a durable future. Annu Rev Phytopathol 51:291–319. https://doi.org/10.1146/annurev-phyto-082712-102334
Minh BQ, Schmidt HA, Chernomor O et al (2020) IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37:1530–1534. https://doi.org/10.1093/molbev/msaa015
Muthusamy S, Sivalingam P, Sridhar J et al (2017) Biotic stress inducible promoters in crop plants-a review. J Agric Ecol. 04:14–24. https://doi.org/10.53911/JAE.2017.4202
Nasir B, Ijaz S, Awan FS, Haq IU (2021) Genome-wide probing of NBS-LRR encoding genes in red clover (Trifolium pratense L) for the identification of resistance gene analogs in Trifolium alexandrinum L. Science Asia 47:425. https://doi.org/10.2306/scienceasia1513-1874.2021.054
Nelson R, Wiesner-Hanks T, Wisser R, Balint-Kurti P (2018) Navigating complexity to breed disease-resistant crops. Nat Rev Genet 19:21–33. https://doi.org/10.1038/nrg.2017.82
Okuyama Y, Kanzaki H, Abe A et al (2011) A multifaceted genomics approach allows the isolation of the rice Pia -blast resistance gene consisting of two adjacent NBS-LRR protein genes. Plant J 66:467–479. https://doi.org/10.1111/j.1365-313X.2011.04502.x
Pace CN (1995) Evaluating contribution of hydrogen bonding and hydrophobic bonding to protein folding. Methods Enzymol 259:538–554
Prakash G, Kumar A, Sheoran N et al (2019) First draft genome sequence of a pearl millet blast pathogen, Magnaporthe grisea strain PMg_Dl, obtained using PacBio single-molecule real-time and Illumina NextSeq 500 sequencing. Microbiol Resour Announc 8(20):10–128
Pujar M, Kumar S, Sharma R et al (2023) Identification of genomic regions linked to blast ( Pyricularia grisea ) resistance in pearl millet. Plant Breed 142:506–517. https://doi.org/10.1111/pbr.13111
Pylaeva S, Brehm M, Sebastiani D (2018) Salt bridge in aqueous solution: strong structural motifs but weak enthalpic effect. Sci Rep 8:13626. https://doi.org/10.1038/s41598-018-31935-z
Qiao X, Li Q, Yin H et al (2019) Gene duplication and evolution in recurring polyploidization–diploidization cycles in plants. Genome Biol 20:38. https://doi.org/10.1186/s13059-019-1650-2
Rairdan GJ, Collier SM, Sacco MA et al (2008) The coiled-coil and nucleotide binding domains of the potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell 20:739–751. https://doi.org/10.1105/tpc.107.056036
Rawal HC, Amitha Mithra SV, Arora K et al (2018) Genome-wide analysis in wild and cultivated Oryza species reveals abundance of NBS genes in progenitors of cultivated rice. Plant Mol Biol Report 36:373–386. https://doi.org/10.1007/s11105-018-1086-y
Roy SW, Gilbert W (2005) Rates of intron loss and gain: Implications for early eukaryotic evolution. Proc Natl Acad Sci USA 102:5773–5778. https://doi.org/10.1073/pnas.0500383102
Sagi MS, Deokar AA, Tar’an B (2017) Genetic analysis of NBS-LRR gene family in chickpea and their expression profiles in response to Ascochyta blight infection. Front Plant Sci 8:838. https://doi.org/10.3389/fpls.2017.00838
Shen X-J, Wang Y-Y, Zhang Y-X et al (2018) Overexpression of the wild soybean R2R3-MYB transcription factor GsMYB15 enhances resistance to salt stress and Helicoverpa armigera in transgenic Arabidopsis. Int J Mol Sci 19:3958. https://doi.org/10.3390/ijms19123958
Sheshadri SA, Nishanth MJ, Simon B (2016) Stress-mediated cis-element transcription factor interactions interconnecting primary and specialized metabolism in planta. Front Plant Sci 7:1725. https://doi.org/10.3389/fpls.2016.01725
Sievers F, Wilm A, Dineen D et al (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. https://doi.org/10.1038/msb.2011.75
Singh S, Sharma R, Chandra Nayaka S et al (2021) Understanding pearl millet blast caused by Magnaporthe grisea and strategies for its management. In: Nayaka SC, Hosahatti R, Prakash G et al (eds) Blast disease of cereal crops: evolution and adaptation in context of climate change. Springer, Cham, pp 151–172
Tameling WIL, Elzinga SDJ, Darmin PS et al (2002) The tomato R gene products I-2 and Mi-1 are functional ATP binding proteins with ATPase activity. Plant Cell 14:2929–2939. https://doi.org/10.1105/tpc.005793
van Ooijen G, Mayr G, Kasiem MMA et al (2008) Structure–function analysis of the NB-ARC domain of plant disease resistance proteins. J Exp Bot 59:1383–1397. https://doi.org/10.1093/jxb/ern045
Wang Y, Tang H, Debarry JD et al (2012) MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40:e49. https://doi.org/10.1093/NAR/GKR1293
Wang P, Moore BM, Panchy NL et al (2018) Factors influencing gene family size variation among related species in a plant family, Solanaceae. Genome Biol Evol 10:2596–2613. https://doi.org/10.1093/gbe/evy193
Wang T, Jia Z-H, Zhang J-Y et al (2020) Identification and analysis of NBS-LRR genes in Actinidia chinensis genome. Plants 9:1350. https://doi.org/10.3390/plants9101350
Wang S, Wang X, Zhang R et al (2022) RppM, encoding a typical CC-NBS-LRR protein, confers resistance to southern corn rust in maize. Front Plant Sci 13:951318. https://doi.org/10.3389/fpls.2022.951318
Wang X, Xu Y, Fan H et al (2023) Research progress of plant nucleotide-binding leucine-rich repeat protein. Horticulturae 9:122. https://doi.org/10.3390/horticulturae9010122
Williams SJ, Sornaraj P, deCourcy-Ireland E et al (2011) An autoactive mutant of the M Flax rust resistance protein has a preference for binding ATP, whereas wild-type M protein binds ADP. Mol Plant Microbe Interact 24:897–906. https://doi.org/10.1094/MPMI-03-11-0052
Xu G, Guo C, Shan H, Kong H (2012) Divergence of duplicate genes in exon–intron structure. Proc Natl Acad Sci USA 109:1187–1192. https://doi.org/10.1073/pnas.1109047109
Yin T, Han P, Xi D et al (2023) Genome-wide identification, characterization, and expression profile of NBS-LRR gene family in sweet orange (Citrus sinensis). Gene 854:147117. https://doi.org/10.1016/j.gene.2022.147117
Yu Y, Ma L, Wang X et al (2022) Genome-wide association study identifies a rice panicle blast resistance gene, Pb2, encoding NLR protein. Int J Mol Sci 23:5668. https://doi.org/10.3390/ijms23105668
Yuan B, Zhai C, Wang W et al (2011) The Pik-p resistance to Magnaporthe oryzae in rice is mediated by a pair of closely linked CC-NBS-LRR genes. Theor Appl Genet 122:1017–1028. https://doi.org/10.1007/s00122-010-1506-3
Yuan M, Ngou BPM, Ding P, Xin X-F (2021) PTI-ETI crosstalk: an integrative view of plant immunity. Curr Opin Plant Biol 62:102030. https://doi.org/10.1016/j.pbi.2021.102030
Zhang Z (2022) KaKs_calculator 3.0: calculating selective pressure on coding and non-coding sequences. Genom Proteom Bioinf 20:530–540. https://doi.org/10.1016/J.GPB.2021.12.002
Zhang Z, Xiao J, Wu J et al (2012) ParaAT: a parallel tool for constructing multiple protein-coding DNA alignments. Biochem Biophys Res Commun 419:779–781. https://doi.org/10.1016/j.bbrc.2012.02.101
Zhang W, Yuan Q, Wu Y et al (2022) Genome-wide identification and characterization of the CC-NBS-LRR gene family in cucumber (Cucumis sativus L.). Int J Mol Sci. 23:5048. https://doi.org/10.3390/ijms23095048
Zipfel C (2014) Plant pattern-recognition receptors. Trends Immunol 35(7):345–351
Acknowledgements
Aruljothi Ambalavanan acknowledges The Graduate School at ICAR-IARI and ICAR for providing the Postgraduate Fellowship and Alexander Balamurugan for guiding phenotyping of plants.
Funding
This research was funded by "Editing rice genes through CRISPR/Cas9 technology for enhanced and durable blast resistance in rice" (Grant No. BT/PR32125/AGIIl/103/1147/2019) sponsored by the Department of Biotechnology (DBT), India and Indian Council of Agricultural Research-ICAR-Consortium Research Platform on Genomics (ICAR-G/CRP-Genomics/2015–2720/IARI-12-151). The bioinformatics analysis is supported by the “Network Project on Computational Biology and Agricultural Bioinformatics” (Agril.Edn.14(44)/2014-A&P).
Author information
Authors and Affiliations
Contributions
Aruljothi Ambalavanan: methodology, formal analysis, visualization, writing—original draft. Mallana Gowdra Mallikarjuna: conceptualization, methodology, formal analysis, visualization, writing—review and editing, supervision. Shilpi Bansal: formal analysis, visualization, writing—original draft. Bishnu Maya Bashyal: writing—review and editing, supervision. Sabtharishi Subramanian: writing—review and editing, supervision. Aundy Kumar: writing—review and editing, supervision. Ganesan Prakash: conceptualization, methodology, writing—review and editing, funding acquisition, project administration, supervision.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Dorothea Bartels.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ambalavanan, A., Mallikarjuna, M.G., Bansal, S. et al. Genome-wide characterization of the NBLRR gene family provides evolutionary and functional insights into blast resistance in pearl millet (Cenchrus americanus (L.) Morrone). Planta 259, 143 (2024). https://doi.org/10.1007/s00425-024-04413-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-024-04413-2