Abstract
Main conclusion
Axillary meristems (AMs) located in the leaf axils determine the number of shoots or tillers eventually formed, thus contributing significantly to the plant architecture and crop yields. The study of AM initiation is unavoidable and beneficial for crop productivity.
Abstract
Shoot branching is an undoubted determinant of plant architecture and thus greatly impacts crop yield due to the panicle-bearing traits of tillers. The emergence of the AM is essential for the incipient bud formation, and then the bud is dormant or outgrowth immediately to form a branch or tiller. While numerous reviews have focused on plant branching and tillering development networks, fewer specifically address AM initiation and its regulatory mechanisms. This review synthesizes the significant advancements in the genetic and hormonal factors governing AM initiation, with a primary focus on studies conducted in Arabidopsis (Arabidopsis thaliana L.) and rice (Oryza sativa L.). In particular, by elaborating on critical genes like LATERAL SUPPRESSOR (LAS), which specifically regulates AM initiation and the networks in which they are involved, we attempt to unify the cascades through which they are positioned. We concentrate on clarifying the precise mutual regulation between shoot apical meristem (SAM) and AM-related factors. Additionally, we examine challenges in elucidating AM formation mechanisms alongside opportunities provided by emerging omics approaches to identify AM-specific genes. By expanding our comprehension of the genetic and hormonal regulation of AM development, we can develop strategies to optimize crop production and address global food challenges effectively.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants demonstrate remarkable shoot plasticity to ensure survival and propagation, particularly in the face of challenging external and internal conditions. In seed plants, the architecture of shoots is predominantly influenced by factors such as the number, position, orientation, and size of shoot branches. The precise regulation of shoot branching represents a crucial adaptive strategy orchestrated by a complex regulatory network.
The development of the primary shoot axis originates from the activity of the shoot apical meristem (SAM), a group of mitotic cells formed during embryogenesis. Subsequently, derivatives of this meristem generate all above-ground portions of plants (Bowman and Eshed 2000). The SAM continuously produces aerial organs by adding growth units called phytomere, which typically consist of an internode, a leaf, and an axillary meristem (AM) which is initiated in the leaf axil (Wang et al. 2018). The AM, acting as a new SAM of the secondary growth axis, differentiates into, such as in rice, tiller bud, leaf sheath primordium, and leaf primordium (Yan et al. 2023; Wang 2021). Therefore, the scalable branching across multiple levels is enabled by the specification and activity of AMs, leading to the generation of diverse architectural forms. Notably, AM activity has long been a target of breeding selection due to its significant contribution to crop yield through effects on tiller/branch number, panicle number, and panicle branches (Springer 2010; Wang and Li 2008, 2011; Shao et al. 2019).
Numerous studies over several decades have sought to elucidate AM initiation mechanisms. The prevailing model proposes that main endogenous and developmental cues interact to regulate this process. For instance, the LATERAL SUPPRESSOR (LAS), REVOLUTA (REV), and CUP-SHAPED-COTYLEDON (CUC) genes (Aida et al. 1999; Otsuga et al. 2001; Greb et al. 2003; Hibara et al. 2006) can function together in a regulatory cascade controlling AM initiation. An Auxin minimum niche is required to sustain SHOOT MERISTEMLESS (STM) expression in the boundary zone, thereby maintaining AM identity (Guo et al. 2015). Subsequently, WUSCHEL (WUS) follows the expression of STM and then activates the expression of CLAVATA3 (CLV3) (Shi et al. 2016; Cao and Jiao 2020; Cao et al. 2020), forming a WUS-CLV3 loop in the center of leaf axils to maintain stem cell activity (Xin et al. 2017), indicating the completion of AM initiation and de novo formation of new stem cell niches in the leaf axils (Yang et al. 2023). Following the auxin minimum, a cytokinin pulse occurs in the leaf axil during AM formation (Wang et al. 2014b). In addition to auxin and cytokinin, many other phytohormones (e.g., strigolactones, brassinosteroids, gibberellins, and abscisic acid) participate and interplay in shoot AM formation (Napoli and Ruehle 1996; Beveridge et al. 1997; Beveridge 2000, 2006; Turnbull et al. 2002; Sorefan 2003; Foo et al. 2001; Morris et al. 2001; Zhang et al. 2020b; Chatfield et al. 2000; Reddy et al. 2013). These factors intricately interact and regulate AM initiation through shared or distinct mechanisms.
In this article, we focus on explaining, unifying, and differentiating the intertwined mechanisms of AM development in several plant species, including Arabidopsis (Arabidopsis thaliana), tomato (Solanum lycopersicum), and rice (Oryza sativa). Additionally, we also discuss the challenges of identifying more genes specially involved in AM formation and propose available methodologies suitable for resolving these problems. But outgrowth is not the focus of this present review, and we refer readers to our newly published review on this topic (Yuan et al. 2023). All the genes mentioned in this review are listed in Table 1.
Origin of AM: detached or de novo?
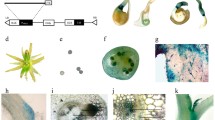
A distinctive characteristic of plants is their remarkable ability for reiterative growth and continuous organogenesis over their lifetimes. Analogous to the SAM, AMs play a pivotal role in initiating the development of lateral organs. This precisely regulated developmental process can result in AM formation, subsequently giving rise to the development of branches/tillers. In crops such as wheat and rice, the branches or tillers originating from AMs ultimately contribute to the formation of panicles, determining the overall grain yield (Fig. 1 and Video S1). However, grain yield loss will occur if plants have defective AM formation (Fig. 1 and Video S1).
IIlustrution of the dynamic developmental process of axillary meristem (AM) initiationof crops and efects comparisons between plants with normal ability to launch AM initiation or not A in the up line depicts the plant with the normality to generate AMs, resulting in more tillers/branches (the top right) if AMs can outgrowth subsequently. AMs arises from the leaf axil framed by black circles. The AM boxed in the shoot base are closed up in the top right. B in the bottom line delineates the plant with severly defect in AM initiation, which can lead to monoculm phenotype with low grain yield (the bottom right). The leaf axil where generate AM are barren. For more detail information, please refer to Video S1
Two alternative models for AM formation have been proposed: the ‘detached meristem’ model and the ‘de novo induction’ model (Fig. 2 and Video S2). The ‘detached meristem’ model posits that AMs form from pluripotent stem cells that bud off from the primary SAM and maintain meristematic potential in the leaf axils as leaf primordia develop (Steeves and Sussex 1989). This is supported by evidence showing that leaf axil cells remain undifferentiated and express the meristem marker STM (Grbic and Bleecker 2000; Long and Barton 2000). Laser ablation experiments have also shown that AMs originate from cells with STM mRNA persistence (Shi et al. 2016). In addition, studies reveal that AM progenitor cells are set aside early in SAM development (Burian et al. 2016).
Dynamic illustration of distinct origins of AM. A The detached meristem model: one axillary meristem emanates from the shoot apical meristem (SAM). This process is represented successively from a group of cells frst derived from the SAM,which then grow up to one tiller bud. B The de novo induction model: In some cases, the tiller forms directly from determinate cells, without derivation from the SAM. When essential genes are disrupted, the tiller can even form far from the leaf axil (indicated by arrow). The enlarged pictures (top right) represent the region framed by boxes. For more detail information of these processes, please refer to Video S2
Alternatively, the ‘de novo induction’ model proposes that a set of differentiated cells equivalent to their neighbors can form an AM given an appropriate localized signal (Long and Barton 2000). Evidence for this includes AMs arising on the underside of leaves in Arabidopsis phabulosa-1d (phb-1d) mutants. (McConnell and Barton 1998). Additionally, ectopic AM occurs in stm mutants that lost SAM characteristics (Endrizzi et al. 1996). Adventitious SAMs can arise from the axils of cotyledons and cultured root explants in pinhead mutants (McConnell and Barton 1995). Ectopic expression of the AM regulator, Super determinant 1A (SDE1), which is confined in leaf axils and regulates AM development, leads to ectopic meristem formation at the distal leaflets even in the shoot away from leaf axils (López et al. 2021). Together, these observations lend credence to the ‘de novo induction’ hypothesis, whereby differentiated cells can acquire meristematic identity.
The ongoing debate between the “detached meristem” and “de novo induction” theories in AM initiation is complex. Moreover, the expression of STM in the interprimordial regions between SAM and leaf axils (Greb et al. 2003; Shuai et al. 2002) complicates this debate, as it challenges clear differentiation between these two concepts. We propose investigating a range of mutants, specifically those with distinct AM initiation but no SAM defects as a strategy, and the second one that knocks out the AM-specific expressed genes by high-through CRISPR-Cas9 system, to bridge the understanding between these two mechanisms. This approach is promising because the genes associated with these mutants might exclusively influence AM or interact with SAM-related genes to trigger AM initiation.
The genetic and epigenetic factors regulate AM initiation
Understanding the molecular mechanisms that govern shoot branching heavily relies on characterizing genes responsible for AM initiation. To this end, multiple genes, including the key genes like rice MONOCULM1 (MOC1) and its orthologues Lateral suppressor (LS) and LAS in tomato and Arabidopsis, STM, and REVOLUTA (REV) in Arabidopsis (Long et al. 1996; Schumacher et al. 1999; Otsuga et al. 2001; Greb et al. 2003; Li et al. 2003), have been identified and thoroughly studied.
Generally, current mutants related to AM development exhibit morphological defects that can be categorized into two classes. The first type comprises mutations affecting AM initiation, leading to a lack of AM formation, as the las mutant exemplifies. The second type enhances AM formation, resulting in a bush phenotype, as observed in supershoot (sps) mutants (Tantikanjana et al. 2001). In the following discussion, we summarize genes associated with AM initiation, accompanied by a critical analysis of their hierarchical relationships, where applicable (Fig. 3).
Summary diagram of components and phytohormones in AM initiation. Blue arrows indicate promotion and red-fat ended lines depict inhibition. The axillary meristem (AM) is circled. The genes acting with several genes are framed framed with circles with blue background. In this model, many factors function at diferent stages, including key hub genes (e.g., CUCs and LAS genes) and phytohormones (e.g., auxin, BR, and CK, etc.). LFY, LEAFY; EXB1, EXCESSIVE BRANCHES1; LOF1/LOF2, LATERAL ORGAN FUSION1/2; RAX1, REGULATOR OF AXILLARY MERISTEM1; BR, Brassinosteroid; BZR1, BRASSINAZOLE-RESISTANT1; BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR1; LOB, LATERAL ORGAN BOUNDARIES; DRN, DORNROSCHEN; CUCs, CUP-SHAPED-COTYLRDON genes; BRC1, BRANCHED1; STM, SHOOT MERISTEMLESS; LAS, LATERAL SUPPRESSOR; SPL9, SQUAMOSA PROMOTER BINDIN PROTEIN-LIKE9; GA, Gibberellin; ATH: ARABIDOPSIS THALIANA HOMEOBOX GENE1; GA2ox4, gibberellin 2- oxidase4; DELLA, aspartic acid–glutamic acid–leucine–leucine–alanine; AXR1, Arabidopsis auxin-resistant 1; ROX, REGULATOR OF AXILLARY MERISTEM FORMATION; REV, REVOLUTA; PAGO10, ARGONAUTE10; BR, Brassinosteroid; MP, monopteros; ARF5, monopteros; IPT, ADENYLATE ISOPENTENYLTRANSFERASE; ARR1, Arabidopsis response regulator1; WUS, WUSCHEL; CLV3: CLAVATA3; PRC, Polycomb repressive complex; DPA4, NGATHA-LIKE transcription factors DEVELOPMENT-RELATED PcG TARGET IN THE APEX4; SOD7, SUPPRESSOR OF DA1-1 7; UBP15, UBIQUITIN-SPECIFIC PROTEASE15; B-ARR, type-B Arabidopsis response regulator
Given that AMs function as new SAMs, generating vital plant structures such as tillers/branches, leaves, flowers, etc., it is getting essential to explore whether the genes instrumental in SAM development also play a role in the initiation of AMs. It is hypothesized that these sorts of genes are involved in the fundamental processes of meristem initiation. This involvement is direct unless the formation of AMs is an indirect result of these mutations. To understand this relationship, we examine key genes like STM, CUCs, REV, PINHEAD, CLV3, and more, aiming to elucidate their functions and the potential hierarchical interactions among them. For example, the sustained expression of STM, a number of the KNOTTED class of homeodomain genes essential for SAM formation, suggests the presence of cells in an indeterminate state (Long et al. 1996). Overexpression of STM can lead to many ectopic SAMs in tobacco plants, indicating a cell fate switch from determinacy to indeterminacy in cell fate (Sinha et al. 1993). Notably, despite the role of STM in SAM, ablation of most cells within the STM-expressing region prevents AM initiation (Shi et al. 2016). These findings indicate that AM and SAM share a comparable molecular regulatory mechanism, with STM also playing a crucial role in AM initiation. However, it is noteworthy that ectopic STM expression is inadequate to activate AM formation from leaf axil cells that have lost STM expression (Shi et al. 2016), suggesting some cells undergo irreversible fate change or require special triggers to reverse them to indeterminate states. As STM has been proposed as an early marker of AM initiation (Long and Barton 2000), a small group of stem cells in the boundaries between the SAM and the emerging leaf primordium will develop into AM expressing STM, suggesting an involvement of STM in AM initiation (Keller et al. 2006). Furthermore, Wang et al. proposed a two-stage model for cell division during AM initiation, associating each stage with distinct STM expression levels (Wang and Jiao 2018b). In this model, maintaining low STM expression is required but insufficient for AM initiation. A subsequent increase in STM induces AM initiation and bulging (Shi et al. 2016). The early low levels of STM expression are presumably needed for stem cell competence, although these cells lack CLV3 or WUS expression, which are also essential for AM formation (Shi et al. 2016). It has been shown that the ARABIDOPSIS THALIANA HOMEOBOX GENE1 (ATH1), encoding a BEL1-like homeodomain (BLH) type three-amino-acid loop extension (TALE) class homeodomain protein, maintains STM expression, thus preserving the meristemic cell fate (Gómez-Mena and Sablowski 2008).
The WUS gene, a homeodomain transcription factor expressed in the SAM of the organing center, defines the stem cell niche (Wang et al. 2017). Despite its role in embryonic SAM formation, WUS in Arabidopsis and its rice ortholog MONOCULM 3 (MOC3) are required to initiate AM (Lu et al. 2015; Tanaka et al. 2015; Wang et al. 2017; Xin et al. 2017). Moreover, WUS expression is repressed by a polypeptide signal encoded by CLV3, acting as a stem cell marker (Schoof et al. 2000). Interestingly, WUS and CLV3 have a feedback relationship during AM initiation (Fig. 3). CLV3 is undetectable in leaf axils in wus mutants, suggesting that WUS can activate CLV3 (Xin et al. 2017). However, WUS expression is highly elevated in the clv3-2 leaf axils, where the AM primordium is larger than that in the wide-type, suggesting CLV3 signaling already restricted WUS expression to enable proper AM size determination in early developmental stages (Xin et al. 2017). WUS expression precedes AM initiation after STM expression in the initial phases (Guo et al. 2020) (Fig. 3). Moreover, Wang et al. 2021 considered that AM initiation concurs with the expression of WUS and CLV3 between leaf primordium 11 (P11) and P13 in Arabidopsis (Wang 2021). This finding is underscored by a similar observation in rice, where AM initiation can be detected at the P3 stage, as evidenced by the expression of MOC3, a rice ortholog of WUS (Lu et al. 2015; Shao et al. 2019).
The REV gene, encoding an HD-ZIPIII transcription factor, is indispensable for forming all lateral meristems in addition to its role in SAM development (Otsuga et al. 2001). Indeed, the loss-of-function of REV mutants leads to the absence of AMs (Tian et al. 2014; Talbert et al. 1995). REV can bind to the STM promoter region, indicating the STM requirement of REV during AM formation (Tian et al. 2014). The ectopic expression of the REV homolog PHAVULOTA (PHV) maintains and further activates ectopic STM expression on the abaxial leaf side, leading to ectopic AM formation (Shi et al. 2016). Histological analysis revealed that REV expression precedes WUS, which indicates that REV activity is epistatic to WUS (Otsuga et al. 2001). Regarding STM expression, no difference exists between wus mutants and wild-type, consistent with STM acting epistatically to WUS (Wang et al. 2017).
The redundant CUP-SHAPED COTYLEDON (CUC) genes (CUC1, CUC2, and CUC3) in Arabidopsis encode NAC transcription factors that significantly contribute to embryonic shoot meristem formation and shoot organ boundary specification. CUC2 and CUC3, but not CUC1, influence AM formation. Further analysis indicates that CUC3 plays a more significant role in regulating shoot branching than CUC2, but the effect is most prominent when CUC2 and CUC3 are combined (Hibara et al. 2006). CUC2 and CUC3 can directly activate the expression of DA1, encoding a ubiquitin-dependent peptidase, while mutations of the DA1 substrate in UBIQUITIN-SPECIFIC PROTEASE15 (UBP15) lead to repression of AM initiation (Li et al. 2020). Two transcription factors, the NGATHA-LIKE transcription factors DEVELOPMENT-RELATED PcG TARGET IN THE APEX4 (DPA4) and SUPPRESSOR OF DA1-1 7 (SOD7) redundantly repress CUC expression in the leaf axil and dpa4-2 sod7-2 double mutants display delayed AM initiation (Nicolas et al. 2022). DRN and its homolog DRNL, which encode AP2-type transcription factor family proteins, are required for AM initiation by directly activating CUC2. Large portions of the leaf axils in single or double dornroschen (drn) and drnlike (drnl) mutants are barren (Tian et al. 2014).
Recessive mutations in the PINHEAD locus of Arabidopsis disrupt the primary shoot meristem and AM formation, resulting in a single leaf or a slender pin-like organ and reduced lateral buds both in axils of cauline and rosette leaves (McConnell and Barton 1995; Ratcliffe et al. 1999; Zhang et al. 2020a). PINHEAD expression coincides in the leaf axil where AMs form and its overexpression occasionally produces more than one AMs per leaf axil (Zhang et al. 2020a), indicating its role in controlling AM formation. STM and REV are both down-regulated and up-regulated in the pinhead and its overexpression mutants, respectively (Fig. 3), suggestive of an epistatic role of PINHEAD to REV and STM (Zhang et al. 2020a).
Plant microRNAs are endogenous, single-stranded, and nontranslated RNA molecules that are highly complementary to their target mRNAs, mediating post-transcriptional gene silencing through mRNA cleavage (Bartel and Bartel 2003). The miR164 genes, comprising miR164A, miR164B, and miR164C (Wang et al. 2004; Bonnet et al. 2004; Reinhart et al. 2002), post-transcriptionally regulate CUC genes (Schwab et al. 2005; Raman et al. 2008). Constitutive overexpression of miR164 phenocopies the branching habits of cuc1 cuc2 double mutants by downregulating CUC1 and CUC2 transcripts (Nemhauser et al. 2004; Mallory et al. 2004). Conversely, the loss of function of miR164 genes can produce more accessory buds (Raman et al. 2008). In contrast, overexpression of miR164 in the cuc3-2 mutant abolishes AMs, indicating that miR164, CUC1, CUC2, and CUC3 play a pivotal role in AM initiation (Raman et al. 2008). Besides miR164, the rescue of AM defects in Arabidopsis argonaute 10 (ago10, also known as pinhead) by sequestering miR165/166 (Zhu et al. 2011), which targets REV, suggests AGO10/PINHEAD acts upstream of the REV and STM through the sequestration of miR165/166.
Collectively, these genes, such as STM, CUCs and REV, are integral in orchestrating the development of both the SAM and AMs. Yet, the precise timing and spatial dynamics governing the initiation of AM development remain topics for further exploration. We hypothesize that these genes depend on additional genes, specifically expressed in the leaf axils, to function as triggers for the initiation of AMs. This activation might occur either preceding or following their expression. Notably, a particular category of genes, known to manifest AM defects by specially regulating AM initiation, includes Arabidopsis LAS and REGULATOR OF AXILLARY MERISTEM (RAX) (Keller et al. 2006), as well as rice MOC1 (Li et al. 2003) and LAX PANICLE1 (LAX1) (Komatsu et al. 2003). Disruption of MOC1 in rice or its orthologous genes (e.g., LS in tomato and LAS in Arabidopsis), transcription factors of the GRAS family, results in the shortage of AMs and, consequently, fewer branches or tillers. MOC1 and LAS are expressed explicitly in the AM initiation zone (Greb et al. 2003; Li et al. 2003; Schumacher et al. 1999). These studies suggest a conserved function of these genes in both monocot and dicot. It is worth noticing that, in Arabidopsis, STM is focused on a group of small and densely cytoplasmic cells near the adaxial center of the primordium border, where it is required for AM initiation. However, these cells fail to develop into a new AM without STM expression in las mutants, suggesting that focused STM expressing denoting meristem organization onset relies on LAS function (Greb et al. 2003). This coincides with our hypothesis that SAM-regulating genes necessitate precise mediation to initiate AM development at an appropriate location. Furthermore, LAS has been proven to be a hub gene that integrates inputs from many upstream genes, as indicated by a leaf axil-enriched gene regulatory network analysis (Tian et al. 2014).
Despite the similarities in expression patterns between LAS and MOC1, notable differences exist. LAS expression regions extend to several layers of SAM beyond the AM, compared with MOC1, which remains undetectable in SAM (Greb et al. 2003; Li et al. 2003). Furthermore, the inflorescence meristems generated by AM were not affected in las mutants but otherwise in moc1 mutants (Chun et al. 2022; Greb et al. 2003; Zhang et al. 2021b). These variations underscore the evolutionary divergence between monocotyledonous and dicotyledonous plants, highlighting distinct regulatory mechanisms in plant architecture development. Likewise, REGULATOR OF AXILARRY MERISTEM FORMATION (ROX) is an Arabidopsis gene encoding bHLH protein, orthologous to the branching regulators LAX1 in rice and BARREN STALK1 (BA1) in maize (Yang et al. 2012; Komatsu et al. 2003; Matthes et al. 2019). Loss-of-function of ROX caused compromised AM formation. Its expression extended to the SAM and the AM, unlike LAX1’s AM location (Oikawa and Kyozuka 2009). In contrast to LAX1 and BA1, flower development was uninfluenced in rox mutants (Yang et al. 2012), further supporting the hypothesis of evolutionary distinctions between monocotyledons and dicotyledons. However, this inference should be pitched out with caution as the differences between las and rox mutants of Arabidopsis and their corresponding wild types became more pronounced when studied under short-day conditions (Yang et al. 2012; Greb et al. 2003). Because Arabidopsis, native to Europe and central Asia, has spread in the temperate climate zones of the five continents and, therefore, originally adapted to long-day conditions (Hsu et al. 2019).
In addition to the reliance of STM on LAS, other SAM-related genes are also affected in las. For example, WUS is undetectable in mutants las, indicating that WUS acts downstream of LAS (Wang et al. 2017). Enriched REV expression in leaf axils relies on LAS, a member of the GRAS family controlling AM initiation (Greb et al. 2003). Substantial upregulation of LAS accumulation was observed in mir164 triple mutants, indicating that miR164 can negatively regulate LAS (Raman et al. 2008). Thus, AM initiation is inhibited by miR164 through restricting CUC1/2 accumulation, which in turn regulates LAS expression (Raman et al. 2008). This is further substantiated by the observed down-regulation of LAS in the double mutant cuc1 cuc2, placing LAS downstream of CUC1 and CUC2 (Hibara et al. 2006) (Fig. 3). The regulation of the AM-specific gene LAS by CUC genes suggests that specific signals from the SAM are necessary to trigger AM initiation. This mechanism underscores the intricate interplay between SAM and AM development.
In addition to LAS, other genes specially expressed in or near AM also mediate SAM-related genes. For example, REGULATOR OF AXILLARY MERISTEMS1 (RAX1), an MYB family gene, promotes AM initiation by specifying the location of the stem cell niche. RAX1 functions redundantly with RAX2 and RAX3 to regulate AM initiation (Keller et al. 2006). RAX1 is initially detectable in a subregion along the boundary between the meristem and leaf primordia, similar to LAS (Keller et al. 2006). LEAFY (LFY), a master regulator of the transition of the reproductive stage, directly activates RAX1 to promote AM initiation (Chahtane et al. 2013). RAX1 also directly enhances CUC2 expression in vivo and in vitro (Tian et al. 2014). LATERAL ORGAN FUSION1 (LOF1) is also an MYB domain gene, which is expressed in the boundary domain of the SAM and leaf primordia. Loss-of-function if lof1 mutants lack AMs and STM expression in the corresponding boundary domain (Lee et al. 2009). Further gene expression analysis in lof1 mutants positioned LOF1 upstream of the RAX1, STM, LAS, and CUC genes (Shuai et al. 2002). However, Gendron et al. (2012) suggested that CUC genes may positively regulate LOF1 and LOF genes (Gendron et al. 2012), a way similar to that of LAS and CUCs. The EXCESSIVE BRANCHES1 (EXB1) gene, encoding the WRKY transcription factor WRKY71, affects AM initiation. EXB1 disruption results in reduced branching, while overexpression of EXB1 in exb1-D gain-of-function mutants leads to severe bushy and dwarf phenotypes (Guo et al. 2015). EXB1 is shown to control AM initiation by positively regulating the transcription of RAX1, RAX2, and RAX3 (Guo et al. 2015). Interestingly, overexpression of rice WRKY72 in Arabidopsis also increases shoot branches (Song et al. 2010), implying evolutionary conservation of EXB1/WRKY71 function in AM formation between monocots and dicots (Guo et al. 2015).
In addition to the precise regulation of SAM-related genes by genes expressed in AMs, epigenetic modulation allows nuanced expression of the same gene in distinct cell types and developmental contexts. Such epigenetic control allows for the diverse expression patterns of genes, which may be broadly expressed yet exhibit distinct functions in various cellular environments and stages of plant development. Thus, epigenetic control is intrinsically involved in all developmental processes, including AM initiation. Since many genes mediating WUS and STM, such as REV and ARABIDOPSIS RESPONSE REGULATOR 1 (ARR1) which is a type-B ARR transcription factor, are not exclusively expressed in leaf axils. Therefore, precisely expressing the genes needed for particular stages is imperative for AM control. The Polycomb Repressive Complex 2 (PRC2) establishes the H3K27me3 mark in plants and animals, providing a docking site for PRC1 to impose repressive chromatin (Zheng and Chen 2011). In mature leaves, where cells are fully differentiated, both WUS and STM exhibit high levels of H3K27me3, indicative of a low abundance of WUS and STM mRNA. In contrast, these two genes have a low concentration of H3K27me3 and a high concentration of H3K4me2/3, a mark associated with active chromatin, in tissues containing the leaf axil STM-expressing cells (Shi et al. 2016; Cao and Jiao 2020). Accordingly, STM and WUS are elevated in prc mutant (Shi et al. 2016; Wang et al. 2017). Moreover, applying histone deacetylation inhibitor trichostatin A induces ectopic WUS expression (Xin et al. 2017). Wang et al. showed that epigenetics contributes to the dynamic of WUS, with expression terminated in leaf axils and then reactivated de novo (Xin et al. 2017). A large abundant H3K27me3 represses the WUS expression in the leaf axil, while histone H3/H4 acetylation (H3/4Ac) is depleted. Before WUS activation, the levels of the H3K27me3 repressive mark decrease, while the levels of the active H3/4Ac mark increase (Wang et al. 2017; Cao and Jiao 2020).
In conclusion, the intricate web of genetic and epigenetic factors orchestrates the initiation of axillary meristems in plants. The collaborative action of genes such as CUC, WUS, STM, REV, LAS, and others, along with the regulatory influence of miRNAs, creates a finely tuned molecular symphony.
How do phytohormones precisely control AM initiation?
Phytohormones regulate diverse developmental processes throughout the plant life cycle. For example, auxin orchestrates developmental responses such as gravitropism and apical dominance, which depend on forming auxin gradients in plant tissues (Casanova-Sáez et al. 2021; Leyser 2018). Cytokinins (CKs) influence agricultural processes, including growth, nutrient responses, and biotic/abiotic stress responses (Kieber and Schaller 2018). Gibberellins (GAs) promote growth by regulating seed germination, root/shoot elongation, flowering, and fruit patterning (Binenbaum et al. 2018). Brassinosteroids (BRs) also stimulate plant growth and development by controlling cell division, elongation, and differentiation (Planas-Riverola et al. 2019). Given the multiple roles of phytohormones in plant development, regulatory mechanisms must exist to precisely control axillary meristem (AM) initiation. Based on current understanding, we synthesize the hormonal control of AM initiation as follows:
Lines of evidence that auxin is involved in AM initiation have been reported. For example, the AGC III kinase PINOID (PID) modulates polar auxin transport by regulating PIN1 localization within the cell (Michniewicz et al. 2007). Severe homozygous pid mutants resemble pin1 mutants and fail to form AMs compared to wide-type plants (Wang et al. 2014a). This highlights the importance of directional auxin transport for AM initiation. In the dominant mutant exb-1D, which displays excessive branching, the auxin biosynthesis gene TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1 (TAA1) and the auxin transport genes (e.g., PIN5), are repressed by EXB1 induction (Guo et al. 2015), suggesting the importance of auxin homeostasis to control shoot branching. Additionally, as in auxin resistance 1 (axr1) mutants, reduced auxin sensitivity enhances AM formation (Stirnberg et al. 1999), implying the significance of auxin in mediating AM initiation. The dynamic requirement of auxin was further investigated. Namely, the auxin minimum is one prerequisite for AM initiation, exemplified by that: PIN1 mediates auxin flow in the adaxial domain away from the leaf axil toward the tip of the leaf primordium, thus establishing the auxin minimum in the leaf axil (Wang et al. 2014b; Bayer et al. 2009). The shoot meristem marker STM is activated during AM formation (Long and Barton 2000; Greb et al. 2003). Conversely, ectopically expressing an auxin biosynthesis gene, indicating higher auxin levels in the leaf axil, decreases STM expression (Wang et al. 2014b). This auxin minimum niche sustaining STM expression in the boundary zone to maintain the AM identity was evidenced by Guo et al. 2015 (Guo et al. 2015). In contrast, an elevated auxin concentration in the stem cell maintenance stage, driven by a specially located leaf axil gene promoter, perturbs the AM initiation (Wang et al. 2014b). Consistent with this, pin-formed 1 (pin1)-like phenotypes are observed in mutants lacking PID function. Because the PID disruption presumably maintains unidirectional auxin apical-basal transport, consequently increasing auxin to inhibit AM formation (Friml et al. 2004). These lines of evidence show that the auxin minimum is the precondition for AM initiation. However, maize auxin biosynthesis mutants display defects in vegetative AM formation (Matthes et al. 2019), implying that auxin is still required at some points during AM initiation. The requirement of auxin during AM initiation was demonstrated by the monopteros (mp) mutant. MP, also known as ARF5, is an auxin response factor that activates downstream signaling in response to auxin. MP expresses in the youngest leaf primordia and ceases its expression at the later AM formation stage when AM starts bulging (Guan et al. 2017; Guo et al. 2020), indicating the existence of a high concentrate auxin in the later AM bulging stage and a positive role for auxin in AM initiation (Zhang et al. 2020a). Furthermore, MP may activate the expression of PINHEAD, whose protein sequesters miR165/166 to release REV and STM, thereby promoting AM initiation (Zhang et al. 2020a). Moreover, rescued AM defects in mpΔ mutants by deleting the ARF5-binding site in the AGO10/PINHEAD promoter indicated auxin signaling is required in late AM initiation stages (Zhang et al. 2020a). In summary, auxin is essential for AM initiation, playing dynamic roles during this process. In the early stages of AM formation, an auxin minimum is required for meristematic cell maintenance, while in the later stages, higher auxin levels promote their activation. Further dissection of auxin synthesis, transport and signaling dynamics will provide deeper insights into its complex regulation of AM development.
Genetic analysis has shown that CK perception and signaling are essential for AM initiation. Several instances have favored the requirement of CK. For instance, AM initiation deficiency of rax1 mutants can be partially rescued by either CK production in leaf axil or exogenous CK treatment (Wang et al. 2014b). Additionally, CK receptors and the signaling detector in the leaf axils are upregulated prior to and during AM initiation, indicating cytokinin's role in AM initiation (Wang et al. 2014b). Mutants defective in CK receptors such as Arabidopsis histidine kinase 2 (ahk2), ahk3, and ahk4 with compromised CK perception and the corresponding double mutants exhibit defects in AM initiation. Moreover, B-type ARABIDOPSIS RESPONSE REGULATOR (ARR) transcription factors, which act downstream of the CK signaling pathway, are also required for AM initiation (Wang et al. 2014b). Furthermore, the SAM-related genes and CKs were mutually regulated to guarantee AM initiation. For example, during AM initiation, STM activates CK biosynthesis in leaf axils (Guo et al. 2015). CK signaling then activates de novo WUS expression in leaf axils (Wang et al. 2017; Guo et al. 2020). Type-BRRs, transcriptional activators in CK signaling, especially ARR1, directly bind to the WUS promoter to activate its expression (Cao and Jiao 2020). ARR1 also directly binds to the LAS promoter to activate its expression (Tian et al. 2014). In addition to the low auxin condition, a subsequent pulse of CK occurs prior to AM initiation (Wang et al. 2014b). No CK signal can be detectable without an auxin minimum in the leaf axil (Wang et al. 2014b), demonstrating the dependence of the leaf axil CK pulse on the auxin minimum. Supporting this, sps mutants, which show a bushy phenotype due to enhanced AM formation and lateral bud release, have elevated levels of CK but decreased levels of auxin (Tantikanjana et al. 2001). Furthermore, in mutants such as las, rax, and rev with compromised AM initiation, the leaf axils lack CK signaling pulse (Wang et al. 2017), indicating that CK is required for AM initiation. Overall, the evidence demonstrates that CK signaling is a key step following the establishment of the auxin minimum niche. CK perception and downstream transcriptional activation of WUS, LAS and other AM regulators promote the activation and bulging of meristematic cells to initialize AM development.
Gibberellic acids (GAs) are growth-promoting hormones that mediate various plant developmental processes throughout the plant life cycle (Yamaguchi 2008). However, exogenous GA application decreases AM formation. Leaf axils ectopically expressing a GA biosynthesis gene showed significantly lower AM formation (Zhang et al. 2020b). Conversely, the GA-deficient mutant ga1-3 displays more AMs, indicating a negative role of GA in AM formation. However, how GA precisely regulates AM initiation, especially regulating AM-specific or SAM-related genes, deserves investigation due to the importance of AMs. DELLA proteins, master repressors of GA signaling, participate in various physiological processes by interacting with various transcription factors, including BRASSINAZOLE-RESISTANT1 (BZR1), ARR1, and ARF6 (Van De Velde et al. 2017). A della pentuple mutant shows defects in AM formation, suggesting that DELLAs play a role in this process. DELLAs interact with SPL9, thus attenuating its repression of LAS. This promotes AM initiation, with LAS then inducing GA deactivation enzyme Gibberellin 2-beta-dioxygenase 4 (GA2ox4) to form a low-GA condition in leaf axils (Zhang et al. 2020b). Thus, the crosstalk and balance between GA metabolism and LAS precisely modulate AM formation spatiotemporally.
In addition to auxin and Cks, BR-responsive genes are highly enriched in organ boundary cells, suggesting these sites are novel centers of BR activity (Tian et al. 2014). BR is an essential plant steroid hormone regulating cell division and expansion (Gendron et al. 2012). As low cell division rates are required in the boundary zone to maintain AM competence, BR accumulation is negatively regulated in leaf axils by LATERAL ORGAN BOUNDARIES (LOB), a key boundary-specific transcription factor. LOB directly upregulates PHYB ACTIVATION TAGGED SUPPRESSOR1 (BAS1), a cytochrome P450 enzyme that inactivates BRs through C-26 hydroxylation, thereby reducing BR levels to decrease cell division and expansion in the boundary zone (Gendron et al. 2012; Bell et al. 2012). Furthermore, the SAM-developmental gene CUC3 was inhibited by the BR-activated gene BRASSINAZOLE-RESISTANT1 (BZR1) by directly binding to the promoter of CUC3, indicating low BR levels in boundary zones are required to activate AM initiation. In summary, LOB restricts BR accumulation in leaf axil boundary zones through BAS1 induction. Low BR levels inhibit cell division and expansion while also alleviating BZR1 repression of CUC3. Fine-tuned crosstalk between BR and key AM regulators like LOB and CUC3 allows proper AM initiation.
While plant hormones, such as auxin, CKs, BRs, and GAs, participate in various development in the entire plant life, precisely where and when they act is the key point for development. Likewise, AM initiation is associated with phytohormones, which must be involved at the right time and location.
Genes regulating AM formation affect grain yield
For crop species, AMs are essential for producing tillers bearing grains, determining the number of seed spikes per plant and the number of seeds per spike—all key factors influencing overall crop yield (Wang and Jiao 2018a), such as in rice, maize, and wheat. These factors are directly determined by branching ability during vegetable and reproductive growth stages. Namely, AM essentially harbors a niche with a group of meristematic cells to influence branching in tilers and panicles. For instance, the LAX1 and MOC1 genes in rice are involved in the formation of both tillers and panicle branches. Mutations in either MOC1 or LAX1 resulted in a reduced number of both tillers and panicle branches (Wang and Li 2011). Further exploring indicated that LAX1 is regulated indirectly by the gene DEFECTIVE STIGMA AND PANICLE (DSP), determining tiller primordium formation and synergistically regulating panicle primordium development (Yu et al. 2023). Likewise, in Helianthus annuus, a dicotyledonous species, the mutated REGULATOR OF AXILLARY MERISTEM FORMATION-LIKE (Ha-ROXL), akin to LAX1, affects both AM initiation and Floral meristems (FMs) (Basile et al. 2019), which suggested a shared role of LAX1 and its orthologs in influencing grain yield across dicots and monocots. However, disruption of ROX in Arabidopsis, an ortholog of LAX1, displays compromised AM formation during the vegetative phase, particularly noticeable under short-day photoperiods (Yang et al. 2012). Again, we must be cautious about drawing definitive conclusions regarding this abnormality observed in Arabidopsis, since Arabidopsis originated in and is generally grown under long-day conditions (Hsu et al. 2019). Furthermore, in las and rax mutants, the AM initiation defects are more easily recognized under short-day conditions than that under long-day conditions (Greb et al. 2003; Keller et al. 2006), concurring with our perspective in making critical sense of AM formation in Arabidopsis. Collectively, further research on AM initiation genes is needed to elucidate the genetic mechanisms underlying tiller and panicle branching. Exploring conserved and specialized regulators of AMs will provide insights to improve branching and optimize crop yields.
Challenges and opportunities regarding isolation genes involved in AM formation
While essential for plant development and agriculture, the molecular mechanisms underlying AM formation stay elusive. The study of AM initiation has been hurdled, mainly due to the shortage of mutants, specially affecting AM development in plants like rice and Arabidopsis. This implies that many unknown AM initiation regulators demand to be identified. Notably, alternative methodologies developed recently could be employed to resolve this problem. For example, Yang et al. suggested the utilization of genetic backgrounds with reduced apical dominance to identify more AM initiation regulators (Yang et al. 2023). Genome editing technologies drive significant advances in life sciences due to precise modifications at target genomic loci (Xing et al. 2023; Doudna and Charpentier 2014). CRISPR/Cas9 systems have been broadly adopted as a targeted genetic manipulation tool that has been applied to many species, such as rice, wheat, tomato, and more (Doudna and Charpentier 2014). This routine technology can also be used to identify and validate new genes that act specifically in AM interactions. The utility of this technology is not limited to model plants, but can further be extended to cultivated crop plants and their wild progenitors, which often have very different architectures. Utilizing this technology allows for discovering AM-specific genes across a diverse range of plant species. Emerging yet thriving omics may also help distinguish new genes involved in AM initiation regulation. For example, single-cell omics technologies reveal the intracellular dynamics of different individual cells and answer biological questions with high-dimensional catalogs of millions of cells, including transcriptomics, genomics, chromatin accessibility, epigenomics, and proteomics data across species (Mo and Jiao 2022). Initially applied in animals, single-cell RNA sequencing (sc-RNA) technologies have been embraced by the field of plants. In Arabidopsis, Zhang et al. carried out Sc-RNA to define the cellular taxonomy of the Arabidopsis vegetative shoot apex at the transcriptome level and found that the shoot apex is composed of highly heterogeneous cells (Zhang et al. 2021a); in maize, sc-RNA analyzed single cells from developing maize ears, helping to identify candidate genes associate crops yield traits (Xu et al. 2021); in rice, analysis of root tips using Sc-RNA provided insight into the transcriptomic landscape of major cell types of rice root tip at single-cell resolution (Wang et al. 2021). In addition, a gene regulatory network-based investigation of trichoblast differentiation in Arabidopsis revealed novel transcription factors and previously unknown feedback loops/mechanisms by harnessing trajectory inference, one algorithm used in Sc-RNA analysis (Denyer et al. 2019). Despite Sc-RNA, single-cell level chromatin has also been practiced in plants, which is essential in AM initiation. For example, scATAC-seq has been applied to profile root tip cells in Arabidopsis, along with sc-RNA data, suggesting a connection between chromatin accessibility and expression dynamics (Farmer et al. 2021). Together, since the efficiency of the omics-based approaches, these techniques are more commonly used to investigate cell identity and fate changes, which also occur in AM initiation. As cell identity and fate changes occur during AM initiation, applying single-cell omics techniques could reveal new genes and networks controlling this process. These emerging approaches may expedite research on AM initiation mechanisms. Combined with clever genetics, single-cell omics technologies provide promising avenues to elucidate the molecular control of AM development.
Concluding remarks
This review has covered significant recent advances in elucidating the intricate molecular mechanisms governing AM initiation. Research over decades has revealed that AM development relies on coordinated regulation by transcriptional, hormonal, and epigenetic factors. Key regulators such as LAS, RAX1, STM, REV, WUS, and CUC2 converge to control gene expression programs activating meristematic fate in leaf axil cells. Intricate crosstalk between auxin, CKs, GAs, and other hormones establishes a niche conducive to AM formation. Moreover, dynamic changes in chromatin modifications facilitate spatiotemporal patterns of AM gene expression. Despite progress, questions remain regarding the developmental origin of AM progenitor cells, limitations for identifying more AM-specific, and integration of the various pathways regulating AM initiation. Key next steps include: (1) Elucidating the developmental relationship between the shoot apical meristem and AM progenitor cells and reconciling detached versus de novo origins during AM initiation; (2) Identifying additional novel regulators and networks of AM formation, combing omics- sequencing and cellular resolution imaging techniques; (3) Exploring divergence and specialization of AM developmental programs between plant species; (4) Leveraging knowledge of AM formation mechanisms to improve crop architecture and yield. Collectively, unraveling the AM initiation process remains an exciting frontier in plant development biology. Translation of these fundamental findings to crop species holds immense promise for agricultural enhancement. We anticipate the next decades would witness transformative discoveries illuminating how plants elaborate their axillary meristems to elaborately branch out their forms.
Data availability
Data sharing does not apply to this article as no datasets were generated or analyzed during the current study.
References
Aida M, Ishida T, Tasaka M (1999) Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126(8):1563–1570. https://doi.org/10.1242/dev.126.8.1563
Bartel B, Bartel DP (2003) MicroRNAs: at the root of plant development? Plant Physiol 132(2):709–717. https://doi.org/10.1104/pp.103.023630
Basile A, Fambrini M, Tani C, Shukla V, Licausi F, Pugliesi C (2019) The Ha-ROXL gene is required for initiation of axillary and floral meristems in sunflower. Genesis 57(9):e23307. https://doi.org/10.1002/dvg.23307
Bayer EM, Smith RS, Mandel T, Nakayama N, Sauer M, Prusinkiewicz P, Kuhlemeier C (2009) Integration of transport-based models for phyllotaxis and midvein formation. Genes Dev 23(3):373–384. https://doi.org/10.1101/gad.497009
Bell EM, Lin WC, Husbands AY, Yu L, Jaganatha V, Jablonska B, Mangeon A, Neff MM, Girke T, Springer PS (2012) Arabidopsis LATERAL ORGAN BOUNDARIES negatively regulates brassinosteroid accumulation to limit growth in organ boundaries. Proc Natl Acad Sci 109(51):21146–21151. https://doi.org/10.1073/pnas.1210789109
Beveridge CA (2000) Long-distance signalling and a mutational analysis of branching in pea. Plant Growth Regul 32(2):193–203. https://doi.org/10.1023/A:1010718020095
Beveridge CA (2006) Axillary bud outgrowth: sending a message. Curr Opin Plant Biol 9(1):35–40. https://doi.org/10.1016/j.pbi.2005.11.006
Beveridge CA, Symons GM, Murfet IC, Ross JJ, Rameau C (1997) The rms1 mutant of pea has elevated indole-3-acetic acid levels and reduced root-sap zeatin riboside content but increased branching controlled by graft-transmissible signal (s). Plant Physiol 115(3):1251–1258. https://doi.org/10.1104/pp.115.3.1251
Binenbaum J, Weinstain R, Shani E (2018) Gibberellin localization and transport in plants. Trends Plant Sci 23(5):410–421. https://doi.org/10.1016/j.tplants.2018.02.005
Bonnet E, Wuyts J, Rouze P, Van De Peer Y (2004) Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc Natl Acad Sci 101(31):11511–11516. https://doi.org/10.1073/pnas.0404025101
Bowman JL, Eshed Y (2000) Formation and maintenance of the shoot apical meristem. Trends Plant Sci 5(3):110–115. https://doi.org/10.1016/s1360-1385(00)01569-7
Burian A, Pierre KC (2016) Patterns of stem cell divisions contribute to plant longevity. Curr Biol 26(11):1385–1394. https://doi.org/10.1016/j.cub.2016.03.067
Cao X, Jiao Y (2020) Control of cell fate during axillary meristem initiation. Cell Mol Life Sci 77(12):2343–2354. https://doi.org/10.1007/s00018-019-03407-8
Cao X, Wang J, Xiong Y, Yang H, Yang M, Ye P, Bencivenga S, Sablowski R, Jiao Y (2020) A self-activation loop maintains meristematic cell fate for branching. Curr Biol 30(10):1893–1904. https://doi.org/10.1016/j.cub.2020.03.031
Casanova-Sáez R, Mateo-Bonmatí E, Ljung K (2021) Auxin metabolism in plants. Cold Spring Harb Perspect Biol 13(3):1–22. https://doi.org/10.1101/cshperspect.a039867
Chahtane H, Vachon G, Le Masson M, Thévenon E, Périgon S, Mihajlovic N, Kalinina A, Michard R, Moyroud E, Monniaux M, Sayou C, Grbic V, Parcy F, Tichtinsky G (2013) A variant of LEAFY reveals its capacity to stimulate meristem development by inducing RAX1. Plant J 74(4):678–689. https://doi.org/10.1111/tpj.12156
Chatfield SP, Stirnberg P, Forde BG, Leyser O (2000) The hormonal regulation of axillary bud growth in Arabidopsis. Plant J 24(2):159–169. https://doi.org/10.1046/j.1365-313x.2000.00862.x
Chun Y, Kumar A, Li X (2022) Genetic and molecular pathways controlling rice inflorescence architecture. Front Plant Sci 13:1010138–1010155. https://doi.org/10.3389/fpls.2022.1010138
Denyer T, Ma X, Klesen S, Scacchi E, Nieselt K, Timmermans MCP (2019) Spatiotemporal developmental trajectories in the Arabidopsis root revealed using high-throughput single-cell RNA Sequencing. Dev Cell 48(6):840–852. https://doi.org/10.1016/j.devcel.2019.02.022
Doudna JA, Charpentier E (2014) Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346 (6213):1258096. https://doi.org/10.1126/science.1258096
Endrizzi K, Moussian B, Haecker A, Levin JZ, Laux T (1996) The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J 10(6):967–979. https://doi.org/10.1046/j.1365-313x.1996.10060967.x
Farmer A, Thibivilliers S, Ryu KH, Schiefelbein J, Libault M (2021) Single-nucleus RNA and ATAC sequencing reveals the impact of chromatin accessibility on gene expression in Arabidopsis roots at the single-cell level. Mol Plant 14(3):372–383. https://doi.org/10.1016/j.molp.2021.01.001
Foo E, Turnbull CGN, Beveridge CA (2001) Long-distance signaling and the control of branching in the rms1 mutant of pea. Plant Physiol 126(1):203–209. https://doi.org/10.1104/pp.126.1.203
Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk PB, Ljung K, Sandberg G (2004) A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306(5697):862–865. https://doi.org/10.1126/science.1100618
Gendron JM, Liu JS, Fan M, Bai MY, Wenkel S, Springer PS, Barton MK, Wang ZY (2012) Brassinosteroids regulate organ boundary formation in the shoot apical meristem of Arabidopsis. Proc Natl Acad Sci 109(51):21152–21157. https://doi.org/10.1073/pnas.1210799110
Gómez-Mena C, Sablowski R (2008) ARABIDOPSIS THALIANA HOMEOBOX GENE1 establishes the basal boundaries of shoot organs and controls stem growth. Plant Cell 20(8):2059–2072. https://doi.org/10.1105/tpc.108.059188
Grbic V, Bleecker AB (2000) Axillary meristem development in Arabidopsis thaliana. Plant J 21(2):215–223. https://doi.org/10.1046/j.1365-313x.2000.00670.x
Greb T, Clarenz O, Schäfer E, Müller D, Herrero R, Schmitz G, Theres K (2003) Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev 17(9):1175–1187. https://doi.org/10.1101/gad.260703
Guan C, Wu B, Yu T, Wang Q, Krogan NT, Liu X, Jiao Y (2017) Spatial auxin signaling controls leaf flattening in Arabidopsis. Curr Biol 27(19):2940–2950. https://doi.org/10.1016/j.cub.2017.08.042
Guo D, Zhang J, Wang X, Han X, Wei B, Wang J, Li B, Yu H, Huang Q, Gu H, Qu L-J, Qin G (2015) The WRKY transcription factor WRKY71/EXB1 controls shoot branching by transcriptionally regulating RAX genes in Arabidopsis. Plant Cell 27(11):3112–3127. https://doi.org/10.1105/tpc.15.00829
Guo Y, Tian C, Jiao Y, Wang Y (2020) Multifaceted functions of auxin in vegetative axillary meristem initiation. J Genet Genom 47(9):591–594. https://doi.org/10.1016/j.jgg.2020.10.001
Hibara K-I, Karim MR, Takada S, Taoka K-I, Furutani M, Aida M, Tasaka M (2006) Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 18(11):2946–2957. https://doi.org/10.1105/tpc.106.045716
Hsu CW, Lo CY, Lee CR (2019) On the postglacial spread of human commensal Arabidopsis thaliana: journey to the East. New Phytol 222(3):1447–1457. https://doi.org/10.1111/nph.15682
Keller T, Abbott J, Moritz T, Doerner P (2006) Arabidopsis REGULATOR OF AXILLARY MERISTEMS1 controls a leaf axil stem cell niche and modulates vegetative development. Plant Cell 18(3):598–611. https://doi.org/10.1105/tpc.105.038588
Kieber JJ, Schaller GE (2018) Cytokinin signaling in plant development. Development 145(4):149344–149351. https://doi.org/10.1242/dev.149344
Komatsu K, Maekawa M, Ujiie S, Satake Y, Furutani I, Okamoto H, Shimamoto K, Kyozuka J (2003) LAX and SPA: major regulators of shoot branching in rice. Proc Natl Acad Sci 100(20):11765–11770. https://doi.org/10.1073/pnas.1932414100
Lee D-K, Geisler M, Springer PS (2009) LATERAL ORGAN FUSION1 and LATERAL ORGAN FUSION2 function in lateral organ separation and axillary meristem formation in Arabidopsis. Development 136(14):2423–2432. https://doi.org/10.1242/dev.031971
Leyser O (2018) Auxin signaling. Plant Physiol 176(1):465–479. https://doi.org/10.1104/pp.17.00765
Li X, Qian Q, Fu Z, Wang Y, Xiong G, Zeng D, Wang X, Liu X, Teng S, Hiroshi F (2003) Control of tillering in rice. Nature 422(6932):618–621. https://doi.org/10.1038/nature01518
Li Y, Xia T, Gao F, Li Y (2020) Control of plant branching by the CUC2/CUC3-DA1-UBP15 regulatory module. Plant Cell 32(6):1919–1932. https://doi.org/10.1105/tpc.20.00012
Long J, Barton MK (2000) Initiation of axillary and floral meristems in Arabidopsis. Dev Biol 218(2):341–353. https://doi.org/10.1006/dbio.1999.9572
Long JA, Moan EI, Medford JI, Barton MK (1996) A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379(6560):66–69. https://doi.org/10.1038/379066a0
López H, Schmitz G, Thoma R, Theres K (2021) Super determinant1A, a RAWUL domain-containing protein, modulates axillary meristem formation and compound leaf development in tomato. Plant Cell 33(7):2412–2430. https://doi.org/10.1093/plcell/koab121
Lu Z, Shao G, Xiong J, Jiao Y, Wang J, Liu G, Meng X, Liang Y, Xiong G, Wang YJJOG (2015) MONOCULM 3, an ortholog of WUSCHEL in rice, is required for tiller bud formation. J Genet Genom 42(2):71–78. https://doi.org/10.1016/j.jgg.2014.12.005
Mallory AC, Dugas DV, Bartel DP, Bartel B (2004) MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr Biol 14(12):1035–1046. https://doi.org/10.1016/j.cub.2004.06.022
Matthes MS, Best NB, Robil JM, Malcomber S, Gallavotti A, McSteen P (2019) Auxin evodevo: conservation and diversification of genes regulating auxin biosynthesis, transport, and signaling. Mol Plant 12(3):298–320. https://doi.org/10.1016/j.molp.2018.12.012
McConnell JR, Barton MK (1995) Effect of mutations in the PINHEAD gene of Arabidopsis on the formation of shoot apical meristems. Dev Genet 16(4):358–366. https://doi.org/10.1002/dvg.1020160409
McConnell JR, Barton MK (1998) Leaf polarity and meristem formation in Arabidopsis. Development 125(15):2935–2942. https://doi.org/10.1242/dev.125.15.2935
Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, Schwab R, Weigel D, Meyerowitz EM, Luschnig C, Offringa R, Friml J (2007) Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130(6):1044–1056. https://doi.org/10.1016/j.cell.2007.07.033
Mo Y, Jiao Y (2022) Advances and applications of single-cell omics technologies in plant research. Plant J 110(6):1551–1563. https://doi.org/10.1111/tpj.15772
Morris SE, Turnbull CG, Murfet IC, Beveridge CA (2001) Mutational analysis of branching in pea. Evidence that Rms1 and Rms5 regulate the same novel signal. Plant Physiol 126(3):1205–1213. https://doi.org/10.1104/pp.126.3.1205
Napoli C, Ruehle J (1996) New mutations affecting meristem growth and potential in Petunia hybrida Vilm. J Hered 87(5):371–377. https://doi.org/10.1093/oxfordjournals.jhered.a023016
Nemhauser JL, Mockler TC, Chory J (2004) Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol 2(9):e258. https://doi.org/10.1371/journal.pbio.0020258
Nicolas A, Maugarny-Calès A, Adroher B, Chelysheva L, Li Y, Burguet J, Bågman AM, Smit ME, Brady SM, Li Y, Laufs P (2022) De novo stem cell establishment in meristems requires repression of organ boundary cell fate. Plant Cell 34(12):4738–4759. https://doi.org/10.1093/plcell/koac269
Oikawa T, Kyozuka J (2009) Two-step regulation of LAX PANICLE1 protein accumulation in axillary meristem formation in rice. Plant Cell 21(4):1095–1108. https://doi.org/10.1105/tpc.108.065425
Otsuga D, DeGuzman B, Prigge MJ, Drews GN, Clark SE (2001) REVOLUTA regulates meristem initiation at lateral positions. Plant J 25(2):223–236. https://doi.org/10.1046/j.1365-313x.2001.00959.x
Planas-Riverola A, Gupta A, Betegón-Putze I, Bosch N, Ibañes M, Caño-Delgado AI (2019) Brassinosteroid signaling in plant development and adaptation to stress. Development 146 (5):dev151894. https://doi.org/10.1242/dev.151894
Raman S, Greb T, Peaucelle A, Blein T, Laufs P, Theres K (2008) Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J 55(1):65–76. https://doi.org/10.1111/j.1365-313x.2008.03483.x
Ratcliffe OJ, Bradley DJ, Coen ES (1999) Separation of shoot and floral identity in Arabidopsis. Development 126(6):1109–1120. https://doi.org/10.1242/dev.126.6.1109
Reddy SK, Holalu SV, Casal JJ, Finlayson SA (2013) Abscisic acid regulates axillary bud outgrowth responses to the ratio of red to far-red light. Plant Physiol 163(2):1047–1058. https://doi.org/10.1104/pp.113.221895
Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) MicroRNAs in plants. Genes Dev 16(13):1616–1626. https://doi.org/10.1101/gad.1004402
Schoof H, Lenhard M, Haecker A, Mayer KFX, Jürgens G, Laux T (2000) The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL Genes. Cell 100(6):635–644. https://doi.org/10.1016/s0092-8674(00)80700-x
Schumacher K, Schmitt T, Rossberg M, Schmitz G, Theres K (1999) The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc Natl Acad Sci 96(1):290–295. https://doi.org/10.1073/pnas.96.1.290
Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D (2005) Specific effects of MicroRNAs on the plant transcriptome. Dev Cell 8(4):517–527. https://doi.org/10.1016/j.devcel.2005.01.018
Shao G, Lu Z, Xiong J, Wang B, Jing Y, Meng X, Liu G, Ma H, Liang Y, Chen F, Wang Y, Li J, Yu H (2019) Tiller Bud formation regulators MOC1 and MOC3 cooperatively promote tiller bud outgrowth by activating FON1 Expression in rice. Mol Plant 12(8):1090–1102. https://doi.org/10.1016/j.molp.2019.04.008
Shi B, Zhang C, Tian C, Wang J, Wang Q, Xu T, Xu Y, Ohno C, Sablowski R, Heisler MG, Theres K, Wang Y, Jiao Y (2016) Two-step regulation of a meristematic cell population acting in shoot branching in Arabidopsis. PLoS Genet 12(7):e1006168. https://doi.org/10.1371/journal.pgen.1006168
Shuai B, Reynaga-PeñA CG, Springer PS (2002) The LATERAL ORGAN BOUNDARIES gene defines a novel, plant-specific gene family. Plant Physiol 129(2):747–761. https://doi.org/10.1104/pp.010926
Sinha NR, Williams RE, Hake S (1993) Overexpression of the maize homeo box gene, KNOTTED-1, causes a switch from determinate to indeterminate cell fates. Genes Dev 7(5):787–795. https://doi.org/10.1101/gad.7.5.787
Song Y, Chen L, Zhang L, Yu D (2010) Overexpression of OsWRKY72 gene interferes in the abscisic acid signal and auxin transport pathway of Arabidopsis. J Biosci 35(3):459–471. https://doi.org/10.1007/s12038-010-0051-1
Sorefan K (2003) MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev 17(12):1469–1474. https://doi.org/10.1101/gad.256603
Springer N (2010) Shaping a better rice plant. Nat Genet 42(6):475–476. https://doi.org/10.1038/ng0610-475
Steeves TA, Sussex IM (1989) Patterns in plant development. Cambridge University Press, Cambridge
Stirnberg P, Chatfield SP, Leyser HMO (1999) AXR1 acts after lateral bud formation to inhibit lateral bud growth in Arabidopsis. Plant Physiol 121(3):839–847. https://doi.org/10.1104/pp.121.3.839
Talbert PB, Adler HT, Parks DW, Comai L (1995) The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development 121(9):2723–2735. https://doi.org/10.1242/dev.121.9.2723
Tanaka W, Ohmori Y, Ushijima T, Matsusaka H, Matsushita T, Kumamaru T, Kawano S, Hirano H-Y (2015) Axillary meristem formation in rice requires the WUSCHEL Ortholog TILLERS ABSENT1. Plant Cell 27(4):1173–1184. https://doi.org/10.1105/tpc.15.00074
Tantikanjana T, Yong JWH, Letham DS, Griffith M, Hussain M, Ljung K, Sandberg G, Sundaresan V (2001) Control of axillary bud initiation and shoot architecture in Arabidopsis through the SUPERSHOOT gene. Genes Dev 15(12):1577–1588. https://doi.org/10.1101/gad.887301
Tian C, Zhang X, He J, Yu H, Wang Y, Shi B, Han Y, Wang G, Feng X, Zhang C, Wang J, Qi J, Yu R, Jiao Y (2014) An organ boundary‐enriched gene regulatory network uncovers regulatory hierarchies underlying axillary meristem initiation. Mol Syst Biol 10 (10):755–773. https://doi.org/10.15252/msb.20145470
Turnbull CGN, Booker JP, Leyser HMO (2002) Micrografting techniques for testing long-distance signalling in Arabidopsis. Plant J 32(2):255–262. https://doi.org/10.1046/j.1365-313x.2002.01419.x
Van De Velde K, Ruelens P, Geuten K, Rohde A, Van Der Straeten D (2017) Exploiting DELLA signaling in cereals. Trends Plant Sci 22(10):880–893. https://doi.org/10.1016/j.tplants.2017.07.010
Wang Y (2021) Stem cell basis for fractal patterns: axillary meristem initiation. Front Plant Sci:3075
Wang Y, Jiao Y (2018a) Auxin and above-ground meristems. J Exp Bot 69(2):147–154. https://doi.org/10.1093/jxb/erx299
Wang Y, Jiao Y (2018b) Axillary meristem initiation—a way to branch out. Curr Opin Plant Biol 41:61–66. https://doi.org/10.1016/j.pbi.2017.09.001
Wang Y, Li J (2008) Molecular basis of plant architecture. Annu Rev Plant Biol 59(1):253–279. https://doi.org/10.1146/annurev.arplant.59.032607.092902
Wang Y, Li J (2011) Branching in rice. Curr Opin Plant Biol 14(1):94–99. https://doi.org/10.1016/j.pbi.2010.11.002
Wang X-J, Reyes JL, Chua N-H, Gaasterland T (2004) Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol 5(9):1–15. https://doi.org/10.1186/gb-2004-5-9-r65
Wang Q, Kohlen W, Rossmann S, Vernoux T, Theres K (2014a) Auxin depletion from the leaf axil conditions competence for axillary meristem formation in Arabidopsis and tomato. Plant Cell 26(5):2068–2079. https://doi.org/10.1105/tpc.114.123059
Wang Y, Wang J, Shi B, Yu T, Qi J, Meyerowitz EM, Jiao Y (2014b) The stem cell niche in leaf axils is established by auxin and cytokinin in Arabidopsis. Plant Cell 26(5):2055–2067. https://doi.org/10.1105/tpc.114.123083
Wang J, Tian C, Zhang C, Shi B, Cao X, Zhang T-Q, Zhao Z, Wang J-W, Jiao Y (2017) Cytokinin signaling activates WUSCHEL expression during axillary meristem initiation. Plant Cell 29(6):1373–1387. https://doi.org/10.1105/tpc.16.00579
Wang B, Smith SM, Li J (2018) Genetic regulation of shoot architecture. Annu Rev Plant Biol 69:437–468. https://doi.org/10.1146/annurev-arplant-042817-040422
Wang Y, Huan Q, Li K, Qian W (2021) Single-cell transcriptome atlas of the leaf and root of rice seedlings. J Genet Genom 48(10):881–898. https://doi.org/10.1016/j.jgg.2021.06.001
Xin W, Wang Z, Liang Y, Wang Y, Hu Y (2017) Dynamic expression reveals a two-step patterning of WUS and CLV3 during axillary shoot meristem formation in Arabidopsis. J Plant Physiol 214:1–6. https://doi.org/10.1016/j.jplph.2017.03.017
Xing S, Sun Y, Li B, Li H, Zhao KT, Chen K, Gao C (2023) Anthocyanin-assisted Agrobacterium infiltration for the rapid evaluation of genome editing efficiencies across multiple plant species. Natl Sci Open 2(5):20220052. https://doi.org/10.1360/nso/20220052
Xu X, Crow M, Rice BR, Li F, Harris B, Liu L, Demesa-Arevalo E, Lu Z, Wang L, Fox N, Wang X, Drenkow J, Luo A, Char SN, Yang B, Sylvester AW, Gingeras TR, Schmitz RJ, Ware D, Lipka AE, Gillis J, Jackson D (2021) Single-cell RNA sequencing of developing maize ears facilitates functional analysis and trait candidate gene discovery. Dev Cell 56(4):557-568.e556. https://doi.org/10.1016/j.devcel.2020.12.015
Yamaguchi S (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59(1):225–251. https://doi.org/10.1146/annurev.arplant.59.032607.092804
Yan Y, Ding C, Zhang G, Hu J, Zhu L, Zeng D, Qian Q, Ren D (2023) Genetic and environmental control of rice tillering. Crop J:1287–1302.
Yang F, Wang Q, Schmitz G, Müller D, Theres K (2012) The bHLH protein ROX acts in concert with RAX1 and LAS to modulate axillary meristem formation in Arabidopsis. Plant J 71(1):61–70. https://doi.org/10.1111/j.1365-313x.2012.04970.x
Yang T, Jiao Y, Wang Y (2023) Stem cell basis of shoot branching. Plant Cell Physiol 64(3):291–296. https://doi.org/10.1093/pcp/pcac165
Yu L, Yao M, Mao L, Ma T, Nie Y, Ma H, Shao K, An H, Zhao J (2023) Rice DSP controls stigma, panicle and tiller primordium initiation. Plant Biotechnol J 21(11):2358–2373. https://doi.org/10.1111/pbi.14137
Yuan Y, Khourchi S, Li S, Du Y, Delaplace P (2023) Unlocking the multifaceted mechanisms of bud outgrowth: advances in understanding shoot branching. Plants 12(20):3628–3652. https://doi.org/10.3390/plants12203628
Zhang C, Fan L, Le BH, Ye P, Mo B, Chen X (2020a) Regulation of ARGONAUTE10 expression enables temporal and spatial precision in axillary meristem initiation in Arabidopsis. Dev Cell 55(5):603–616. https://doi.org/10.1016/j.devcel.2020.10.019
Zhang QQ, Wang JG, Wang LY, Wang JF, Wang Q, Yu P, Bai MY, Fan M (2020b) Gibberellin repression of axillary bud formation in Arabidopsis by modulation of DELLA-SPL9 complex activity. J Integr Plant Biol 62(4):421–432. https://doi.org/10.1111/jipb.12818
Zhang TQ, Chen Y, Wang JW (2021a) A single-cell analysis of the Arabidopsis vegetative shoot. Dev Cell 56(7):1056–1074. https://doi.org/10.1016/j.devcel.2021.02.021
Zhang Z, Sun X, Ma X, Xu B, Zhao Y, Ma Z, Li G, Khan NU, Pan Y, Liang Y (2021b) GNP6, a novel allele of MOC1, regulates panicle and tiller development in rice. Crop J 9(1):57–67. https://doi.org/10.1016/j.cj.2020.04.011
Zheng B, Chen X (2011) Dynamics of histone H3 lysine 27 trimethylation in plant development. Curr Opin Plant Biol 14(2):123–129. https://doi.org/10.1016/j.pbi.2011.01.001
Zhu H, Hu F, Wang R, Zhou X, Sze SH, Liou LW, Barefoot A, Dickman M, Zhang X (2011) Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell 145(2):242–256. https://doi.org/10.1016/j.cell.2011.03.024
Funding
This work was supported in part by the National Key Research and Development Program of China (Grant No. 2021YFD1200601-08).
Author information
Authors and Affiliations
Contributions
Yundong Yuan wrote the initial draft and revised the manuscript. Yan Fang Du and Pierre Dalaplace thoroughly reviewed the manuscript and provided insightful feedbacks.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no competing interests to declare that are relevant.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Communicated by Gerhard Leubner.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file 1 (MP4 13633 kb)
Supplementary file 2 (MP4 21716 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yuan, Y., Du, Y. & Delaplace, P. Unraveling the molecular mechanisms governing axillary meristem initiation in plants. Planta 259, 101 (2024). https://doi.org/10.1007/s00425-024-04370-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-024-04370-w