Abstract
Main conclusion
NO enhances the resistance of tomato seedlings to salt stress through protein S-nitrosylation and transcriptional regulation, which involves the regulation of MAPK signaling and carbohydrate metabolism.
Abstract
Nitric oxide (NO) regulates various physiological and biochemical processes and stress responses in plants. We found that S-nitrosoglutathione (GSNO) treatment significantly promoted the growth of tomato seedling under NaCl stress, indicating that NO plays a positive role in salt stress resistance. Moreover, GSNO pretreatment resulted in an increase of endogenous NO level, S-nitrosothiol (SNO) content, S-nitrosoglutathione reductase (GSNOR) activity and GSNOR expression under salt stress, implicating that S-nitrosylation might be involved in NO-alleviating salt stress. To further explore whether S-nitrosylation is a key molecular mechanism of NO-alleviating salt stress, the biotin-switch technique and liquid chromatography/mass spectrometry/mass spectrometry (LC–MS/MS) were conducted. A total of 1054 putative S-nitrosylated proteins have been identified, which were mainly enriched in chloroplast, cytoplasm and mitochondrion. Among them, 15 and 22 S-nitrosylated proteins were involved in mitogen-activated protein kinase (MAPK) signal transduction and carbohydrate metabolism, respectively. In MAPK signaling, various S-nitrosylated proteins, SAM1, SAM3, SAM, PP2C and SnRK, were down-regulated and MAPK, MAPKK and MAPKK5 were up-regulated at the transcriptional level by GSNO treatment under salt stress compared to NaCl treatment alone. The GSNO pretreatment could reduce ethylene production and ABA content under NaCl stress. In addition, the activities of enzyme identified in carbohydrate metabolism, their expression at the transcriptional level and the metabolite content were up-regulated by GSNO supplication under salt stress, resulting in the activation of glycolysis and tricarboxylic acid cycle (TCA) cycles. Thus, these results demonstrated that NO might beneficially regulate MAPK signaling at transcriptional levels and activate carbohydrate metabolism at the post-translational and transcriptional level, protecting seedlings from energy deficiency and salinity, thereby alleviating salt stress-induced damage in tomato seedlings. It provides initial insights into the regulatory mechanisms of NO in response to salt stress.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Salt stress is one of the major environmental stressors that remarkably inhibits crop productivity in many regions worldwide. Salinity destroys the processes of plant growth and development by decreasing the ability of plants to uptake water and nutrients and by enhancing the cytotoxicity due to excessive uptake of the sodium and chloride (Isayenkov et al. 2019). In addition, plant physiological physiognomies are extensively susceptible to high salt toxicity. High salt prevents photosynthetic activity and respiration, decreases enzyme activities, protein and nucleic acid synthesis and organic solute accumulation, and disturbs ion homeostasis, osmotic and hormonal balance, which impairs the seed germination, seedling growth, plant life and crop productivity (Hussain et al. 2017). Furthermore, salinity can promote the generation of reactive oxygen species, such as hydrogen peroxide, superoxide anion radicals and hydroxyl free radicals, thus resulting in oxidative stress and lipid peroxidation in plants. Salinity can also affect endogenous hormone levels and lead to metabolism disorders, thereby affecting plant growth and development (Hussain et al. 2017). Therefore, enhancing the resistance of crop plants to salinity has currently been an urgent requirement.
Gaseous signaling molecules play vitally important roles in various physiological processes of plants (Niu et al. 2019; Wei et al. 2021). Free radical nitric oxide (NO), a small ubiquitous signaling molecule, is produced via non-enzymatic and enzymatic pathways in plants. As a key physiological regulator, NO can be involved in the regulation of various plant developmental processes, including seed germination, seedling development (Khan et al. 2020), adventitious root development, stomatal closure, photomorphogenesis and postharvest senescence (Niu et al. 2019). In addition, NO interacts with hormone molecules to modulate plant development processes and respond to abiotic stresses (Wei et al. 2020). The responses of NO in various abiotic stresses have been indicated in multiple studies (Sehar et al. 2019; Wei et al. 2020). Therefore, as a multifunctional signaling molecule, the roles of NO in plant growth and abiotic stress responses may be a great topic of interest.
Accumulating evidences indicated that NO could also exert its biological functions through NO-dependent post-translational modification, protein S-nitrosylation (Wei et al. 2020). NO groups covalently link cysteine residues of target proteins to form S-nitrosothiols (SNOs) (Feng et al. 2013a). At present, S-nitrosylation has emerged as an important mechanism involved in regulating plant growth, development and stress stimulus.
Mitogen-activated protein kinase (MAPK) cascades are recognized as major signal transduction modules in eukaryotes, which include three protein kinases and mediate sequential phosphorylation reactions, as follows: a MAPKKK phosphorylates and activates a MAPKK, which, in turn, activates a MAPK by phosphorylation. Recent studies also pointed out that there was a complex crosstalk between the MAPK cascades and NO signal transduction. Asai et al. (2008) indicated that MAPK signaling regulated NO bursts in Nicotiana benthamiana. Zhang et al. (2007) found that abscisic acid (ABA) and hydrogen peroxide could induce NO production in corn leaves and then activate a 46 kDa MAPK. Subsequently, data suggested that MAPK signaling was indispensable for salt stress responses. Xu et al. (2008) revealed that activation of MAPK kinase 9 induced ethylene biosynthesis, and then enhanced the sensitivity to salt stress in Arabidopsis. These studies illustrated that there might be an intricate crosstalk network between NO and MAPKs in response to stress stimulus.
Carbohydrates provide nutrition and energy in plants. NO plays a vital role in regulating the process of carbohydrates metabolism. For example, Sami et al. (2020) found that sodium nitroprusside (SNP) treatment activated carbohydrate metabolism and further improved the photosynthetic efficiency in mustard plants. The studies mentioned above indicated that there might be a complex relationship between NO and MAPK signal transduction or carbohydrates metabolism, which might play an essential role in plant growth and stress responses. However, whether NO responds to salt stress by regulating MAPK signal transduction or carbohydrates metabolism at the post-translational level (protein S-nitrosylation) has not been investigated.
Recently, proteomics have been directly used to cope with the large-scale determination of genes and cellular function at the protein level. In addition, various S-nitrosylated proteins have been detected by proteomic analyses in plants (He et al. 2018; Niu et al. 2019). Certain target proteins of NO have been recognized as significant for the regulation of physiological and pathological cellular processes. Thus, in this work, the S-nitrosylated proteome was conducted to decipher the effects and mechanisms of NO under salt stress in tomato seedlings. We found that NO plays a positive role in enhancing the salt resistance and explored the possible effects of protein S-nitrosylation enriched in MAPK signaling pathway and carbohydrates metabolism in response to salt stress, which would further improve our understanding of NO signaling transduction in molecular mechanisms and provide initial insights into the regulatory role of NO in response to salt stress.
Materials and methods
Plant materials and treatment
The tomatoes “Micro-Tom” were used as experimental material. Tomato seeds were kindly supplied by Plenty Gardening Co., Ltd. (Nanjing, China). First, the seeds were disinfected with 1% of NaClO and transferred to 1/2 Hoagland solution for 7 days after germination, then cultivated in Hoagland solution for another 21 days. Then, uniform seedlings were treated with 150 mM NaCl, 10 μM S-nitrosoglutathione (GSNO) (Cayman, Ann Arbor, MI, USA) or 10 μM NO scavenger, 2-(4-carboxy-2-phenyl)-4, 4, 5, 5-tetramethylimidazoline-1-oxyl-3-oxide (cPTIO) (Cayman) after adding to plants in Hoagland solution for 7 days, respectively. The seedlings treated with the Hoagland solution with no extra compounds served as controls. The concentrations of NaCl, GSNO and cPTIO based on the results of a preliminary experiment. The culture solution of different treatments was replaced every 2 days to keep the consistent supplement of extra compounds. The brown bottles as containers for growing plants were used to prevent the effect of light on the root system and Hoagland solution containing GSNO. The bottle mouth was properly sealed with a sealing film to prevent NO volatilizing released from GSNO. The experimental environment was kept at 16 h light at 250 μmol m−2 s−1 photons irradiance at 26 °C and 8 h dark at 20 °C, and 60% relative humidity.
Measurement of morphological indexes
For the morphological phenotype detection, leaf area and total root length of tomato seedlings were measured after treatment with the corresponding solution for 7 days. Leaf area was measured by a leaf area scanner (YMJ-C, Zhejiang Topp Co., Ltd., Zhejiang, China) on the whole single plant. After removing the aboveground part of treated seedlings, the roots were scanned with a root scanner (STD4800, Regent Instruments, Inc., Quebec City, Canada). The total root length of each plant was analyzed by software Win RHIZO 5.0 (Regent Instruments).
Measurement of physiological indexes
The aboveground part of tomato seedlings was used as material to determine the fresh weight (FW) and dry weight (DW). The FW was measured by an electronic balance (SQP, Sartorius, Yihuaxin Electronics Co., Ltd., Shenzhen, China), and samples were then dried at 80 °C to constant weight to obtain its DW. Then, the root physiological activity was measured by the 2,3,5-triphenyl tetrazoliumchloride (TTC) method as described by Lindström and Nyström (1987). Briefly, 0.5 g of fresh root samples was cut into 2 cm from the root tips and incubated for 1 h with a mixture of 5 mL 0.4% (w/v) TTC and 5 mL phosphate buffer (pH 7.5) at 37 °C. The experiment was terminated by adding 2 mL of 1 M H2SO4. Roots were dried with filter paper, and ethyl acetate extraction was performed. The absorbance of the extract at 485 nm was recorded. In addition, a calibration curve was drawn. Root activity was expressed as the amount of TTC deoxidized to triphenylformazan per g fresh weight per hour (µg g−1 h−1) (Tang et al. 2016).
Determination of endogenous SNO content, NO production, and GSNOR activity
Total SNO content was measured using the fluorimetric-based method with minor modifications (Mioto et al. 2017; Pan et al. 2021). Briefly, leaf samples (about 0.2 g) were ground to powder with liquid nitrogen and extracted in extracted buffer (50 mM phosphate buffer, pH 7.2, 80 mM S-methyl methanethiosulfonate) and were then centrifuged at 50,000g for 20 min at 4 °C. The supernatants were collected and mixed with 3 mL chilled acetone. Next, samples were incubated at – 20 °C for 60 min and centrifuged at 50,000g for 20 min at 4 °C. Acetone was discarded and the pellets were resuspended in extracted buffer. The operations above were all conducted in the dark. The protein concentration was measured by the Bradford method. To measure the total SNO content, an extract of 200 µL was mixed with 1.5 µL of 5 mM diaminorhodamine-4 M solution in DMSO. The mixture was divided into two aliquots of 100 µL. One of the aliquots remained at room temperature in absolute darkness for 10 min and the other aliquot was exposed to UV light for 10 min. The volume of both aliquots was adjusted to 1 mL with phosphate buffer (50 mM, pH 7.2). The fluorescence was detected by a spectrofluorometer (LS-55, Perkin-Elmer, Waltham, MA, USA) with 560 nm excitation and 575 nm emission. Total SNO content was calculated according to the difference in fluorescence and GSNO standard curve.
GSNOR activity was detected using the method of Niu et al. (2019). Samples were homogenized with 20 mM Tris–HCl (pH 8.0, 0.2 mM NADH, and 0.5 mM EDTA) and centrifuged at 10,000g for 20 min at 4 °C. The reaction was started by adding 0.4 mM GSNO and the absorbance of the sample was measured at 340 nm.
NO content was measured according to the Griess reagent method with some modifications (Xuan et al. 2012). The detection principle is that NO is easily oxidized in vivo or in the aqueous solution to form NO2−. Therefore, the NO content is measured by concentrations of nitrite in plant extracts. Briefly, a quantity of 0.5 g of leaves was finely frozen in liquid nitrogen with the extract mixture (4 mL of 50 mM ice-cold acetic acid buffer, containing 4% (w/v) zinc acetate, preparing with mortar and pestle). After that, centrifugation was performed at 10,000 g for 15 min at 4 °C, and then the supernatants were collected together in a new tube with 0.1 g charcoal. The supernatants were filtered and collected again, and 1 mL of the mixture was pipetted into 1 mL of Griess reagent. The absorbance of the incubation solution was detected at 540 nm. NO content was assayed by comparison to a standard curve of NaNO2.
Unlike animal systems, nitrite cannot be considered a reliable measure of 'NO production' in plant cells due to highly variable nitrogen metabolism and nitrite/nitrate sources in plant nutrition. Now, 4-amino-5-methylamino-2´, 7´-diaminofluoresceindiacetate (DAF-FM DA) (Beyotime, Jiangsu, China) is a fluorescent probe for the quantitative detection of NO with higher detection sensitivity. It has only a weak fluorescence itself, but can generate strong fluorescence after reacting with NO. Furthermore, the level of endogenous NO can be measured by the NO fluorescent probe DAF-FM DA (Niu et al. 2019). The root tips were loaded with 5 µM DAF-FM DA for 30 min at 37 °C in the dark. Then, the samples were washed three times with fresh buffer. DAF-FM DA fluorescence was visualized using a laser scanning confocal microscope (Leica TCS SL; Leica Microsystems, Wetzlar, Germany). The excitation wavelength was 488 nm and the emission wavelength was 515 nm.
Biotin-switch assay and identification of S-nitrosylated proteins
The identification of the S-nitrosylated proteins was performed by biotin-switch assay as previously described (Feng et al. 2013a; Niu et al. 2019) with some modifications. Leaves of tomato seedlings were ground in liquid nitrogen, extracted by HEN-2 buffer (250 mM Hepes–NaOH, 1 mM EDTA, 0.1 mM neocuproine and 1% proteinase inhibitor), and followed by centrifugation at 13,000 g for 10 min at 4 °C. The supernatant was collected, and the concentrated protein extract was diluted to 1 mg mL− 1 with HEN-2 buffer. Then, 500 µL of the protein solutions was blocked using blocking buffer containing 250 mM Hepes (pH 7.7), 1 mM EDTA, 0.1 mM neocuproine, 2.5% (w/v) SDS, 25 mM S-methylmethane thiosulfonate (MMTS), and incubated at 50 °C for 30 min with frequent vortexing. Subsequently, the sample was precipitated by adding two volumes of acetone. The pellet was washed three times with 70% acetone and dissolved with 170 µL HEN-1 buffer (250 mM Hepes, PH 7.7, 1 mM EDTA, 1% SDS) before the addition of 10 µL of 1 M sodium ascorbate and 20 µL of 4 mM biotin-HPDP (Sigma, St Louis, MO, USA). After labeling for 90 min at room temperature, the reacted proteins were precipitated and washed with acetone, and continued to be incubated in the solution containing 400 μL HENS buffer (250 mM Hepes, pH 7.7, 1 mM EDTA, 1% SDS), 800 μL neutralization buffer (25 mM Hepes, pH 7.7, 100 mM NaCl, 1 Mm EDTA, 0.5% (v/v) Triton X-100), and 30 μL streptavidin agarose beads (Thermo Fisher) at 4 °C overnight on a temperate vortex. The operations above were all conducted in the dark with occasional dim light. Afterward, the beads were mildly washed three times with washing buffer containing 25 mM Hepes (pH 7.7), 600 mM NaCl, 1 mM EDTA, and 0.5% (v/v) Triton X-100. The proteins were then eluted in 50 μL elution buffer (250 mM Hepes, pH 7.7, 1 mM EDTA, 1% (v/v) β-mercaptoethanol). Finally, the biotin-labeled products were analyzed by 15% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie brilliant blue or immunoblotting with anti-biotin antibody (Agrisera, Vännäs, Sweden).
The S-nitrosylated proteins were identified by LC–MS/MS (Feng et al. 2013a; Niu et al. 2019). LC–MS/MS technology was used to identify the proteins obtained from gel strips. After the biotin-labeled products were analyzed by SDS-PAGE, the target band was excised, washed twice with water and subsequently washed with 50% acetonitrile until transparent. The gel slices were first dehydrated in 50% acetonitrile for 15 min and then completely dehydrated in 100% acetonitrile. After drying, 20 µL of 10 ng µL− 1 trypsin was added and kept for 30 min on ice. After the enzyme solution was completely absorbed by the micelles, 100 mM TEAB buffer was added to a total volume of 100 µL and digested overnight at 37 °C. The supernatant was collected by low-speed centrifugation, 100 µL of 0.1% formic acid was added to the gel, and shaken at room temperature for 5 min. After low-speed centrifugation, the supernatant was collected, and the enzymatic samples were combined, centrifuged at 12,000 g for 5 min at room temperature. The supernatant was taken and slowly passed through a C18 desalting column, and then washed three times continuously using 1 mL of washing solution (0.1% formic acid, 4% acetonitrile). 0.4 mL of eluent (0.1% formic acid, 75% acetonitrile) was added for 2 consecutive elutions and eluted samples were pooled and lyophilized. The fragments after enzymolysis were loaded for analysis. The data from MS and MS/MS were used to search uniprot-Solanum_lycopersicum.fasta. The search parameters were set to detect four differential modifications: 57 amu on cysteine (indicating carboxyamidomethylation), 71 amu on cysteine (indicating propionamidation), 428 amu on cysteine (indicating biotinylation) and 16 amu on methionine (indicating oxidation). The peptides matching criteria of a cross correlation score (Xcorr) were 1.9, 2.2 and 3.75 for + 1, +2 and +3 charged peptides, respectively (Feng et al. 2013a). The subcellular localization information was annotated by using the Cell-mPLOC 2.0, which covers a wide range of annotation information, including 6 databases: eukaryotic, human, plant, Gram-positive bacteria, Gram-negative bacteria and viruses (Chou and Shen 2010).
Total RNA extraction, cDNA synthesis and quantitative real-time PCR (qRT-PCR) analysis
Total RNA extraction, cDNA synthesis, and qRT-PCR were conducted as described by Gao et al. (2021). All the primers used for RT-PCR were designed using prime 5 software, as shown in Table S1.
Determination of S´adenosyl-L-methionine (SAM) content, ethylene production and ABA content
Ethylene production was determined according to the method of Wang et al. (2020) with some modifications. The four fully expanded upper leaves beneath the growing tip of the plant were sampled, and then placed in a 795 mL desiccative airtight container and incubated at 25 °C for 12 h. About 1 mL of headspace gas from each container was collected using a gas-tight hypodermic syringe and injected into a gas chromatograph (GC-17A, Shimadzu, Kyoto, Japan) immediately for ethylene concentration measurement. The gas chromatograph was equipped with a flame ionization detector and an activated alumina column.
ABA content was determined according to Yan et al. (2022). Briefly, leaves (1 g) were ground with a mortar and pestle in 3 mL of cooled 80% (v/v) methanol solution. After incubation for 12 h at 4 °C, the extract was centrifuged at 8000 g for 15 min. The supernatant liquid was extracted and the aqueous phase was collected. The solution sample was eluted through columns and prewashed with methyl alcohol. The supernatant was collected and dried in a vacuum at 40 °C. The supernatants were dissolved in methyl alcohol at a constant volume, a microporous membrane (0.22 μM) was established to filter the products and high-performance liquid chromatography (HPLC) was carried out.
SAM contents were determined as described by Roeder et al. (2009) using the 5,5′-dithiobis-(2-nitrobenzoic acid)-GR recycling procedure.
Determination of glucose (Glc), glucose-6-phosphate (G6P), fructose-6-phosphate (F6P) and pyruvate content
Glucose, G6P, F6P and pyruvate content were analyzed as described by Zhao et al. (2021). The relative content (compared to the control) is shown in Fig. 6.
Determination of malic acid and citric acid content
Fresh sample (2 g) was added into a 30 mL centrifuge tube, and deionized water was added to make up the volume to 25 mL, and ultrasonically extracted for 60 min. After that, the extract was centrifuged at 10,000 g for 10 min. The supernatants were filtered through a 0.22 μm membrane and the HPLC was carried out. Chromatography conditions were as follows: chromatographic column: Hi-PiexH (300 mm × 7.7 mm, 5 μm); detector: UV detector; detection wavelength: 210 nm; mobile phase: 0.2 mmol L−1 sodium dihydrogen phosphate; column temperature: 25 °C; flow rate: 0.5 mL min − 1; injection volume: 10 μL. The relative content (compared to the control) is shown in Fig. 6.
Enzyme activity assays
The activity of pyruvate kinase (PK) was measured and initiated by the addition of ADP (Baud et al. 2007). The rate of oxidation of NADH at 30 °C was measured at 340 nm and an NADH standard curve was used to calculate the activity in the sample. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) activity was assayed using spectrophotometric quantification (340 nm) of NADH according to the method described previously (Guo et al. 2014). Tomato samples (about 0.5 g) were collected and ground in a 1.5-mL centrifuge tube by a pestle. Total protein was extracted from samples using chilled buffer A (50 Tris–HCl, pH 7.5, 10 mM KCl, 1 mM EDTA, 0.5 mM PMSF, and 2 mM DTT). The homogenate was centrifuged at 10,000g for 20 min and the supernatant was used for each activity assay. One unit (U) is defined as the amount of enzyme that catalyzes the consumption of 1 µmol min−1 NADH under each specified assay condition. Citrate synthase (CS) activity was assayed according to the method described by Tilbrook et al. (2014). One unit (U) of enzyme activity was defined as the catalytic production of 1 nmol TNB per minute per mg of tissue protein in the reaction system at 25 °C. The extinction coefficient of DTNB at 412 nm is 13.6 × 10−3 mL nmol−1 cm−1. The fructose-bisphosphate aldolase (FBA) activity was measured based on Boyer’s modification of the hydrazine assay (Sehrawat et al. 2013a). Malate dehydrogenase (MDH) activity was determined by monitoring the increase in absorbance at 340 nm due to NADH production (Xin et al. 2014). The relative activity (compared to the control) is shown in Fig. 6.
Statistical analysis
The experimental design was a completely randomized design with three replicates. Each replicate contained 15 plants. The data are normally distributed using Shapiro–Wilk test, and satisfy the homogeneity of variance by Levene’s Test. Then, the data were further analyzed using one-way analysis of variance (ANOVA) of SPSS 22.0 software (SPSS, Inc., Chicago, IL, USA). Significant analyses were performed using Duncan’s multiple range test at P < 0.05 level. Where indicated, the value of data was expressed as mean ± standard error (SE).
Results
NO alleviated the salt toxicity on the growth of tomato seedlings
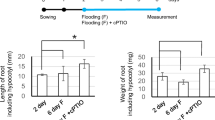
Since NO seems to be a vital participant in abiotic stress response in plants, we first studied the possible roles of NO in salt-stressed tomato seedlings. As shown in Table 1, in comparison with the control, NaCl treatment obviously inhibited the growth of tomato seedlings, while GSNO treatment alone significantly promoted seedling growth. The GSNO treatment alone obviously increased the leaf dry weight, fresh weight, leaf area and root activity by 12.5, 22.85, 10.09 and 13.67%, respectively, but had no obvious effect on the total root length in comparison to the control, indicating the positive role of GSNO in promoting the growth of tomato seedlings. The co-treatment of NaCl + GSNO significantly alleviated NaCl stress. Compared with NaCl treatment alone, NaCl + GSNO treatment increased leaf fresh weight and dry weight by 48.3% and 20.0%, respectively (Table 1). The NaCl + GSNO treatment increased leaf area more than NaCl treatment alone, indicating that GSNO has also a positive biological effect on the growth of tomato leaves under NaCl stress. Moreover, NaCl + GSNO treatment significantly increased the total root length and root activity in tomato seedlings and the obtained increment was 19.46% and 13.33% compared with NaCl treatment alone (Table 1). In addition, compared to the control, leaves became yellowed, smaller and curled, and roots became shorter under NaCl stress, yet were properly alleviated in NaCl + GSNO treatment (Fig. 1). These results, together with the growth data of tomato seedlings, indicated that GSNO could alleviate the salt stress in tomato seedlings.
Effect of NO on the total S-nitrosylation levels during the growth of tomato seedlings under salt stress
To further elucidate whether S-nitrosylation was involved in the process of NO-alleviating salt stress, SNO content, endogenous NO levels, GSNOR activity and GSNOR expression were measured in tomato seedlings (Fig. 2). Compared with the control, NaCl and GSNO treatment alone significantly increased SNO and NO content (Figs. 2a and b). Meanwhile, NaCl + GSNO application caused an increase in SNO and NO content by 62.04 and 70.52%, respectively, compared to NaCl treatment alone. In addition, NaCl and GSNO treatment alone significantly enhanced GSNOR activity compared to the control, and the highest GSNOR activity was obtained in the co-treatment of NaCl + GSNO (Fig. 2c). Similarly, compared with NaCl treatment alone, NaCl + GSNO treatment significantly up-regulated GSNOR expression by 88.87% (Fig. 2d). The fluorescence intensity of NO production was enhanced under salt stress. Application of GSNO also significantly enhanced the fluorescence intensity of NO production in root tips of tomato seedlings under salt stress in comparison with NaCl treatment alone (Fig. 2e). However, compared with the control, the fluorescence intensity of NO production was weakened in treatment with NO scavenger cPTIO.
Effect of GSNO on the levels of endogenous NO (a, e), total S-nitrosothiol (SNO) (b), S-nitrosoglutathione reductase (GSNOR) activity (c), and GSNOR expression levels (d) during the growth of tomato seedlings under salt stress. The values (means ± SE) are the averages of three-independent experiments (n = 15). Bars not sharing the same letters indicate statistically significant differences by Duncan’s multiple range test (P < 0.05)
Identification of S-nitrosylated proteins and subcellular location
In order to further confirm whether there exist possible mechanisms for S-nitrosylation during GSNO alleviating salt stress in tomato seedlings, biotin-switch detection was performed, and S-nitrosylated proteins were screened using the LC–MS/MS proteomic approach. As shown in Fig. 3a, NaCl and NaCl + GSNO treatments obviously increased the S-nitrosylation of proteins in tomato seedlings compared to the control. Interestingly, among these treatments, there were obvious protein bands of protein S-nitrosylation around 75 kDa and 63 kDa. The band of potential candidates for S-nitrosylation around 63 kDa in the NaCl treatment was shallower than that in NaCl + GSNO treatment (Fig. 3a). A total of 1054 putative S-nitrosylated proteins have been identified in the control (549), NaCl (788), and NaCl + GSNO (680) treatments (Fig. 3). Among them, 31.3% (349) of proteins were constitutively S-nitrosylated in all three treatments (Fig. 3b; Supplementary Table S2). In addition, 6.4% (67) of S-nitrosylated proteins were identified only in control; 22.5% (237) of S-nitrosylated proteins were identified only in NaCl treatment alone and 12.9% (136) of S-nitrosylated proteins were identified only in NaCl + GSNO treatment alone (Fig. 3b).
Analysis of S-nitrosylated proteins identified from tomato seedlings of different treatments using the LC–MS/MS. a Change of protein S-nitrosylation in different treatments in tomato seedlings. S-Nitrosylation was detected using a biotin-switch assay with anti-biotin antibody. b Overlap of the S-nitrosylated proteins of tomato seedlings in different treatments displayed by a Venn diagram. c–e The subcellular localization of the S-nitrosylated proteins identified in the control (c), under NaCl treatment (d), under GSNO + NaCl treatment (e)
The subcellular localization analysis found that the S-nitrosylated proteins were mainly enriched in chloroplasts, cytoplasm, mitochondrion, followed by nucleus and cell membrane in all three treatments (Fig. 3c–e). The control, NaCl and NaCl + GSNO treatments enriched 172 (29.05%), 198 (23.10%), 183 (25.85%) S-nitrosylated-proteins located in the chloroplast, respectively; 115 (19.43%), 178 (20.77%), and 150 (21.19%) proteins were located in the cytoplasm, respectively; 81 (13.68%), 107 (12.49%), and 94 (13.28%) proteins were located in the mitochondria, respectively; 51 (8.61%). 92 (10.74%), and 66 (9.32%) proteins were located in the nucleus, respectively; and 34 (5.74%), 67 (7.82%), and 50 (7.06%) proteins were located in the cell membrane, respectively. Only a small part of S-nitrosylated protein was located in the endoplasmic reticulum, peroxisome, cell membrane, vacuole, plastid and Golgi apparatus.
KEGG pathway annotation of S-nitrosylated proteins
To further elucidate the key pathway of S-nitrosylated protein in different treatments, the KEGG pathway annotation of S-nitrosylated protein was performed. Figure 4 shows that S-nitrosylated proteins were involved in the main biochemical metabolic pathways and signal transduction pathways. KEGG biological process analyses revealed that enriched S-nitrosylated proteins mainly participated in the metabolism, genetic information processing, transport and catabolism, signal transduction and environmental adaptation. We found that proteins enriched in the signal transduction pathway were involved in active oxygen scavenging, ATP synthesis and MAPK signaling pathway. In addition, the S-nitrosylated proteins identified in the metabolic pathway were mainly related to the carbohydrate metabolism, amino acid metabolism and energy metabolism (Fig. 4).
S-Nitrosylated proteins enriched in MAPK signaling transduction and carbohydrate metabolism
The analyses of KEGG pathway annotation revealed that proteins enriched in signal transduction were involved in the MAPK signaling transduction. Thus, we further analyzed these proteins identified as targets of S-nitrosylation and enriched in MAPK signaling pathway (Table 2). As shown in Table 2, 15 S-nitrosylated proteins were involved in MAPK signaling transduction, which were significantly enriched in cytoplasm (33.33%) and nucleus (13.33%), whereas the subcellular localizations of 53.33% proteins were unknown (Fig. 5a). These S-nitrosylated proteins might have 1 to 4 sites of S-nitrosylation modification (Table 2). Five S-nitrosylated proteins [serine/threonine-protein phosphatase (G8Z285); MAP kinase kinase (MAPKK), MAPK (A0A3Q7GX91), 14-3-3 protein 9 and S-adenosylmethionine synthase 3 (SAM3)] were identified in the whole stage of salt stress response in tomato seedlings, however, their S-nitrosylation levels may differ. MAP kinase kinase 5 (MAPKK5), mitogen-activated protein kinase (MAPK), serine/threonine-protein kinase (SnRK) and S-adenosylmethionine synthase (SAM) were S-nitrosylated in both NaCl and NaCl + GSNO treatments. Interestingly, 14-3-3 protein 8 and serine/threonine-protein phosphatase (A0A3Q7FED8) were found to undergo S-nitrosylation only in NaCl treatment. Furthermore, the functions of S-nitrosylated proteins involved in MAPK signaling were analyzed (Table 2). We found that two proteins, serine/threonine-protein phosphatases (G8Z285 and A0A3Q7J0U6), functioned in the brassinolide-mediated signaling pathway. MAPKK 5 and MAPKK in the protein kinase signal transduction cascade activated by stress were identified. Moreover, MAPK (A0A3Q7GX91) participated in gene expression and intracellular signal transduction and had MAP kinase activity. PP2C was related to protein serine/threonine phosphatase activity. Three proteins (SAM 1, SAM 3, SAM) were related to the biosynthesis of the ethylene precursor S-adenosylmethionine. These results indicated various functions of S-nitrosylated proteins enriched in MAPK signaling transduction, which might participate in salt stress response in tomato seedlings.
S-Nitrosylated proteins enriched in carbohydrate metabolism
A total of 22 S-nitrosylated proteins were enriched in carbohydrate metabolism (Table 3). The subcellular localizations analysis revealed that these proteins were significantly enriched in the cytosol (45.45%), followed by mitochondrion (31.82%), cell wall (13.64%) and chloroplast (4.55%), and the localization of 4.55% proteins was unknown (Fig. 5b; Table 3). In addition, these S-nitrosylated proteins identified in carbohydrate metabolism were involved in glycolysis, tricarboxylic acid (TCA) cycle, degradation of carbohydrates and gluconeogenesis (Table 3). For example, various glycolytic enzymes including fructose-bisphosphate aldolase (FBA), pyruvate kinase (PK), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), glucose-6-phosphate isomerase (GPI) and enolase were found to be S-nitrosylated (Table 3). Aconitate hydratase (ACN), isocitrate dehydrogenase (ICDH), citrate synthase (CS), malate dehydrogenase (MDH) and malic enzyme (ME) in TCA cycle were detected to be S-nitrosylated (Table 3). Of these proteins, FBA, PK, GAPDH and enolase in glycolysis and CAN, ICDH and ME in TCA cycle were identified in all three treatments (Table 3). Two key enzymes, MDH and CS, in TCA cycle were S-nitrosylated only in NaCl + GSNO treatment (Table 3). Besides, several key enzymes in gluconeogenesis, degradation of pectin and sucrose were also found to undergo S-nitrosylation (Table 3). These results highlighted a possible role for the S-nitrosylation of essential proteins in carbohydrate metabolism in response to salt stress.
GSNO regulates MAPK signaling transduction under salt stress
To gain insight into the exact roles of NO-induced protein S-nitrosylation in MAPK signaling transduction under salt stress, S-nitrosylated proteins were validated at the transcription level (Fig. 6). Three proteins (SAM1, SAM3, SAM) related to the biosynthesis of ethylene were S-nitrosylated. Compared with the control, NaCl treatment remarkably promoted SAM1, SAM3, SAM expression levels. However, NaCl + GSNO significantly decreased their relative expression compared with NaCl treatment alone (Fig. 6a). Interestingly, compared with NaCl treatment alone, GSNO supplement under salt stress also decreased SAM content and ethylene production (Fig. 6b, d), indicating that NO decreased the expression levels of genes related to the biosynthesis of ethylene and SAM content and then inhibited ethylene biosynthesis in response to salt stress. In addition, two key negative regulators of ABA signaling, PP2C and SnRK, underwent S-nitrosylation in both NaCl and NaCl + GSNO treatments. Compared with the control, the transcriptional expressions of PP2C and SnRK were down-regulated under salt stress. Meanwhile, GSNO treatment further down-regulated the expression levels of PP2C and SnRK (Fig. 6c) and decreased ABA content under salt stress (Fig. 6f). In comparison with NaCl treatment, NaCl + GSNO treatment enhanced the transcriptional levels of stress-activated proteins MAPK, MAPKK and MAPKK5 (Fig. 6e). These results indicated that GSNO could regulate the transcriptional levels of proteins enriched in MAPK signaling, and reduce ethylene biosynthesis and ABA content in response to salt stress.
Effect of GSNO on MAPK signaling transduction. a, c and e The expression pattern of genes in MAPK signaling transduction subjected to different treatments using qRT-PCR. b SAM content. d Ethylene production. f ABA content. The values (means ± SE) are the averages of three-independent experiments (n = 15). Bars not sharing the same letters indicate statistically significant differences by Duncan’s multiple range test (P < 0.05)
GSNO regulates carbohydrate metabolism under salt stress
To decipher the exact roles of protein S-nitrosylation in carbohydrate metabolism under salt stress, enzyme activities undergone S-nitrosylation and their gene expressions in the carbohydrate metabolism pathway were assayed (Fig. 7). Key proteins (FBA, GAPDH and PK) in glycolysis were found to be S-nitrosylated in the control, NaCl and NaCl + GSNO treatments. Their activities drastically decreased in NaCl treatment, whereas GSNO treatment enhanced their activities under salt stress, implying the activation of FBA, PK and GAPDH by NO under salt stress. Similarly, the relative expressions of S-nitrosylated proteins [FBA (A0A3Q7HX95, A0A3Q7ETK4 and A0A3Q7EYM6), GAPDH and enolase] at the transcriptional level were regulated by GSNO under salt stress. For example, the expression levels of FBA1, FBA2, FBA3, GAPDH and enolase were significantly up-regulated by NaCl + GSNO treatment in comparison with the NaCl treatment alone. Compared to NaCl treatment alone, NaCl + GSNO treatment remarkably increased the content of four metabolites (glucose, glucose-6-phosphate, fructose-6-phosphate and pyruvate) in glycolysis.
Effect of GSNO on carbohydrate metabolism. a Relative activity of FBA, GAPDH and PK. b and c The expression pattern of genes in glycolysis. d Relative content of glucose (Glc), glucose-6-phosphate (G6P), fructose-6-phosphate (F6P) and pyruvate. e Relative activity of MDH and CS. f and g The expression pattern of genes in TCA cycles. h Relative content of malic acid and citric acid. The values (means ± SE) are the averages of three-independent experiments (n = 15). Bars not sharing the same letters indicate statistically significant differences by Duncan’s multiple range test (P < 0.05)
In addition, two key enzymes (MDH and CS) in the TCA cycle also underwent S-nitrosylation when GSNO appeared. In addition, exogenous GSNO significantly enhanced MDH and CS activities under salt stress compared to the NaCl treatment alone (Fig. 7e). Compared to the NaCl treatment alone, NaCl + GSNO treatment significantly increased both the expression levels of MDH, MDH2 and CS and the contents of malic acid and citric acid (Fig. 7f, h), which implied that NO might activate the expression levels and activities of MDH and CS and then increase related metabolite content under salt stress. In addition, the relative expression of other key proteins (ME, ICDH and ACO) in the TCA at the transcriptional level was up-regulated by GSNO under salt stress (Fig. 7g). Thus, these results indicated that NO might increase the activity of these enzymes in carbohydrate metabolism and up-regulate their relative expression at the transcriptional level, thereby increasing metabolite contents and promoting carbohydrate metabolism under salt stress.
Discussion
NO, a ubiquitous bioactive gas, plays an important role in the processes of plant growth and development and in response to multiple stress stimulus. The results presented herein indicated that GSNO treatment promoted the growth of tomato seedlings and further alleviated NaCl treatment-induced salt toxicity in tomato seedling (Table 1; Fig. 1), implying a molecular mechanism of NO on alleviating salt stress. In rice, the increase of endogenous NO production could not only alleviate Cd-induced inhibition of adventitious root elongation and lateral root formation, but also reduced Cd-caused increase of lignin deposition, which further altered the root system formation (Piacentini et al. 2020). Protein S-nitrosylation is one of the important ways by which NO exerts its biological function when plants respond to various abiotic stresses (Niu et al. 2019; Wei et al. 2020). To investigate the potential regulation of NO to physiological processes by modifying cysteine residues of proteins, the changes of total S-nitrosylation level in response to salt stress were analyzed (Fig. 2). Application of GSNO strikingly increased the level of endogenous SNO and NO production under salt stress compared to NaCl treatment (Fig. 2a and b). In addition, GSNO treatment alone induced a positive increase of endogenous NO content compared to the control, which was reversed by applying NO scavenger cPTIO (Fig. 2e). This indicated that GSNO pretreatment-induced increase of endogenous NO level could function in response to salt stress. Since NO and SNO content are closely related to endogenous S-nitrosylation levels, the regulation of endogenous S-nitrosylation levels might be an essential mechanism of NO on the alleviation of salt stress. In addition, it is known that GSNOR can function in modulating the global S-nitrosylation levels (Niu et al. 2019; Wei et al. 2020). Moreover, transgenic rice with overexpressed OsGSNOR was identified with lower SNO levels, indicating that GSNOR might exert its function in regulating SNO homeostasis (Lin et al. 2012). That is to say, NO may inhibit GSNOR activity to prevent S-nitrosoglutathione scavenging (Frungillo et al. 2014). However, in our study, highest GSNOR expression and GSNOR activity were detected in GSNO + NaCl treatment (Fig. 2d). GSNOR itself could be S-nitrosylated by a NO-dependent way and S-nitrosylated GSNOR made itself inactivate (Pan et al. 2021). Thus, this might lead to a higher level of GSNOR in the process of NO-alleviating salt stress. Consequently, this indicated that GSNO supplement might enhance the tolerance of tomato seedlings to salt stress by regulating the levels of endogenous S-nitrosylation.
For a deeper insight, the candidates of S-nitrosylated proteins were identified and analyzed. Compared to the control, both NaCl and NaCl + GSNO treatments obviously increased nitrosylation of proteins (Fig. 3a). In our results of mass spectrometry analysis, a total of 1054 S-nitrosylated proteins were identified, the number of S-nitrosylated proteins under NaCl treatment was the highest, 788 proteins, followed by NaCl + GSNO, with 680 identified (Fig. 3b). The number of S-nitrosylated proteins in the control was the least with 549 proteins. In addition, among them, 349 of proteins were constitutively S-nitrosylated in all three treatments (Table S2; Fig. 3). Obviously, compared to the control, salt stress could induce more S-nitrosated proteins in tomato seedlings (Fig. 3). It has been indicated that S-nitrosation level and the number of S-nitrosated proteins change significantly under biotic or abiotic stresses. For example, an increase of S-nitrosated proteins was previously identified in Arabidopsis under pathogen stimulation (Maldonado-Alconada et al. 2011) and in the apoplast of Brassica juncea seedlings under cold stress (Sehrawat and Deswal 2014). Interestingly, various studies showed that S-nitrosylated proteins significantly increased after salt stress stimulus, revealing the possible roles of NO target proteins in response to salt stress (Fares et al. 2011; Qi et al. 2020). In Arabidopsis, 926 proteins were identified to undergo S-nitrosylation and a large number of proteins involved in the stress responses were identified to be S-nitrosylated (Hu et al. 2015). Thus, it was shown that NO-induced protein S-nitrosylation could participate in regulating the stress responses of salt throughout plant development and during plant responses to different stress conditions and that the regulation of protein S-nitrosation is not similar in different cell types or physiological conditions.
Interestingly, our results indicated that the number of S-nitrosylated proteins slightly decreased after adding GSNO under salt stress compared with NaCl treatment alone (Fig. 3), which might be due to that NO affected the turnover ratio of S-nitrosylation under salt stress via regulating S-nitrosoglutathione reductase (Gong et al. 2015). The GSNOR activity and its expression level were further increased in NaCl + GSNO treatment compared to the NaCl treatment alone. The degree of S-nitrosation of any protein depends on the rate of S-nitrosation and de-nitrosation. GSNOR is considered as not only the main pathway of irreversible degradation of low molecular S-nitrosothiols, namely GSNO, to oxidized glutathione, but also as an essential enzyme that indirectly affects S-nitrosation level. GSNOR could control the level of protein S-nitrosothiols, which are formed in transnitrosylation reactions of protein and GSNO (Petřivalský et al. 2015). It is worth noting that loss-of-function mutations in AtGSNOR1 have been proposed to result in increased total SNO levels, whereas mutations that enhanced AtGSNOR1 activity decreased cellular SNO concentrations. Importantly, S-nitrosylation, as a prototypic redox-based, post-translational modification, is the addition of an NO moiety to a protein cysteine (Cys) thiol to form an SNO (Yun et al. 2016). Thus, these results parallel our findings for elevated GSNOR activity, decreased S-nitrosylated proteins in NaCl + GSNO treatment in comparison with NaCl stress alone, further implying that GSNO-enhanced GSNOR activity resulted in slightly decreased S-nitrosylated proteins under salt stress.
Interestingly, GSNOR itself could also undergo S-nitrosylation in a NO-dependent way and its activity was inhibited by itself S-nitrosylation (Pan et al. 2021). Thus, this might also lead to the slightly decreased S-nitrosylated proteins after adding GSNO under salt stress because of the destabilization of GSNOR caused by the S-nitrosylation modification on itself. In addition, herein identified S-nitrosylated proteins were mainly enriched in the cytoplasm, chloroplast and mitochondria (Fig. 3), followed by the nucleus and cell membrane. Similarly, a previous study has shown that 191 S-nitrosylated proteins identified in the multiple metabolic pathways of tea were located in chloroplasts, cytoplasm and mitochondria (Qiu et al. 2019). This suggested that most proteins underwent S-nitrosylation modification might exert their biological functions and respond to adversity mainly in the chloroplast, cytoplasm or mitochondria. Hu et al. (2015) also revealed that the most abundant fraction was found to be localized in the chloroplast/plastid, implying that the physiological activities associated with the photosynthetic organelle are extensively modulated by S-nitrosylation modification. However, it is worth noting that both, our research results and previous studies (Hu et al. 2015; He et al. 2018; Qiu et al. 2019), showed that the level of S-nitrosylated proteins enriched in the chloroplast and mitochondria is significantly higher than that one located in the nucleus or cell membrane. On the one hand, this may be due to that these proteins underwent S-nitrosylation modification function rather on various metabolic pathway in these organelles. On the other hand, extracting procedures for enrichment of S-nitrosylated proteins might result in significant bias in respect to the representation of proteins from different subcellular compartments, especially cell membrane proteins and nuclear proteins. In future studies, this issue about methodological limitation needs to be further addressed and explored.
Besides, various S-nitrosylated proteins have been indicated to function in carbon and energy metabolism, genetic information, photosynthesis pathway, stress response and cellular signaling transduction, and so on (He et al. 2018; Niu et al. 2019). In our study, the S-nitrosylated proteins were mainly involved in the metabolism, genetic information processing, transport and catabolism, signal transduction and environmental adaptation (Fig. 4). As a consequence, these results indicated that these S-nitrosylated proteins could function in various pathways, which might play a vital role in the resistance of tomato seedlings to salt stress.
MAPK cascades play an important role in transduction extracellular signals to cellular responses. Further, as a key signal regulator, NO has been used to induce the activation of MAPK signal. Interestingly, in our study, 15 S-nitrosylated proteins were identified in the MAPK signal transduction pathway, which mainly were enriched cytoplasm and nucleus (Table 2; Fig. 5a). The SAMs, methionine (Met) and 1-aminocyclopropane carboxylic acid (ACC) were recognized as important precursors for ethylene synthesis. SNP treatment inhibited ethylene biosynthesis by regulating the activities of ACC synthesis and ACC oxidase during the postharvest freshness of cut flowers (Liao et al. 2013). MET synthase was a candidate for S-nitrosylation in Kalanchoe pinnata, although this has not been linked to a loss in enzymatic activity (Abat et al. 2010). In our study, three SAMs (SAMs1, SAMs3 and SAMs) proteins have been found to be S-nitrosylated in tomato seedlings for the first time (Table 2). Moreover, among them, SAMs3 was identified in all treatments, SAMs was S-nitrosylated in both NaCl and NaCl + GSNO treatments, whereas SAMs1 was S-nitrosylated only in NaCl + GSNO treatment, implying the involvement of SAMs in salt stress through S-nitrosylation. The SAM content and ethylene production were decreased by GSNO + NaCl treatment compared to NaCl treatment alone (Fig. 6b, d). An earlier study indicated that MAT was S-nitrosylated, which caused the decrease of MAT activity (Lindermayr et al. 2006). Since the inhibition of MAT activity by S-nitrosylation could lead to the decreased SAM it may also well explain the reason of the reduction of ethylene production caused by GSNO supplement (Fig. 6d). Furthermore, the transcriptional expression of SAMs1, SAMs3 and SAMs was down-regulated by GSNO treatment under salt stress compared to NaCl treatment alone (Fig. 6), suggesting that NO significantly decreased the expression of SAMs at the transcriptional level, thereby alleviating salt stress in tomato seedlings. Singh et al. (2017) indicated that NO-mediated transcriptional modulation enhanced plant adaptive responses to arsenic stress. Since SAMs can catalyze the synthesis of the ethylene precursor and NO is known to modulate the biosynthesis of ethylene, SAMs probably mediate the crosstalk between NO signaling and ethylene biosynthesis (He et al. 2018). Thus, these results clarified that NO could regulate the relative expression of SAMs mainly through transcriptional regulation, thereby regulating the ethylene production in MAPK signaling, which probably lead to the alleviation of salt damage to the growth of tomato seedlings.
The core ABA signaling is composed of PYR/PYL/RCAR receptors, PP2C phosphatases and SnRK2 kinases. The ABA could bound PYLs to form complexes with the clade A PP2C, which allows the release of the inhibition of SnRK2 protein kinases by PP2C. In the absence of ABA, PP2C interacts with and inhibits SnRK2 to prevent ABA signaling (Chen et al. 2020). The data presented herein demonstrated that PP2C underwent S-nitrosylation modification in all three treatments and SnRK was S-nitrosylated only in NaCl and NaCl + GSNO treatments in tomato seedlings (Table 2). Especially, S-nitrosylated PP2C was reported for the first time in our study. A previous study reported that SnRK2.6 and SnRK2.2 could be S-nitrosylated and thus inactivated, thereby negatively regulating ABA signaling in the processes of germination and early seedling growth (Wang et al. 2015a). In addition, another member of the SnRK2 family, SnRK2.6 (OST1) was found to be inactivated by S-nitrosylation at cysteine residue 137 in the guard cell, which resulted in the negative regulation of ABA signaling and contributed to ABA insensitivity in stomatal closure (Wang et al. 2015b). Thus, we hypothesized that the S-nitrosylated SnRK might inactivated in our study, especially in the appearance of NO, which will require further verification in the future. Plants accumulate quickly ABA, which in turn activated multiple stress responses when subjected to various abiotic stresses. When environments are optimal, ABA is decreased to a basal level to promote optimal growth. Thus, the regulation of ABA levels in tissues and cells is critical for balancing defense and growth processes when plants faced to non-optimal environments (Chen et al. 2020). In our results, GSNO treatment also further reduced ABA content under salt stress compared with NaCl treatment (Fig. 6f). This indicated that the regulation of ABA levels plays a key role in the process of NO-alleviating salt stress. Moreover, our results indicated that the relative expressions of PP2C and SnRK were significantly down-regulated in GSNO-treated tomato seedlings under salt stress compared to NaCl treatment alone (Fig. 6c), implying that the expressions of PP2C and SnRK were inhibited at transcriptional levels by NO under salt stress. Accordingly, it could be speculated that, the key proteins SnRK and PP2C in ABA signaling pathway underwent NO-induced post-translational modification, S-nitrosylation, and NO affected their relative expressions at transcriptional levels, which resulted in a reduction in ABA level to a basal level and consequently alleviated salt toxicity in tomato seedlings.
MAPKs are the key proteins involving in plant stress response, and loss-of-function mutations affecting MAPK signals can lead to the increase of salt sensitivity (Yoo et al. 2008). In our study, two MAPKK proteins [MAPKK5 (S-nitrosylated in NaCl treatment) and MAPKK (S-nitrosylated in control, NaCl and NaCl + GSNO treatments)] and two MAPK proteins [MAPK (K4AZN6) (S-nitrosylated in NaCl and NaCl + GSNO treatments), MAPK (A0A3Q7GX91) (S-nitrosylated in all treatments)] were identified in tomato seedlings (Table 2). Feng et al. (2013b) indicated that extracellular signal-regulated kinase MAPK3/MAPK1 underwent S-nitrosylation by GSNO pretreatment and thus inhibited its phosphorylation, which provided critical insights into the control of apoptosis and tumor development. Hence, the function of S-nitrosylation about MAPK protein prompted us to study how this post-translational modification works its phosphorylation under salt stress. Meanwhile, the expression levels of these transcriptional genes (MKK2, MEK1 and MAPK5) of MAPKK5, MAPKK and MAPK (accession number: K4AZN6), respectively, were significantly up-regulated by GSNO treatment under salt stress compared to NaCl treatment alone (Fig. 6e). In addition, MAPK5 and MEK1 overexpression also enhanced the resistance to the stress stimulus in plants (Zhang et al. 2022), which further confirmed our results (Fig. 6e). Thus, NO might enhance the resistance of tomato seedlings to salt stress by increasing the expression of various MAPKs at transcriptional levels, whereas whether the S-nitrosylated MAPK proteins identified in our study affect their phosphorylation levels remains to be established in future. Taken together, these results mentioned above clarified that various S-nitrosylated proteins were identified in the MAPK signal transduction pathway and NO could regulate MAPK signaling mainly by modulating ethylene production and ABA signaling through transcriptional regulation in response to salt stress, thereby reducing ABA level and ethylene biosynthesis and alleviating salt toxicity in tomato seedlings.
Early studies showed that various proteins in the carbohydrate metabolism were modified by S-nitrosylation under cold stress (Sehrawat et al. 2013b) and during the aging of elm seeds (He et al. 2018). He et al. (2018) reported that 11 S-nitrosylated proteins were involved in carbohydrate metabolism during the aging of elm seeds, revealing the regulatory mechanism of post-translational modification on the carbohydrate metabolism. In our study, 22 proteins in the carbohydrate metabolic pathway were found to undergo S-nitrosylation, which mainly were enriched in cytosol, mitochondrion and cell wall (Table 3; Fig. 5b). Of these proteins, FBA, PK, GAPDH and enolase in glycolysis were identified to undergo S-nitrosylation in all three treatments (Table 3). Previously, Wang et al. (2017) found that the S-nitrosylation of GAPDH induced by SNP treatment decreased its activity, leading to the inhibition of root growth. However, in Nicotiana tabacum, GAPDH S-nitrosylation did not affect the total GAPDH activity under salt stress and only a small proportion of GAPDH appeared to be regulated by S-nitrosylation in response to salt stress (Wawer et al. 2010). This implies that the regulation of GADPH activity after S-nitrosylation might be different due to different plant tissues or stimuli conditions. Previous studies found that FBA could undergo S-nitrosylation and function in non-aged and aging seeds of elm (He et al. 2018), GSNO-treated leaf of Kalanchoe pinnata (Abat et al. 2010) and the development of adventitious roots of cucumber (Niu et al. 2019). In addition, Chen et al. (2013) found that S-nitrosylated FBA was inactivated after seed aging compared to normal seeds. Meanwhile, FBA played important roles in response to salt stress in Sesuvium portulacastrum (Fan et al. 2009). This implied that S-nitrosylated FBA might function in response to salt stress. The activities of FBA, GAPDH and PK were enhanced by GSNO treatment under salt stress compared to the NaCl treatment alone (Fig. 7a). In fact, the level of cellular protein S-nitrosylation is regulated by NO levels and this type of post-translational modification can affect protein/enzyme activity (Kovacs and Lindermayr 2013). In our results, high NO and SNO levels were detected after adding GSNO under salt stress, which could highlight the high possibility of S-nitrosylation. Thus, to some extent, there is a possibility that S-nitrosylation of glycolytic enzymes mentioned above may be activated and function in response to salt stress, which would be a fruitful area for future work in investigating whether S-nitrosylation of glycolytic enzyme members is involved in the glycolytic pathway and consequently regulates plant growth and development as well as stress responses. In addition, the expression levels of these enzymes S-nitrosylated at transcriptional level and the contents of related metabolites in glycolysis were significantly increased by GSNO under salt stress, which further confirmed the activation of glycolysis by NO in another way. This illustrated that NO responded to salt stress by modulating gene expressions and inducing metabolic changes in glycolysis pathway. Intriguingly, [(z)-1-[2-aminoethyl]-N-[2-ammonioethyl] amino] diazen-1-ium-1, 2 diolate (DETA-NO) has been found to protect plants from ATP reduction by accelerating the rate of glucose consumption (Almeida et al. 2001). Therefore, NO might alleviate salt toxicity via glycolytic activation and consequent ATP supplementation by post-translational modification, transcriptional and metabolic regulation, which is consistent with the data from He et al. (2018), who found that GSNO-mediated the activation of glycolysis by protein S-nitrosylation and transcriptional regulation, thereby protecting the seeds from energy deficiency.
Glycolysis and cycle can cause oxidative degradation of carbohydrates produced by photosynthesis, thereby providing plant cells with more energy and carbon sources (Li et al. 2015). In our study, the activities of key enzymes (MDH and CS) underwent S-nitrosylation in NaCl + GSNO treatment. Besides, ME, ICDH and ACO in the TCA cycle were found to be S-nitrosylated in all three treatments (Table 3) and their transcriptional levels were up-regulated by GSNO under salt stress (Fig. 7). Previously, the S-nitrosylated MDH was detected to participate in the aging of elm seeds (He et al. 2018). Moreover, MDH also underwent S-nitrosylation during GSNO-induced adventitious rooting in cucumber (Niu et al. 2019). Interestingly, in the cotyledons supernatant of sunflower seedlings subjected to salt stress, ACO, ICDH, CS and MDH were S-nitrosylated (Jain et al. 2018), implying that S-nitrosylation modification of these enzymes might be involved in growth and development of plant and adverse condition response. In our results, the contents of NO and SNO were detected when applying GSNO under salt stress (Fig. 2), implying the high possibility of S-nitrosylation. In addition, the level of cellular protein S-nitrosylation was modulated by NO and S-nitrosylation, the covalent attachment of an NO moiety to a reactive cysteine thiol of a protein to form an SNO, could affect protein activity, subcellular localization and protein–protein interaction as well as induce protein conformational changes (Kovacs and Lindermayr 2013). The activities of MDH and CS were obviously enhanced by applying GSNO under salt stress (Fig. 7). Thus, it is possible that MDH and CS might be activated through NO-induced S-nitrosylation and act as essential regulators in TCA cycles under salt stress. However, in contrast to this assumption, MDH was inactivated by S-nitrosylation in pea leaves (Ortega-Galisteo et al. 2012). The difference in results could probably be due to the different species and tissues used, and the concentrations, type and the expiration time of NO donors. Thus, further studies need be done to confirm the direct function of S-nitrosylated MDH and CS on themselves activity.
Besides, the transcriptional levels of proteins underwent S-nitrosylation and the contents of related metabolites in TCA cycle were increased by GSNO treatment under salt stress compared to NaCl treatment alone (Fig. 7). Interestingly, Bailey et al. (2019) explained that classical activation of macrophages (M (LPS + IFNƴ)) eliciting the expression of inducible NO synthase (iNOS) and generating large amounts of NO, elevated the levels of critical TCA cycle metabolites citrate and succinate in the pro-inflammatory macrophage. In addition, pyruvate, the final product of glycolysis and substrate in TCA cycle metabolism, is transported into the mitochondrial matrix by a specific transport protein on the mitochondrial inner membrane. The rate of pyruvate transport is vitally important to the respiratory metabolism of plants (Li et al. 2015). In our study, both the pyruvate content and PK activity were increased by GSNO treatment under salt stress (Fig. 7). These results implied that the TCA cycle might be activated by NO through S-nitrosylation modification, transcriptional and metabolic regulation in the process of NO-alleviating salt stress. The TCA cycle is composed by a set of enzymes through the mitochondrial respiratory chain. During plant growth and development, energy and appropriate building blocks are a basic requirement and mitochondrial respiration is an important source for growth and development of plants (Schwarzländer and Finkemeier 2013). Mitochondria participate in diverse processes such as metabolism of organic acids, amino acids, photorespiration, maintenance of reactive oxygen species homeostasis and programmed cell death (Gupta et al. 2011). In addition, mitochondria are the main target for oxidative damage in plant leaves (Bartoli et al. 2004). More importantly, mitochondria represent another essential source of NO in lower and higher plants, and leaf mitochondria may also produce NO (Gupta et al. 2011). As NO can reversely bind to cytochrome c oxidase and affect respiration, mitochondria are recognized not as only sources, but also important targets of NO (Gardner 2005). An earlier study showed that in plant cells NO affected mitochondrial functionality and decreased total cell respiration due to the inhibition of the cytochrome pathway (Zottini et al. 2002). However, plants contain an NO resistant alternative oxidase (AOX) in mitochondria to maintain non-ATP-coupled electron transport even in the presence of NO (Millar and Day 1996). Thus, this implied that the effect of NO on alleviating salt stress in tomato seedlings might also depend on targeting to mitochondria and then maintaining mitochondrial ATP production. In our results, we inferred that NO might activate the mitochondrial TCA cycle by S-nitrosylation modification to alleviate salt stress. Similarly, He et al. (2018) suggested that NO alleviated cell death through protein S-nitrosylation and transcriptional regulation during the aging of elm seeds. Thus, combining our findings, we inferenced that NO promoted glycolysis and activated the TCA cycle at post-translational and transcriptional levels, which produced more energy and glucose entered into the TCA cycle, thereby promoting the growth of tomato seedlings under salt stress. However, the identification of specific S-nitrosylated sites of proteins that were identified to undergo S-nitrosylation in carbohydrate metabolism in our study and the impact of mutations at site mutation on protein activity or salt tolerance needs to be explored in the future.
Conclusion
In conclusion, we found that NO may promote the growth of tomato seedlings during salt stress. In this process, endogenous S-nitrosylation levels and protein S-nitrosylation were regulated. Importantly, 15 and 22 S-nitrosylated proteins were involved in MAPK signal transduction and carbohydrate metabolism, respectively. Various proteins in MAPK signaling were modulated by NO through the regulation of ethylene production and ABA signaling at transcriptional regulation in response to salt stress, thereby reducing ABA level and ethylene biosynthesis and then alleviating salt toxicity in tomato seedlings (Fig. 8). In addition, NO might promote glycolysis and activate the TCA cycle through S-nitrosylation and transcriptional regulation, which reduced energy deficiency and protected the tomato seedlings from salt toxicity (Fig. 9). A possible model is that NO beneficially regulated MAPK signaling at transcriptional levels and activated carbohydrate metabolism by protein S-nitrosylation modification, transcriptional and metabolic regulation, protecting tomato seedlings from salt toxicity, thus promoting the growth of tomato seedlings (Figs. 8 and 9). Thus, future research about the function of NO-mediated protein S-nitrosylation and the mutation validation of specific S-nitrosylation sites in response to salt stress needs to be provided.
Diagram of the NO-induced metabolic changes in MAPK signaling under salt stress. The C (control), N (NaCl) and N + G (NaCl + GSNO) treatments were illustrated using three linked squares. The square is filled blue if one protein was S-nitrosylated at the indicated treatment, and the square remained blank if S-nitrosylation was not detected. A GSNO-mediated decrease in gene expressions under salt stress is indicated with a green arrow. The decrease in the metabolite levels is indicated with green filled arrowhead
Diagram of the NO-induced metabolic changes in carbohydrate metabolism by S-nitrosylation under salt stress. The C (control), N (NaCl) and N + G (NaCl + GSNO) treatments were illustrated using three linked squares. The square is filled blue if one protein was S-nitrosylated at the indicated treatment, and the square remained blank if S-nitrosylation was not detected. A GSNO-mediated increase in gene expression under salt stress is indicated with a red arrow. The increase in the metabolite levels is indicated with red filled arrowhead, and a GSNO-mediated increase in enzyme activity under salt stress is indicated with red plus signs
Author contribution statement
Conception and design: Weibiao Liao designed the study. Lijuan Wei wrote the manuscript. Lijuan Wei, Jing Zhang, Shouhui Wei, Yuzheng Deng, Huwei Liu, Dongliang Hu, Wenting Gong and Ying Pan performed the experiments, analyzed, and interpreted the data. Lijuan Wei, Weibiao Liao and Chunlei Wang participated in data interpretation and revised the manuscript.
Data availability
All data generated or analyzed during this study are included in this manuscript.
Abbreviations
- GSNO:
-
S-Nitrosoglutathione
- SNO:
-
S-Nitrosothiol
- MAPK:
-
Mitogen-activated protein kinase
- TCA:
-
Tricarboxylic acid cycle
- cPTIO:
-
2-(4-Carboxy-2-phenyl)-4, 4, 5, 5-tetramethylimidazoline-1-oxyl-3-oxide
- PK:
-
Pyruvate kinase
- GAPDH:
-
Glyceraldehyde-3-phosphate dehydrogenase
- CS:
-
Citrate synthase
- FBA:
-
Fructose-bisphosphate aldolase
- MDH:
-
Malate dehydrogenase
- SnRK:
-
Serine/threonine-protein kinase
- ICDH:
-
Isocitrate dehydrogenase
- ME:
-
Malic enzyme
- SAM:
-
S´Adenosyl-L-methionine
References
Abat JK, Mattoo AK, Deswal R (2010) S-nitrosylated proteins of a medicinal CAM plant Kalanchoe pinnata -ribulose-1, 5-bisphosphate carboxylase/oxygenase activity targeted for inhibition. FEBS J 275:2862–2872. https://doi.org/10.1111/j.1742-4658.2008.06425.x
Almeida A, Almeida J, Bolaños JP, Moncada S (2001) Different responses of astrocytes and neurons to nitric oxide: the role of glycolytically generated ATP in astrocyte protection. Proc Natl Acad Sci USA 98:15294–15299. https://doi.org/10.1073/pnas.261560998
Asai S, Ohta K, Yoshioka H (2008) MAPK signaling regulates nitric oxide and NADPH oxidase-dependent oxidative bursts in Nicotiana benthamiana. Plant Cell 20:1390–1406. https://doi.org/10.1105/tpc.107.055855
Bailey JD, Diotallevi M, Nicol T, McNeill E, Shaw A, Chuaiphichai S, Crabtree MJ (2019) Nitric oxide modulates metabolic remodeling in inflammatory macrophages through TCA cycle regulation and itaconate accumulation. Cell Rep 28:218–230. https://doi.org/10.1016/j.celrep.2019.06.018
Bartoli CG, Gómez F, Martinez DE, Guiamet JJ (2004) Mitochondria are the main target for oxidative damage in leaves of wheat (Triticum aestivum L.). J Exp Bot 55:1663–1669. https://doi.org/10.1093/jxb/erh199
Baud S, Wuillème S, Dubreucq B, De Almeida A, Vuagnat C, Lepiniec L, Rochat C (2007) Function of plastidial pyruvate kinases in seeds of Arabidopsis thaliana. Plant J 52:405–419. https://doi.org/10.1111/j.1365-313X.2007.03232.x
Chen H, Osuna D, Colville L, Lorenzo O, Graeber K, Küster H, Leubner-Metzger G, Kranner I (2013) Transcriptome-wide mapping of pea seed ageing reveals a pivotal role for genes related to oxidative stress and programmed cell death. PLoS ONE 8:e78471. https://doi.org/10.1371/journal.pone.0078471
Chen K, Li GJ, Bressan RA, Song CP, Zhu JK, Zhao Y (2020) Abscisic acid dynamics, signaling, and functions in plants. J Integr Plant Biol 62:25–54. https://doi.org/10.1111/jipb.12899
Chou KC, Shen HB (2010) Cell-PLoc 2.0: an improved package of web-servers for predicting subcellular localization of proteins in various organisms. Nat Sci 2:1090. https://doi.org/10.4236/ns.2010.210136
Fan W, Zhang Z, Zhang Y (2009) Cloning and molecular characterization of fructose-1, 6-bisphosphate aldolase gene regulated by high-salinity and drought in Sesuvium portulacastrum. Plant Cell Rep 28:975–984. https://doi.org/10.1007/s00299-009-0702-6
Fares A, Rossignol M, Peltier JB (2011) Proteomics investigation of endogenous S-nitrosylation in Arabidopsis. Biochem Biophys Res Comm 416:331–336. https://doi.org/10.1016/j.bbrc.2011.11.036
Feng J, Wang C, Chen Q, Chen H, Ren B, Li X, Zuo J (2013a) S-nitrosylation of phosphotransfer proteins represses cytokinin signaling. Nat Commun 4:1–9. https://doi.org/10.1038/ncomms2541
Feng X, Sun T, Bei Y, Ding S, Zheng W, Lu Y, Shen P (2013b) S-nitrosylation of ERK inhibits ERK phosphorylation and induces apoptosis. Sci Rep 3:1–6. https://doi.org/10.1038/ncomms2541
Frungillo L, Skelly MJ, Loake GJ, Spoel SH, Salgado I (2014) S-nitrosothiols regulate nitric oxide production and storage in plants through the nitrogen assimilation pathway. Nat Commun 5:5401. https://doi.org/10.1038/ncomms6401
Gao R, Luo Y, Yun F, Wu X, Wang P, Liao W (2021) Genome-wide identification, expression profile, and alternative splicing analysis of CAMTA family genes in cucumber (Cucumis sativus L.). Agronomy 11:1827. https://doi.org/10.3390/agronomy11091827
Gardner PR (2005) Nitric oxide dioxygenase function and mechanism of flavohemoglobin, hemoglobin, myoglobin and their associated reductases. J Inorg Biochem 99:247–266. https://doi.org/10.1016/j.jinorgbio.2004.10.003
Gong B, Wen D, Wang X, Wei M, Yang F, Li Y, Shi Q (2015) S-nitrosoglutathione reductase-modulated redox signaling controls sodic alkaline stress responses in Solanum lycopersicum L. Plant Cell Physiol 56:790–802. https://doi.org/10.1093/pcp/pcv007
Guo L, Ma F, Wei F, Fanella B, Allen DK, Wang X (2014) Cytosolic phosphorylating glyceraldehyde-3-phosphate dehydrogenases affect Arabidopsis cellular metabolism and promote seed oil accumulation. Plant Cell 26:3023–3035. https://doi.org/10.1105/tpc.114.126946
Gupta KJ, Igamberdiev AU, Manjunatha G, Segu S, Moran JF, Neelawarne B, Kaiser WM (2011) The emerging roles of nitric oxide (NO) in plant mitochondria. Plant Sci 181:520–526. https://doi.org/10.1016/j.plantsci.2011.03.018
He Y, Xue H, Li Y, Wang X (2018) NO alleviated cell death through protein S-nitrosylation and transcriptional regulation during the ageing of elm seeds. J Exp Bot 142:1–15. https://doi.org/10.1093/jxb/ery270
Hu J, Huang X, Chen L, Sun X, Lu C, Zhang L, Zuo J (2015) Site-specific nitrosoproteomic identification of endogenously S-nitrosylated proteins in Arabidopsis. Plant Physiol 167:1731–1746. https://doi.org/10.1104/pp.15.00026
Hussain S, Junhua Z, Chu Z, Lian-Feng Z, Xiao-Chuang C, Sheng-Miao YU (2017) Effects of salt stress on rice growth, development characteristics, and the regulating ways: a review. J Integr Agr 16:2357–2374
Isayenkov SV, Maathuis FJM (2019) Plant salinity stress: many unanswered questions remain. Front Plant Sci 66:1489–1498. https://doi.org/10.3389/fpls.2019.00080
Jain P, Toerne CV, Lindermayr C, Bhatla SC (2018) S-nitrosylation/denitrosylation as a regulatory mechanism of salt stress sensing in sunflower seedlings. Physiol Plant 162:49–72. https://doi.org/10.1111/ppl.12641
Khan MN, Alamri S, Al-Amri AA, Alsubaie QD, Siddiqui MH (2020) Effect of nitric oxide on seed germination and seedling development of tomato under chromium toxicity. J Plant Growth Regul 40:2358–2370. https://doi.org/10.1007/s00344-020-10212-2
Kovacs I, Lindermayr C (2013) Nitric oxide-based protein modification: formation and site-specificity of protein S-nitrosylation. Front Plant Sci 4:137. https://doi.org/10.3389/fpls.2013.00137
Li L, Xing WW, Shao QS, Shu S, Sun J, Guo SR (2015) The effects of grafting on glycolysis and the tricarboxylic acid cycle in Ca(NO3)2- stressed cucumber seedlings with pumpkin as rootstock. Acta Physiol Plant 37:1–10. https://doi.org/10.1007/s11738-015-1978-5
Liao WB, Zhang ML, Yu JH (2013) Role of nitric oxide in delaying senescence of cut rose flowers and its interaction with ethylene. Sci Hortic 155:30–38. https://doi.org/10.1016/j.scienta.2013.03.005
Lin A, Wang Y, Tang J, Xue P, Li C, Liu L, Hu B, Yang F, Loake GJ, Chu C (2012) Nitric oxide and protein S-nitrosylation are integral to hydrogen peroxide-induced leaf cell death in rice. Plant Physiol 158:451–464. https://doi.org/10.1104/pp.111.184531
Lindermayr C, Saalbach G, Bahnweg G, Durner J (2006) Differential inhibition of Arabidopsis methionine adenosyltransferases by protein S-nitrosylation. J Biol Chem 281:4285–4291. https://doi.org/10.1074/jbc.M511635200
Lindström A, Nyström C (1987) Seasonal variation in root hardiness in container grown Scots pine, Norway spruce, and Lodgepole pine seedlings. Can J Forest Res 17:787–793. https://doi.org/10.1139/x87-126
Maldonado-Alconada AM, Echevarría-Zome S, Lindermayr C, Redondo- López I, Durner J, Jorrin-Novo JV (2011) Proteomic analysis of Arabidopsis protein S-nitrosylation in response to inoculation with Pseudomonas syringae. Acta Physiol Plant 33:1493–1514. https://doi.org/10.1007/s11738-010-0688-2
Millar AH, Day DA (1996) Nitric oxide inhibits the cytochrome oxidase but not the alternative oxidase of plant mitochondria. FEBS Lett 398:155–158. https://doi.org/10.1016/S0014-5793(96)01230-6
Mioto PT, Rodríguez-Ruiz M, Mot AC, Zuccarelli R, Corpas FJ, Freschi L, Mercier H (2017) Alternative fluorimetric-based method to detect and compare total S-nitrosothiols in plants. Nitric Oxide 68:7–13. https://doi.org/10.1016/j.niox.2017.03.001
Niu L, Yu J, Liao W, Xie J, Yu J, Lv J, Wu Y (2019) Proteomic investigation of S-nitrosylated proteins during NO-induced adventitious rooting of cucumber. Int J Mol Sci 20:5363. https://doi.org/10.3390/ijms20215363
Ortega-Galisteo AP, Rodríguez-Serrano M, Pazmiño DM, Gupta DK, Sandalio LM, Romero-Puertas MC (2012) S-Nitrosylated proteins in pea (Pisum sativum L.) leaf peroxisomes: Changes under abiotic stress. J Exp Bot 63:2089–2103. https://doi.org/10.1093/jxb/err414
Pan CL, Li X, Yao SC, Luo SZ, Liu SY, Wang AQ, Xiao D, Zhan J, He LF (2021) S-nitrosated proteomic analysis reveals the regulatory roles of protein S-nitrosation and S-nitrosoglutathione reductase during Al-induced PCD in peanut root tips. Plant Sci 308:110931
Petřivalský M, Kubienová L, Tichá T, Luhová L (2015) S-nitrosoglutathione reductase: a key regulator of S-nitrosylation in plant development and stress responses. In: Gupta KJ, Igamberdiev AU (eds) Reactive oxygen and nitrogen species signaling and communication in plants. Springer, Cham, pp 283–299
Piacentini D, Ronzan M, Fattorini L, Della Rovere F, Massimi L, Altamura MM, Falasca G (2020) Nitric oxide alleviates cadmium-but not arsenic-induced damages in rice roots. Plant Physiol Biochem 151:729–742. https://doi.org/10.1016/j.plaphy.2020.04.004
Qi Q, Dong Y, Liang Y, Li K, Xu H, Sun X (2020) Overexpression of SlMDHAR in transgenic tobacco increased salt stress tolerance involving S-nitrosylation regulation. Plant Sci 299:110609. https://doi.org/10.1016/j.plantsci.2020.110609
Qiu C, Sun J, Wang Y, Sun L, Xie H, Ding Y, Ding Z (2019) First nitrosoproteomic profiling deciphers the cysteine S-nitrosylation involved in multiple metabolic pathways of tea leaves. Sci Rep 9:17525. https://doi.org/10.1038/s41598-019-54077-2
Roeder S, Dreschler K, Wirtz M, Cristescu SM, van Harren FJ, Hell R, Piechulla B (2009) SAM levels, gene expression of SAM synthetase, methionine synthase and ACC oxidase, and ethylene emission from N. suaveolens flowers. Plant Mol Biol 70:535–546. https://doi.org/10.1007/s11103-009-9490-1
Sami F, Siddiqui H, Hayat S (2020) Nitric oxide-mediated enhancement in photosynthetic efficiency, ion uptake and carbohydrate metabolism that boosts overall photosynthetic machinery in mustard plants. J Plant Growth Regul 40:1088–1110. https://doi.org/10.1007/s00344-020-10166-5
Schwarzländer M, Finkemeier I (2013) Mitochondrial energy and redox signaling in plants. Antioxid Redox Signal 18:2122–2144. https://doi.org/10.1089/ars.2012.5104
Sehar Z, Masood A, Khan NA (2019) Nitric oxide reverses glucose-mediated photosynthetic repression in wheat (Triticum aestivum L.) under salt stress. Environ Exp Bot 161:277–289. https://doi.org/10.1016/j.envexpbot.2019.01.010
Sehrawat A, Deswal R (2014) S-nitrosylation analysis in Brassica juncea apoplast highlights the importance of nitric oxide in cold-stress signaling. J Proteome Res 13:2599–2619. https://doi.org/10.1021/pr500082u
Sehrawat A, Abat JK, Deswal R (2013a) RuBisCO depletion improved proteome coverage of cold responsive S-nitrosylated targets in Brassica juncea. Front Plant Sci 4:342. https://doi.org/10.3389/fpls.2013.00342
Sehrawat A, Gupta R, Deswal R (2013b) Nitric oxide–cold stress signaling cross-talk, evolution of a novel regulatory mechanism. Proteomics 13:1816–1835. https://doi.org/10.1002/pmic.201200445
Singh PK, Indoliya Y, Chauhan AS, Singh SP, Singh AP, Dwivedi S, Chakrabarty D (2017) Nitric oxide mediated transcriptional modulation enhances plant adaptive responses to arsenic stress. Sci Rep 7:3592. https://doi.org/10.1038/s41598-017-03923-2
Tang B, Yin CY, Wang YJ, Sun YY, Liu Q (2016) Positive effects of night warming on physiology of coniferous trees in late growing season: leaf and root. Acta Oecol 73:21–30. https://doi.org/10.1016/j.actao.2016.02.002
Tilbrook K, Poirier Y, Gebbie L, Schenk PM, McQualter RB, Brumbley SM (2014) Reduced peroxisomal citrate synthase activity increases substrate availability for polyhydroxyalkanoate biosynthesis in plant peroxisomes. Plant Biotechnol J 12:1044–1052. https://doi.org/10.1111/pbi.12211
Wang P, Du Y, Hou YJ, Zhao Y, Hsu CC, Yuan F, Zhu JK (2015a) Nitric oxide negatively regulates abscisic acid signaling in guard cells by S-nitrosylation of OST1. Proc Natl Acad Sci USA 112:613–618. https://doi.org/10.1073/pnas.1423481112
Wang P, Zhu JK, Lang Z (2015b) Nitric oxide suppresses the inhibitory effect of abscisic acid on seed germination by S-nitrosylation of SnRK2 proteins. Plant Signal Behav 10:e1031939. https://doi.org/10.1080/15592324.2015.1031939
Wang J, Wang Y, Lv Q, Wang L, Du J, Bao F, He YK (2017) Nitric oxide modifies root growth by S-nitrosylation of plastidial glyceraldehyde-3-phosphate dehydrogenase. Biochem Biophys Res Comm 488:88–94. https://doi.org/10.1016/j.bbrc.2017.05.012
Wang C, Fang H, Gong T, Zhang J, Niu L, Huang D, Liao W (2020) Hydrogen gas alleviates postharvest senescence of cut rose ‘Movie star’ by antagonizing ethylene. Plant Mol Biol 102:271–285. https://doi.org/10.1007/s11103-019-00946-3
Wawer I, Bucholc M, Astier J, Anielska-Mazur A, Dahan J, Kulik A, Wendehenne D (2010) Regulation of Nicotiana tabacum osmotic stress-activated protein kinase and its cellular partner GAPDH by nitric oxide in response to salinity. Biochem J 429:73–83. https://doi.org/10.1042/BJ20100492
Wei L, Zhang M, Wei S, Zhang J, Liao W (2020) Roles of nitric oxide in heavy metal stress in plants: cross-talk with phytohormones and protein S-nitrosylation. Environ Pollut 259:113943. https://doi.org/10.1016/j.envpol.2020.113943
Wei L, Wang C, Liao W (2021) Hydrogen sulfide improves the vase life and quality of cut roses and chrysanthemums. J Plant Growth Regul 40:2532–2547. https://doi.org/10.1007/s00344-021-10312-7
Xin X, Tian Q, Yin G, Chen X, Zhang J, Ng S, Lu X (2014) Reduced mitochondrial and ascorbate–glutathione activity after artificial ageing in soybean seed. J Plant Physiol 171:140–147. https://doi.org/10.1016/j.jplph.2013.09.016
Xu J, Li Y, Wang Y, Liu H, Ren D (2008) Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J Biol Chem 283:26996–27006. https://doi.org/10.1074/jbc.M801392200
Xuan W, Xu S, Li M, Han B, Zhang B, Zhang J, Lin Y, Huang J, Shen W, Cui J (2012) Nitric oxide is involved in hemin-induced cucumber adventitious rooting process. J Plant Physiol 169:1032–1039. https://doi.org/10.1016/j.jplph.2012.02.021
Yan M, Yao Y, Mou K, Dan Y, Li W, Wang C, Liao W (2022) The involvement of abscisic acid in hydrogen gas-enhanced drought resistance in tomato seedlings. Sci Hortic 292:110631. https://doi.org/10.1016/j.scienta.2021.110631
Yoo SD, Cho YH, Tena G, Xiong Y, Sheen J (2008) Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signaling. Nature 451:789–795. https://doi.org/10.1038/nature06543
Yun BW, Skelly MJ, Yin M, Yu M, Mun BG, Lee SU, Loake GJ (2016) Nitric oxide and S-nitrosoglutathione function additively during plant immunity. New Phytol 211:516–526. https://doi.org/10.1111/nph.13903
Zhang A, Jiang M, Zhang J, Ding H, Xu S, Hu X, Tan M (2007) Nitric oxide induced by hydrogen peroxide mediates abscisic acid-induced activation of the mitogen-activated protein kinase cascade involved in antioxidant defense in maize leaves. New Phytol 175:36–50. https://doi.org/10.1111/j.1469-8137.2007.02071.x
Zhang Y, Long Y, Liu Y, Yang M, Wang L, Liu X, Luo Y (2022) MAPK5 and MAPK10 overexpression influences strawberry fruit ripening, antioxidant capacity and resistance to Botrytis cinerea. Planta 255:19. https://doi.org/10.1007/s00425-021-03804-z
Zhao Z, Li C, Liu H, Liao W (2021) The involvement of glucose in hydrogen gas-medicated adventitious rooting in cucumber. Plants 10:1937. https://doi.org/10.3390/plants10091937
Zottini M, Formentin E, Scattolin M, Carimi F, Lo Schiavo F, Terzi M (2002) Nitric oxide affects plant mitochondrial functionality in vivo. FEBS Lett 515:75–78. https://doi.org/10.1016/S0014-5793(02)02438-9
Acknowledgements
This study was supported by the National Key Research and Development Program (2018YFD1000800); the National Natural Science Foundation of China (Nos. 32072559, 31860568, 31560563 and 31160398); the Research Fund of Higher Education of Gansu, China (Nos. 2018C-14 and 2019B-082); the Post-Doctoral Foundation of China (Nos. 20100470887 and 2012T50828) and the Natural Science Foundation of Gansu Province, China (Nos. 1606RJZA073 and 1606RJZA077).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Dorothea Bartels.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wei, L., Zhang, J., Wei, S. et al. Nitric oxide alleviates salt stress through protein S-nitrosylation and transcriptional regulation in tomato seedlings. Planta 256, 101 (2022). https://doi.org/10.1007/s00425-022-04015-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-022-04015-w