Abstract
Main conclusion
Distinct plastid types and ultrastructural changes are associated with differences in carotenoid pigment profiles in differently coloured carrots, and a variant of the OR gene, DcOR3Leu is vital for chromoplast biogenesis.
Abstract
Accumulation of different types and amounts of carotenoids in carrots impart different colours to their taproots. In this study, the carotenoid pigment profiles, morphology, and ultrastructure of plastids in 25 carrot varieties with orange, red, yellow, or white taproots were investigated by ultra-high performance liquid chromatography as well as light and transmission electron microscopy. α-/β-Carotene and lycopene were identified as colour-determining carotenoids in orange and red carrots, respectively. In contrast, lutein was identified as the colour-determining carotenoid in almost all tested yellow and white carrots. The latter contained only trace amounts of lutein as a unique detectable carotenoid. Striking differences in plastid types that coincided with distinct carotenoid profiles were observed among the differently coloured carrots. Microscopic analysis of the different carotenoid pigment-loaded plastids revealed abundant crystalloid chromoplasts in the orange and red carrots, whereas amyloplasts were dominant in most of the yellow and white carrots, except for the yellow carrot ‘Yellow Stone’, where yellow chromoplasts were observed. Plastoglobuli and crystal remnants, the carotenoid sequestering substructures, were identified in crystalloid chromoplasts. Crystal remnants were often associated with a characteristic undulated internal membrane in orange carrots or several undulated membranes in red carrots. No crystal remnants, but some plastoglobuli, were observed in the plastids of all tested yellow and white carrots. In addition, the presence of chromoplast in carrot taproots was found to be associated with DcOR3Leu, a natural variant of DcOR3, which was previously reported to be co-segregated with carotene content in carrots. Knocking out DcOR3Leu in the orange carrot ‘Kurodagosun’ depressed chromoplast biogenesis and led to the generation of yellow carrots. Our results support that DcOR3Leu is vital but insufficient for chromoplasts biogenesis in carrots, and add to the understanding of the formation of chromoplasts in carrots.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Carotenoids are a subgroup of isoprenoid pigments that play important roles in plants, animals, fungi, algae, and bacteria (Tapiero et al. 2004; Kim et al. 2021). In plants, carotenoids are common components of cell photosystems and serve as precursors for the biosynthesis of strigolactones and abscisic acids, which are essential for development and growth (Auldridge et al. 2006; Walter and Strack 2011; Al-Babili and Bouwmeester 2015). Plant-derived carotenoids, when they are consumed as a part of the diet, also affect human nutrition and health. For instance, dietary α-/β-carotene is converted into vitamin A for the visual system in the human body (Tanumihardjo and Arscott Sherry 2010), whereas lutein and zeaxanthin are important for visual performance and the prevention of chronic eye-related diseases (Koushan et al. 2013).
In plants, carotenoids are deposited in various plastids, including amyloplasts, etioplasts, chloroplasts, and chromoplasts, among which chromoplasts are the main organelles for carotenoid biosynthesis and storage (Lopez-Juez and Pyke 2005; Howitt and Pogson 2006; Jarvis and López-Juez 2013; Li et al. 2016). The considerable amounts of carotenoids stored in chromoplasts often impart the orange, red, and yellow colours to plants' fruits, flowers, and roots. Plant carotenoids are often deposited in different types of chromoplasts (Kim et al. 2010; Jeffery et al. 2012). For instance, red papaya and tomato fruit predominantly containing lycopene are characterized by chromoplasts of crystalloid types, whereas yellow papaya predominantly accumulates β-cryptoxanthin esters in round-shaped plastids (Schweiggert et al. 2011). In orange carrots cells, carotenoid accumulation mainly occurs in crystalloid chromoplasts (Frey-Wyssling and Schwegler 1965). The abundance of α-and β-carotene in orange carrots (Daucus carota L. var. sativa) imparts orange colour to their taproots and makes carrots an important source of provitamin A in the human diet (Maiani et al. 2010). Compared to orange carrots, non-orange carrots, including red, yellow, and white carrots, accumulate different types and amounts of carotenoids in their taproots, where lycopene and lutein impart red and yellow colours, respectively (Just et al. 2009; Matthieu et al. 2015). However, whether carotenoids from non-orange carrots are sequestered in crystalloid chromoplasts is unknown; furthermore, the ultrastructural differences in plastids among differently coloured carrots remain to be elucidated.
The Orange (OR) gene, encoding a DnaJ Cys-rich zinc finger motif-containing protein, plays an important role in carotenoid accumulation (Lu et al. 2006; Sun et al. 2019). Furthermore, OR could increase the protein levels of phytoene synthase (PSY), the rate-limiting enzyme in the carotenoid biosynthetic pathway, to enhance carotenoid biosynthesis in Arabidopsis (Arabidopsis thaliana) (Zhou et al. 2015). A natural insertion of a long-terminal repeat retrotransposon in OR and a ‘golden’ single-nucleotide polymorphism (SNP) causing a change from Arg108 to His108 in OR protein, designated as ORHis, was individually identified in cauliflower (Brassica oleracea var. botrytis L.) (Lu et al. 2006) and melon (Cucumis melo) (Tzuri et al. 2015). These two types of mutations in OR trigger chromoplast biogenesis, the storage sink for carotenoid accumulation, and dramatically increase carotene levels. Artificial ORHis also trigger chromoplast biogenesis and promote carotenoid overaccumulation in Arabidopsis and sweet potato [Ipomoea batatas (L.) Lam. cv. Yulmi] (Yuan et al. 2015; Kim et al. 2019).
In a previous study, three OR genes, DcOR1 (DCAR_020166), DcOR2 (DCAR_009463), and DcOR3 (DCAR_009172), were identified in the carrot genome (Wang et al. 2021). DcOR1 and DcOR2 are homologous to AtOR, whereas DcOR3 is homologous to AtOR-like. Although chromoplasts were abundant in orange carrot cells, the conserved ‘golden’ SNP substitution from Arg to His was not found within the three DcORs. DcOR3 is located in a QTL associated with the accumulation of several carotenoids (Ellison et al. 2018; Coe et al. 2021). The SNP C551 to T in the coding sequence of DcOR3 causes a mutation from Ser184 (designated as DcOR3Ser, also known as Orw) to Leu184 (designated as DcOR3Leu, also known as Orc) (Ellison et al. 2018). DcOR3Leu co-segregated with high carotene content in carrots, whereas DcOR3Ser was present in carrots that accumulated low levels of carotenes (Coe et al. 2021). However, whether DcOR3Leu is correlated with chromoplast biogenesis in carrots remains unknown. This study aimed to examine the relationship between distinct carotenogenesis and different plastidal structures in various coloured carrot taproots, and the correlation between the DcOR3 genotype and chromoplast biogenesis in differently coloured carrot taproots.
Materials and methods
Plant materials and growth conditions
Ten orange carrot cultivars (Kurodagosun, Interceptor, Mini NO. 2, Emperor 3179, Sanhongqicun, Xiaodingbacun, Hongguan 100, Hongyubahao, Fushiqicun, and Adelaide), five red carrot cultivars (Benhongjinshi, Dayumeirenzhi, Beta Fruit, Meiguihong, and Zihongjian), eight yellow carrot cultivars (Yellow Stone, Qitouhuang, Xiyanghuangsheng, Huanghuluobo, Huangbacun, Jingdingguan 100, Huangshanhu, and Longxiahuang), one white carrot cultivar (White Satin), and white wild carrot variety (Queen Anne’s Lace, Daucus carota L. var. carota) were selected for this study. The carrots were sown and grown at the experimental station of Nanjing Agricultural University. The carrots were harvested 120 d after germination and washed in cold tap water before further analysis.
Carotenoid pigments’ measurement
Fifteen taproots from each carrot cultivar or variety were selected for carotenoid pigment analysis. Fresh carrot taproots were cut into small cubes, pulverized in liquid nitrogen, and lyophilised under vacuum. Dried carrot powder was used for carotenoid pigment extraction using previously described method (Wang et al. 2020). The carotenoid pigments were separated and identified on a Hedera ODS-2 C18 analytical column (250 mm × 4.6 mm; 5 μm nominal particle size; Shimadzu, Tokyo, Japan) using a Waters ACQUITY UPLC H-class system (Waters, Milford) according to a previously described method (Tomasz et al. 2018).
Isolation of protoplasts and light microscopy
Protoplasts were isolated from fresh carrot taproots using a protocol developed by Wan (Wan et al. 1987). In brief, carrot taproots were sliced and digested in an enzyme solution for 12 h in the dark with gentle shaking. The protoplasts were collected, loaded onto a concave glass slide, and observed under an Olympus BX-53 microscope (Olympus, Tokyo, Japan).
Transmission electron microscopy
Phloem tissues of fresh carrot taproots were cut into small sections (ca. 0.5 × 1.0 × 2.0 mm3) and fixed overnight at 4 °C in 2.5% glutaraldehyde (0.1 M phosphate buffer, pH 7.0). The samples were subsequently processed according to a previously described method (Wen et al. 2020) and observed by transmission electron microscopy (TEM) using a Hitachi JEM-1230 (Hitachi Ltd., Tokyo, Japan).
Genomic DNA and total RNA extractions and gene cloning
Genomic DNA or total RNA was isolated from the carrot taproots using a DNA secure Plant Kit or an RNA simple Total RNA Kit (PUDI, Shanghai, China), respectively, according to the manufacturer’s instructions. First-strand cDNA synthesis was conducted using the HiScript II Q RT SuperMix for qPCR kit (Vazyme Biotech Co. Ltd., Nanjing, China), following the manufacturer’s protocol. DcOR3 was amplified using PrimeSTAR HS DNA polymerase (Takara, Dalian, China) and a pair of primers, 5′-CTGTCCACCCTCTCTATCTTTCTTG-3′ and 5′- GCAGTAGCACTATCTTCATCCATTG -3′. The PCR products were sequenced using Sanger sequencing (GENERAL BIOL, Chuzhou, China).
Generation of DcOR3-knockout mutant carrots
The target sites of DcOR3 were identified using the CRISPR-GE online tool (http://skl.scau.edu.cn/targetdesign/) for CRISPR/Cas9-mediated genome editing. Two of the output target sites in the first exon region were selected to design the sgRNA expression cassettes, which were individually driven by AtU3d and AtU6-29 (Supplementary Materials). Two sgRNA expression cassettes were assembled into the pYLCRISPR/Cas9Pubi-H vector to generate the pYLCRISPR/Cas9Pubi-H-DcOR3 construct (Fig. S1). The construct was then transformed into orange carrot ‘Kurodagosun’ to produce a DcOR3-knockout mutant carrot by Agrobacterium-mediated method (Xu et al. 2019a). As previously described, transgenic carrots were grown in artificial climatic chambers (Xu et al. 2019b). To validate the mutation of DcOR3 in transgenic carrot plants, genomic DNA was extracted from taproots to amplify the DNA fragment of the target site using a pair of primers, 5′-CTTGAAGATTCATGAGAAGCAA-3′ and 5′-CAGCACTTCACATCAAGAACTA-3′. The PCR products were directly sequenced to determine the mutations within the target sites.
Gene expression analyses
Quantitative RT-PCR (qRT-PCR) assays were performed to determine the transcript levels of DcOR3Leu in the wild-type and CRISPR knockout carrots using a previously described method (Wang et al. 2021). The relative transcript abundance was normalized to DcActin1 and calculated using the 2−ΔΔCt method (Schmittgen and Livak 2008). The primers used for qRT-PCR assays of DcOR3 and DcActin1 were the same as previously described (Wang et al. 2021).
Data analysis
Carotenoid pigment measurements and qRT-PCR assays for each carrot were performed in triplicate. The results are expressed as mean ± standard deviation (SD).
Accession numbers
Sequence data from this article can be found in GenBank databases under the following accession numbers: DcOR1 (XM_017399272), DcOR2 (MW116211), DcOR3 (XM_017387585), DcActin1 (XM_017371101), and pYLCRISPR/Cas9Pubi-H vector (KR029109.1).
Results
Profiles of carotenoid pigments in differently coloured carrots
A total of 24 different carrot cultivars and one wild carrot were chosen for this study (Fig. 1a). The 25 carrot samples were divided into four colour groups according to the view of skin and transverse section of carrot taproots: the orange carrot group included ‘Kurodagosun’ (KRD), ‘Interceptor’ (INTC), ‘Mini NO. 2’ (MN2), ‘Emperor 3179’ (EP3179), ‘Sanhongqicun’ (SHQC), ‘Xiaodingbacun’ (XDBC), ‘Hongguan 100’ (HG100), ‘Hongyubahao’ (HYBH), ‘Fushiqicun’ (FSQC), and ‘Adelaide’ (ADL); the red carrot group included ‘Benhongjinshi’ (BHJS), ‘Dayumeirenzhi’ (DYMRZ), ‘Beta Fruit’ (BF), ‘Meiguihong’ (MGH), and ‘Zihongjian’ (ZHJ); the yellow carrot group included ‘Yellow Stone’ (YST), ‘Qitouhuang’ (QTH), ‘Xiyanghuangsheng’ (XYHS), ‘Huanghuluobo’ (HHLB), ‘Huangbacun’ (HBC), ‘Jingdingguan 100’ (JDG100), ‘Huangshanhu’ (HSH), and ‘Longxiahuang’ (LXH); the white carrot group included ‘White Satin’ (WST) and wild carrot (Queen Anne’s Lace, QAL).
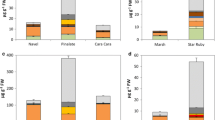
Carotenoid pigment profiles in differently coloured carrots at the 120-day-old stage. a Photographs of outer and cross sections of 25 carrot varieties with orange (top-left), red (top-right), yellow (bottom-left), and white (bottom-right) taproots. Cultivar/variety abbreviations: KRD, Kurodagosun; INTC, Interceptor; MN2, Mini NO. 2; EP3179, Emperor 3179; SHQC, Sanhongqicun; XDBC, Xiaodingbacun; HG100, Hongguan 100; HYBH, Hongyubahao; FSQC, Fushiqicun; ADL, Adelaide; BHJS, Benhongjinshi; DYMRZ, Dayumeirenzhi; BF, Beta Fruit; MGH, Meiguihong; ZHJ, Zihongjian; YST, Yellow Stone; QTH, Qitouhuang; XYHS, Xiyanghuangsheng; HHLB, Huanghuluobo; HBC, Huangbacun; JDG100, Jingdingguan 100; HSH, Huangshanhu; LXH, Longxiahuang; WST, White Satin; QAL, Queen Anne’s Lace. b Carotenoid pigments’ content in taproots of different orange-, red-, yellow-, and white-coloured carrots. Data are means of three replicates with error bars representing ± SD
Carotenoid pigments in 120-day-old taproots of 25 carrot cultivars/varieties were examined. The types and amounts of carotenoid pigments varied significantly between the four carrot groups (Fig. 1b). Four kinds of carotenoids, α-/β-carotene, lycopene, and lutein, which are responsible for carrot colour, were identified within these differently coloured carrots. Lutein was identified in all the orange, red, yellow, and white carrots. Lycopene was abundant in all the five red carrot cultivars, but was undetectable in all the orange, yellow, and white carrots. α-Carotene was abundant in all the orange and red carrot cultivars, except for the red carrot cultivar BHJS, where α-carotene was undetectable. α-Carotene was undetectable in all the yellow and white carrots. β-Carotene was abundant in all the orange and red carrot cultivars, but was scarce in the yellow carrot cultivars YST and QTH and almost undetectable in the other yellow and white carrot cultivars/varieties.
Concerning the amount of carotenoid pigments, the orange and red carrots showed much higher carotenoid pigments than the yellow and white carrots. In the ten orange carrot cultivars, α- and β-carotene were the most abundant carotenoids, followed by lutein. All the orange carrot cultivars showed higher α-carotene content than the other coloured carrot cultivars/varieties. In addition, most orange carrots showed a relatively higher β-carotene content than red carrots, but the content was much higher than that in yellow and white carrots. Among the five red carrot cultivars, β-carotene and lycopene were the most abundant carotenoids, followed by α-carotene and lutein; α-carotene was not identified in the red carrot BHJS. Compared with the orange and red carrot cultivars, all the yellow carrot cultivars showed much lower carotenoid levels and mostly accumulated lutein as the primary carotenoid. In the white carrot varieties, extremely low levels of lutein were identified as the only detectable carotenoids.
Structural diversity of plastids in differently coloured carrots
Plastids act as storage compartments for carotenoid accumulation, imparting red, orange, or yellow hue. Light microscopy and TEM were used to examine carotenoid sequestering structures in differently coloured carrots at the 120-day-old stage to determine whether the differences in plastid types were associated with different carotenoids storage. Striking differences in plastids were observed among the differently coloured carrots (Fig. 2). Visible orange or red crystalloid plastids were frequently observed in the cells of all the orange and red carrots (Fig. 2a, b), whereas only grey globe-shaped plastids were observed in all the yellow and white carrots (Fig. 2c, d), except for the yellow carrot YST, where yellow plastids were found (Fig. 2c).
The orange- or red-coloured plastids in orange and red carrot cells were confirmed to be chromoplasts by TEM analysis (Fig. 3). Two types of chromoplasts, crystalloid and membranous, were found within the cells of orange carrots (Fig. 3a, b). Crystalloid chromoplasts were abundant in all the orange carrots and often contained numerous plastoglobuli, and crystal remnants associated with a characteristic undulated internal membrane (Fig. 3a). In addition, starch granules remained in some crystalloid chromoplasts of the orange carrots KRD, INTC, EP3179, XDBC, and FSQC (Fig. S2a). However, membranous chromoplasts containing membranous structures and plastoglobuli were only observed in the orange carrots MN2, SHQC, and ADL (Fig. 3b). Membranous chromoplasts are less common than crystalloid chromoplasts in orange carrots. No crystal remnants were observed within membranous chromoplasts. Only crystalloid chromoplasts were identified within the cells of red carrots (Fig. 3c). Their crystalloid chromoplasts contained numerous plastoglobuli and crystal remnants. The crystal remnants were often surrounded by one or more undulated membranes and showed different shapes when compared to that of the orange carrot cells. Small starch granules were also found within some crystalloid chromoplasts of BHJS, BF, and ZHJ (Fig. S2b).
Electron micrographs of chromoplasts in orange and red carrots. a Ultrastructure of crystalloid chromoplasts in ten orange carrot cultivars. b Ultrastructure of membranous chromoplasts in orange carrots MN2, SHQC, and ADL. c Ultrastructure of crystalloid chromoplasts in five red carrot cultivars. cr, carotenoid crystal; pg, plastoglobule; cw, cell wall; white arrow, undulated internal membrane
TEM analysis revealed that the yellow plastids of yellow carrot YST were filled with spindle-like plastoglobuli, which comprised plastoglobuli and tubular elements (Fig. 4a). Thus, the yellow plastids in YST were confirmed to be chromoplasts. No crystal remnants, but small starch granules, were found in the chromoplasts of YST (Fig. S2c). Grey globular plastids were identified as amyloplasts filled with large starch granules in the yellow carrots QTH, XYHS, and HSH, and the white carrots WST and QAL (Fig. 4a, b). Starch granules were also found in the globular plastids of the yellow carrot cultivars HHLB, HBC, JDG100, and LXH, but were relatively smaller in size (Fig. 4a). Therefore, we identified the globular plastids of yellow carrots HHLB, HBC, JDG100, and LXH as amyloplasts, as they displayed a grey colour and contained starch granules.
Electron micrographs of plastids in yellow and white carrots. a Ultrastructure of chromoplasts in yellow carrot YST and amyloplasts in other seven yellow carrot cultivars. b Ultrastructure of amyloplasts in white carrots WST and QAL. pg, plastoglobule; cw, cell wall; s, starch; white arrow, tubular element
Relationship between DcOR3 genotype and chromoplast presence
DcOR3 was cloned from all 25 carrot cultivars/varieties for deduced amino acid sequences analysis to test whether DcOR3Leu is correlated with chromoplast biogenesis in differently coloured carrots (Fig. 5). Homozygous DcOR3Leu was found in all the orange and red carrot cultivars, and in the yellow carrot YST, which is known to contain chromoplasts. However, homozygous DcOR3Leu alleles were present in carrots that did not contain chromoplasts in their cells, including XYHS, HBC, JDG100, HSH, and WST. All other non-chromoplast-containing carrots were homozygous for DcOR3Ser. Alignment analysis identified another SNP in DcOR3 of some orange carrot cultivars that caused an Asp to Tyr change (D309Y) (Fig. 5). Nevertheless, this variation appears not to be associated with chromoplast biogenesis or carotenoid accumulation in the different carrot cultivars. Additionally, the conserved ‘golden’ SNP substitution from Arg to His was not found within the DcOR3 from all 25 carrot cultivars/varieties (Fig. 5).
Alignment analysis of the deduced amino acid sequences of DcOR3 from 16 chromoplast-containing carrot cultivars and nine non-chromoplast-containing carrot cultivars/varieties. Identical sequences are shaded in grey. The amino acid sequences of DcOR3 from 16 chromoplast-containing carrot cultivars and nine non-chromoplast-containing carrot cultivars/varieties are enclosed in a red box and a black box, respectively. The two single amino acid changes at positions 184 (Ser-to-Leu) and 309 (Asp to Tyr) of DcOR3 are individually indicated with a red and a black asterisk (✱) below the alignment. The conserved ‘golden’ SNP locus at position 95 (Arg) is indicated with a solid orange circle below the alignment
Knocking out DcOR3 Leu in orange carrot KRD
The CRISPR/Cas9-based genome editing system was used to knock out DcOR3Leu in orange carrot KRD. PCR amplification and sequencing analyses revealed that biallelic mutations occurred at target site 2 (targeted by AtU6-29-driven sgRNA) of one carrot line but not at target site 1 (Fig. 6a, b). The plants of this carrot line carried an allele with a 1-bp deletion and another allele with a 1-bp insertion at target site 2, which could lead to the generation of a frameshift mutant within DcOR3Leu (DcOR3Leu-knockout). Regarding the phenotype, the taproots of DcOR3Leu-knockout carrots presented a yellow phenotype (Fig. 6c). Light microscopy revealed that chromoplast biogenesis was suppressed in the taproot cells of DcOR3Leu-knockout carrots (Fig. 6d). QRT-PCR analysis revealed that the transcript levels of DcOR3Leu in DcOR3Leu-knockout KRD carrots decreased less than twofold compared with that in the wild-type KRD carrots (Fig. 6e).
Knocking out DcOR3Leu in orange carrot KRD by CRISPR/Cas9. a Schematic representation of the two sgRNAs targeting DcOR3Leu, conserved ‘golden’ SNP locus in exon 2, and Leu184 locus in exon 5. The black boxes and broken lines indicate individual exons and introns, respectively. b Representative target site sequences of mutant alleles identified in DcOR3Leu-knockout KRD plants. c Wild-type and DcOR3Leu-knockout KRD carrots. d Representative protoplasts isolated from taproots of DcOR3Leu-knockout carrots. e Relative transcript levels of DcOR3Leu in taproots of wild-type and DcOR3Leu-knockout KRD carrots. Data are means of three replicates with error bars representing ± SD
Discussion
In higher plants, carotenoids are synthesized and stored exclusively in different types of plastids, including chromoplasts, chloroplasts, amyloplasts, and etioplasts (Sun et al. 2018). These different types of plastids exhibit dramatic differences in their capacity to synthesize and accumulate carotenoids, leading to diversity in carotenoid content and composition among various plant species (Sun et al. 2018). Chromoplasts provide a high capacity for carotenoid biosynthesis and storage, as well as bright yellow, orange, or red hues to various plants' flowers, fruits, and roots. In carrots, the orange colouration of taproots resulted from the massive accumulation of carotenoids (Ma et al. 2017; Perrin et al. 2017; Wang et al. 2020). The pigments from orange carrots predominantly comprised α-/β-carotene, which is mainly deposited in crystalloid chromoplasts (Kim et al. 2010; Tomasz et al. 2018). α-/β-Carotene is an important dietary source of provitamin A, and elucidating its storage in chromoplasts is of great interest. Significant differences in carotenoid composition and content were noted among orange, red, yellow and white carrots (Clotault et al. 2008; Ma et al. 2017; Wang et al. 2020). However, the comparative study of carotenoid profile and plastid substructure between these differently coloured carrots have not been conducted. Therefore, the deposition of carotenoid pigments in plastids of these non-orange carrots merits further investigation.
The present study determined carotenoid pigment profiles and plastidal ultrastructures of orange, red, yellow, and white carrots. In agreement with previous studies (Ma et al. 2017), carotenoid pigments from all the orange carrots comprised large amounts of α-/β-carotene and relatively lower amounts of lutein, the former two of which were predominantly responsible for the orange colour. In orange carrots, carotenoids are mainly deposited in crystalloid chromoplasts, and occasionally in membranous chromoplasts. High amounts of carotenes sequestered in chromoplasts can gradually aggregate and finally be deposited in a crystalline form as crystal structures, leading to distortion of the shape of the chromoplast to an elongated crystalline shape (Schweiggert et al. 2011). This was confirmed in the present study, where typical elongated-shaped carotene crystal remnants appeared in the crystalloid chromoplasts but not in the membranous chromoplasts of the orange carrot cells. Crystal remnants were also found in the crystalloid chromoplasts of all the red carrot cells but showed differences in the shape and number of attached undulated membranes compared with those of orange carrots. Similar structures were previously linked to lycopene accumulation in red Citrus fruits and tomatoes (Schweiggert et al. 2011; Zeng et al. 2015). These differences were caused by exclusive lycopene accumulation and crystallization in the chromoplasts of the red carrots. Plastoglobuli, the site for carotenoid storage, were observed within the chromoplasts of orange and red carrot cells. In addition, starch granules were observed in the chromoplasts of some orange and red carrots, supporting previous findings that chromoplasts were converted from amyloplasts in carrots (Kim et al. 2010).
In contrast to the abundant accumulation of carotenes in orange or red carrots, yellow or white carrots mainly accumulate lutein as the primary carotenoid pigment, which is primarily stored in amyloplasts and rarely in globular chromoplasts. Almost all the tested yellow or white carrots contained amyloplasts in their taproot cells, except for the yellow carrot YST, which had chromoplasts in their taproot cells. Globular plastoglobuli were identified in all the yellow and white carrots, while spindle-like plastoglobuli were only identified in the chromoplasts of YST. The spindle-like plastoglobuli were similar to those described for papaya, comprising globular plastoglobuli and attached tubular elements (Schweiggert et al. 2011). The plastoglobuli from yellow carrots were bigger than those from white carrots, partly explaining the higher amounts of carotenoids in yellow than in white carrots.
OR is involved in regulation of carotenoid biosynthesis in many plants. The ‘golden’ SNP, which increases carotene levels and trigger chromoplast biogenesis, has been identified in some plant species (Tzuri et al. 2015; Zhou et al. 2015; Kim et al. 2021). This conserved ‘golden’ SNP was not found in any of the three OR genes of carrots (Wang et al. 2021). However, a Ser-to-Leu amino acid substitution (Ser184Leu) in the DcOR3 protein which previously had been described as influencing carotenoid accumulation was identified in domesticated carrots (Ellison et al. 2018). Previous study has also revealed that DcOR3Leu co-segregated with high carotenoids content and may contribute to the formation of chromoplasts in carrot (Coe et al. 2021). However, the functionality of DcOR3Leu on chromoplasts presence was not verified. In the present study, homozygous DcOR3ser appeared fixed in the non-chromoplast-containing carrot cultivars, whereas all the chromoplast-containing carrot cultivars were fixed with homozygous DcOR3Leu. Knocking out DcOR3Leu in orange carrot KRD suppressed chromoplast biogenesis and led to the generation of yellow carrots. These results suggest that DcOR3Leu is essential for chromoplast biogenesis in carrots. However, homozygous DcOR3Leu was also present in non-chromoplast-containing carrot cultivars, implying that DcOR3Leu is not the only genetic factor responsible for chromoplast biogenesis in carrot taproots. Future work will focus on verifying the function and molecular mechanism of DcOR3Leu and DcOR3Ser in chromoplast biogenesis and identifying other genetic factors controlling chromoplast biogenesis in carrots.
Conclusion
This study showed that the differences in carotenoid pigment profiles were related to the distinct plastid types and ultrastructures of carrots. Overall, orange or red carrots mainly accumulated substantial amounts of carotenes or lycopene that formed crystals in crystalloid chromoplasts. The yellow and white carrots mainly accumulated lutein stored in plastoglobuli of amyloplasts in most cases or in globular chromoplasts in rare cases. These results also support the previous hypothesis that chromoplasts differentiated from amyloplasts. We also determined that allelic variation at DcOR3, generating DcOR3Leu, was associated with chromoplast biogenesis in carrot taproots. Knocking out DcOR3Leu in orange carrot KRD suppresses chromoplast biogenesis. The finds of this study support that DcOR3Leu is vital, but insufficient, for chromoplast biogenesis in carrots. This study provides further understanding of chromoplast formation in carrots.
Author contribution statement
ZSX and YMZ initiated and designed the research. YMZ, RHW, LW, YHW, and HL performed the experiments. ZSX and YMZ analyzed the data. ASX contributed reagents/materials/analysis tools. ZSX and YMZ wrote the manuscript. ZSX and YMZ revised the manuscript.
Data availability
All data generated or analyzed during this study are included in this published article and in its online supplemental material.
References
Al-Babili S, Bouwmeester HJ (2015) Strigolactones, a novel carotenoid-derived plant hormone. Annu Rev Plant Biol 66:161–186
Auldridge ME, Mccarty DR, Klee HJ (2006) Plant carotenoid cleavage oxygenases and their apocarotenoid products. Curr Opin Plant Biol 9:315–321
Clotault J, Peltier D, Berruyer R, Thomas M, Briard M, Geoffriau E (2008) Expression of carotenoid biosynthesis genes during carrot root development. J Exp Bot 59:3563–3573
Coe KM, Ellison S, Senalik D, Dawson J, Simon P (2021) The influence of the Or and carotene hydroxylase genes on carotenoid accumulation in orange carrots [Daucus carota (L.)]. Theor Appl Genet 134:3351–3362
Ellison SL, Luby CH, Corak KE, Coe KM, Senalik D, Iorizzo M, Goldman IL, Simon PW, Dawson JC (2018) Carotenoid presence is associated with the Or gene in domesticated carrot. Genetics 210:1497–1508
Frey-Wyssling A, Schwegler F (1965) Ultrastructure of the chromoplasts in the carrot root. J Ultrastruct Res 13:543–559
Howitt CA, Pogson BJ (2006) Carotenoid accumulation and function in seeds and non-green tissues. Plant Cell Environ 29:435–445
Jarvis P, López-Juez E (2013) Biogenesis and homeostasis of chloroplasts and other plastids. Nat Rev Mol Cell Biol 14:787–802
Jeffery J, Holzenburg A, King S (2012) Physical barriers to carotenoid bioaccessibility. ultrastructure survey of chromoplast and cell wall morphology in nine carotenoid-containing fruits and vegetables. J Sci Food Agric 92:2594–2602
Just BJ, Santos C, Yandell BS, Simon PW (2009) Major QTL for carrot color are positionally associated with carotenoid biosynthetic genes and interact epistatically in a domesticated × wild carrot cross. Theor Appl Genet 119:1155–1169
Kim JE, Rensing KH, Douglas CJ, Cheng KM (2010) Chromoplasts ultrastructure and estimated carotene content in root secondary phloem of different carrot varieties. Planta 231:549–558
Kim SE, Kim HS, Wang Z, Ke QB, Lee CJ, Park SU, Lim YH, Park WS, Ahn MJ, Kwak SS (2019) A single amino acid change at position 96 (Arg to His) of the sweetpotato Orange protein leads to carotenoid overaccumulation. Plant Cell Rep 38:1393–1402
Kim SE, Lee CJ, Park SU, Lim YH, Kim HS (2021) Overexpression of the golden SNP-carrying Orange gene enhances carotenoid accumulation and heat stress tolerance in sweetpotato plants. Antioxidants 10:51
Koushan K, Rusovici R, Li W, Ferguson LR, Chalam KV (2013) The role of lutein in eye-related disease. Nutrients 5:1823–1839
Li L, Yuan H, Zeng YL, Xu Q (2016) Plastids and carotenoid accumulation. Subcell Biochem 79:273–293
Lopez-Juez E, Pyke KA (2005) Plastids unleashed: their development and their integration in plant development. Int J Dev Biol 49:557–577
Lu S, Eck JV, Zhou X, Lopez AB, Kupper H (2006) The cauliflower Or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of carotene accumulation. Plant Cell 18:3594–3605
Ma J, Xu ZS, Tan GF, Wang F, Xiong AS (2017) Distinct transcription profile of genes involved in carotenoid biosynthesis among six different color carrot (Daucus carota L.) cultivars. Acta Bioch Bioph Sin 49:817–826
Maiani G, Castón M, Catasta G, Toti E, Schlemmer U (2010) Carotenoids: Actual knowledge on food sources, intakes, stability and bioavailability and their protective role in humans. Mol Nutr Food Res 53:S194–S218
Matthieu J, Séverine G, Cécile D, Mohamed M, Sébastien H, Anita S, Latifa H, Mathilde B, Didier P, Emmanuel G (2015) Carotenoid content and root color of cultivated carrot: a candidate-gene association study using an original broad unstructured population. PLoS ONE 10:e0116674
Perrin F, Hartmann L, Dubois-Laurent C, Welsch R, Huet S, Hamama L, Briard M, Peltier D, Gagne S, Geoffriau E (2017) Carotenoid gene expression explains the difference of carotenoid accumulation in carrot root tissues. Planta 245:737–747
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3(6):1101–1108
Schweiggert RM, Steingass CB, Heller A, Esquivel P, Carle R (2011) Characterization of chromoplasts and carotenoids of red- and yellow-fleshed papaya (Carica papaya L.). Planta 234:1031–1044
Sun TH, Yuan H, Cao HB, Yazdani M, Tadmor Y, Li L (2018) Carotenoid metabolism in plants: The role of plastids. Mol Plant 11:58–74
Sun TH, Zhou F, Huang XQ, Chen WC, Kong MJ, Zhou CF, Zhuang Z, Li L, Lu S (2019) ORANGE represses chloroplast biogenesis in etiolated Arabidopsis cotyledons via interaction with TCP14. Plant Cell 31:2996–3014
Tanumihardjo SA, Arscott Sherry A (2010) Carrots of many colors provide basic nutrition and bioavailable phytochemicals acting as a functional food. Compr Rev Food Sci Food Saf 9:223–239
Tapiero H, Townsend DM, Tew KD (2004) The role of carotenoids in the prevention of human pathologies. Biomed Pharmacother 58:100–110
Tomasz O, Magdalena KC, Anna MH, Maciej Z, Danuta S, Ewa K, Aleksandra B, Jan S, Rafal B (2018) Unique chromoplast organisation and carotenoid gene expression in carotenoid-rich carrot callus. Planta 248:1455–1471
Tzuri G, Zhou X, Chayut N, Yuan H, Tadmor Y (2015) A “golden” SNP in CmOr governs fruit flesh color of melon (Cucumis melo). Plant J 82:267–279
Walter MH, Strack D (2011) Carotenoids and their cleavage products: Biosynthesis and functions. Nat Prod Rep 28:663–692
Wan XS, Wang FD, Song YG (1987) Culture and plant regeneration from carrot root protoplasts. Plant Physiol Commun 6:30–33
Wang YH, Li T, Zhang RR, Khadr A, Tian YS, Xu ZS, Xiong AS (2020) Transcript profiling of genes involved in carotenoid biosynthesis among three carrot cultivars with various taproot colors. Protoplasma 257:949–963
Wang YG, Xiong AS, Xu ZS (2021) Cloning and expression analysis of three Orange genes in Kurodagosun carrot. J Nuclear Agri Sci 35:2482–2492
Wen X, Heller A, Wang KL, Han QY, Ni YY, Carle R, Schweiggert R (2020) Carotenogenesis and chromoplast development during ripening of yellow, orange and red colored Physalis fruit. Planta 251:1–14
Xu ZS, Feng K, Xiong AS (2019a) CRISPR/Cas9-mediated multiply targeted mutagenesis in orange and purple carrot plants. Mol Biotechnol 61:191–199
Xu ZS, Yang QQ, Feng K, Xiong AS (2019b) Changing carrot color: Insertions in DcMYB7 alter the regulation of anthocyanin biosynthesis and modification. Plant Physiol 181:195–207
Yuan H, Owsiany K, Sheeja T, Zhou XJ, Rodriguez C, Li YX, Chayut N, Yang Y, Welsch R, Thannhauser T (2015) A single amino acid substitution in an ORANGE protein promotes carotenoid overaccumulation in Arabidopsis. Plant Physiol 169:421–431
Zeng YL, Du JB, Wang L, Pan ZY, Xu Q, Xiao SY, Deng XX (2015) A comprehensive analysis of chromoplast differentiation reveals complex protein changes associated with plastoglobule biogenesis and remodeling of protein systems in sweet orange flesh. Plant Physiol 168:1648–1665
Zhou XJ, Welsch R, Yang Y, Álvarez D, Riediger M, Yuan H, Fish T, Liu JP, Thannhauser TW, Li L (2015) Arabidopsis OR proteins are the major posttranscriptional regulators of phytoene synthase in controlling carotenoid biosynthesis. Proc Natl Acad Sci U S A 112:3558–3563
Acknowledgements
The research was supported by Jiangsu Agricultural Science and Technology Independent Innovation Fund [CX(21)3029] and Priority Academic Program Development of Jiangsu Higher Education Institutions (Grant No. PAPD).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Anastasios Melis.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, YM., Wu, RH., Wang, L. et al. Plastid diversity and chromoplast biogenesis in differently coloured carrots: role of the DcOR3Leu gene. Planta 256, 104 (2022). https://doi.org/10.1007/s00425-022-04016-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-022-04016-9