Abstract
Main conclusion
Arundo donax ecotypes react differently to salinity, partly due to differences in constitutive defences and methylome plasticity.
Abstract
Arundo donax L. is a C3 fast-growing grass that yields high biomass under stress. To elucidate its ability to produce biomass under high salinity, we investigated short/long-term NaCl responses of three ecotypes through transcriptional, metabolic and DNA methylation profiling of leaves and roots. Prolonged salt treatment discriminated the sensitive ecotype ‘Cercola’ from the tolerant ‘Domitiana’ and ‘Canneto’ in terms of biomass. Transcriptional and metabolic responses to NaCl differed between the ecotypes. In roots, constitutive expression of ion transporter and stress-related transcription factors’ genes was higher in ‘Canneto’ and ‘Domitiana’ than ‘Cercola’ and 21-day NaCl drove strong up-regulation in all ecotypes. In leaves, unstressed ‘Domitiana’ confirmed higher expression of the above genes, whose transcription was repressed in ‘Domitiana’ but induced in ‘Cercola’ following NaCl treatment. In all ecotypes, salinity increased proline, ABA and leaf antioxidants, paralleled by up-regulation of antioxidant genes in ‘Canneto’ and ‘Cercola’ but not in ‘Domitiana’, which tolerated a higher level of oxidative damage. Changes in DNA methylation patterns highlighted a marked capacity of the tolerant ‘Domitiana’ ecotype to adjust DNA methylation to salt stress. The reduced salt sensitivity of ‘Domitiana’ and, to a lesser extent, ‘Canneto’ appears to rely on a complex set of constitutively activated defences, possibly due to the environmental conditions of the site of origin, and on higher plasticity of the methylome. Our findings provide insights into the mechanisms of adaptability of A. donax ecotypes to salinity, offering new perspectives for the improvement of this species for cultivation in limiting environments.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Global warming is strongly exacerbating salinity of arable lands, since water evaporation causes over accumulation of sodium (Na+) and chloride (Cl−) in the soil (Ismail et al. 2014). Giant reed (Arundo donax L.), a perennial weed that is increasing cultivated for ligno-cellulosic biomass production in temperate climates, possesses large environmental plasticity, being able to survive in suboptimal conditions and to give high yields even in harsh environments. The ability of A. donax to tolerate diverse abiotic stresses, and mainly salt stress, has been reported in several studies (Lewandowski et al. 2003; Papazoglou 2007; Nackley and Kim 2015; Sánchez et al. 2015; De Stefano et al. 2017), but the investigation at the genomic and metabolic level are just at the beginning (Malone et al. 2017). Since genetic diversity in this species is reportedly scarce (Ahmad et al. 2008; Mariani et al. 2010; Saltonstall et al. 2010; Pilu et al. 2014; Guarino et al. 2019), also because of its obligatory asexual mode of reproduction, it is still matter of debate whether specimens of A. donax collected in different environments, defined as ecotypes (Haworth et al. 2017c), may have different physiological adaptation to stress in function of genetic differences or of the original living conditions.
Excess salinity reduces soil water potential, thus impacting on plant water uptake and resulting in water deficiency and osmotic stress. Moreover, Na+ and, to a lesser extent, Cl− are toxic to plant cells, causing reduced photosynthesis, oxidative damage, nutritional imbalance, and metabolic changes (Hasegawa et al. 2000; Negrao et al. 2017). Perception of high ionic concentrations activates stress pathways that overhang the normal developmental process and may impair plant growth and productivity or even lead to plant death. Salinity tolerance is a complex trait, triggered by several genes involved in osmoregulation, exclusion of toxic ions and tissue tolerance and controlled by different genetic pathways (DeRose-Wilson and Gaut 2011). A key component of osmotic regulation is the SOS (Salt Overly Sensitive) pathway (Qiu et al. 2002; Yang et al. 2009). Under salt stress, the protein kinases SOS2 and SOS3 activate a Ca2+-dependent signalling cascade, which promotes Na+ efflux from the cells by SOS1 (Na+/H+ antiporter) as well as abscisic acid (ABA) signalling involved in root to shoot communication of salinity stress (Munns and Tester 2008; Zhu 2016). Another crucial salt tolerance mechanism is mediated by the high-affinity plasma membrane K+ channel (HKT), which alleviates Na+ toxicity by promoting Na+ efflux and K+ uptake in the cytoplasm (Davenport et al. 2007; Kobayashi et al. 2017). To avoid negative effects on cell metabolism, excess sodium in the cytoplasm is also directed to the vacuole by the tonoplast Na+/H+ antiporter (NHX), whose activity is positively regulated by SOS2. Lately, several members of the NHX family emerged to have an equally important role as H+/K+ exchanger, thus contributing to maintain higher level of cytosolic K and to preserve K-dependent metabolic processes, such as protein synthesis (Moller et al. 2009).
Besides maintenance of ion homeostasis, stress signals activate ABA-dependent and ABA-independent transcription factors (TFs), mainly of the dehydration-responsive element binding (DREB), NAC, WRKY and MYB families (Golldack et al. 2014), that assist the above-reported pathways but also trigger and orchestrate transduction pathways tightly linked to cell metabolism and adaptive mechanisms under high salinity, including accumulation of antioxidant molecules (Sairam and Tyagi 2004; Gill and Tuteja 2010). Salt stress induces reactive oxygen species (ROS), such as O2−, ∙OH, ∙O2− and H2O2 (Price et al. 1989; Moran et al. 1994; Mittler 2002), which cause oxidative damage of membranes and macromolecules. To scavenge these toxic compounds, plants have evolved robust antioxidant defences through enzymatic mechanisms, including superoxide dismutase (SOD), whose H2O2 production is further scavenged by catalase (CAT) activity, and non-enzymatic mechanisms, e.g. accumulation of ascorbic acid, glutathione, which is regenerated by glutathione reductase (GR), and polyphenolic compounds.
Recently, epigenetic mechanisms such as histone modifications, production of small non-coding RNAs, and DNA methylation have been reported to be central in adaptation processes when plants are challenged by stressful environmental cues (Deinlein et al. 2014; Ferreira et al. 2015) and were proposed to underlie the remarkable phenotypic plasticity of A. donax (Guarino et al. 2019). Growing evidences indicate DNA cytosine methylation as a crucial mechanism helping individuals to cope with abiotic stress, including salinity, through regulation of gene expression. For example, mangrove plants growing in contrasting natural habitats, such as periodic drought and hyper-saline soils, differed with respect to cytosine methylation despite little genetic differentiation (Lira-Medeiros et al. 2010). However, the involvement of methylation/de-methylation processes in salinity stress is still poorly understood.
Here, we aimed to analyse the response under short- and long-term salt treatment of three A. donax ecotypes displaying different sensitivity to NaCl (De Stefano et al. 2017). We characterised local (root) and distal (leaf) morpho-physiological parameters and gene expression of salt stress-responsive key genes and the gain/loss of metabolites notoriously involved in salt tolerance, such as proline, ABA, malondialdehyde (MDA) and antioxidants. Moreover, to understand whether epigenomic changes may have contributed to the observed variability in the NaCl response of these A. donax ecotypes, we characterised the root and leaf changes in total DNA methylation during short and long exposure to NaCl stress. Our results highlighted different morpho-physiological, transcriptional and metabolic responses to NaCl of the three ecotypes, which may account for their different levels of NaCl tolerance. We found that giant reed ecotypes were able to rapidly change DNA methylation (either reducing or increasing) upon stress imposition, displaying differential methylome flexibility.
Materials and methods
Plant growth and experimental design
The plant genetic material screened in this paper was collected in different hydrogeological basins in the Italian Campania region (Supplementary Table S1). The sampled stand of reeds from each collection site, namely ‘Canneto’, ‘Cercola’ and ‘Domitiana’, was considered to be one ecotype. For each experiment, plant material was re-collected in each original site. The data presented correspond to one representative experiment, which was repeated three times with similar results.
Portions of reeds of about 30 cm were put in large plastic trays filled with weekly refreshed tap water in a temperature-controlled greenhouse of the CNR-IBBR Portici (27 °C day/24 °C night). After about 40 days, rooted sprouts produced from the nodes were collected and transferred in vermiculite-filled black-seed trays (4 × 6 cm pots) in hydroponic float system, blowing air with aquarium aerators. Plants were kept in half-strength modified Hoagland nutrient solution (1.23 mS cm−1 electrical conductivity, EC) for about 30 days, followed by four additional days in full-strength-modified nutrient solution (electric conductivity of 1.92 mS cm−1) (Hoagland and Arnon 1950) as described by De Stefano et al. (2017). The obtained plants were then randomly divided between control and NaCl treatments. For the latter, NaCl was added stepwise in 3 consecutive days up to 150 mM (electric conductivity of 18.8 mS cm−1) and maintained for 21 days. A completely randomized block design, with 12-replica plants per thesis (ecotype × treatment) was adopted.
For RNA extraction and biochemical analyses, A. donax leaves and roots were collected after 2 and 21 days of exposure to 0 or 150 mM NaCl.
Evaluation of salt tolerance
Arundo donax response to NaCl stress was evaluated by checking six biometric parameters after the 21-day NaCl treatment, i.e. stem height, leaf number, fourth leaf width, main root length, shoot and root fresh weight (FW). All values are presented as mean ± SE. For mean comparison and statistical significance, pairwise t test was performed between stressed and control plants within each ecotype.
Leaf greenness was measured using a portable chlorophyll meter (SPAD 502 Plus Meter, Konica Minolta, Tokyo, Japan) in exposed to 2 and 21 days of 0 mM (control) or 150 mM NaCl (stress treatment) treatment. SPAD readings were the average of five measurements from the middle of the second or third last fully collared leaf for each plant. For each ecotype/treatment, SPAD values are expressed as mean ± SD of four plants.
A stress susceptibility index (SSI) was calculated according to the following relationship [(1 − (Ys/Yp))]/[(1 − (Xs/Xp))] (Sánchez et al. 2015), where Ys, Yp, represent parameter under stress, parameter under non-stress conditions for each ecotype, and Xs and Xp parameter mean under stress and parameter mean under non-stress conditions for all ecotypes. The results for stem height, leaf number, fourth leaf width, and shoot FW, were averaged and used to classify the ecotypes in terms of tolerance/sensitivity to NaCl stress.
RNA extraction and qRT-PCR
Samples were ground in liquid nitrogen and total RNA was extracted from 100 mg of tissue using the RNeasy kit (Qiagen). After visual check of RNA integrity on agarose gels and quantification using a NanoDrop ND-8000 spectrophotometer (Fisher Scientific, Waltham, MA, USA), 1 μg of total RNA was reversed transcribed using the QuantiTec Reverse Transcription Kit (Qiagen), according to the manufacturer’s instructions. Real-time qRT-PCR was performed with Platinum® SYBR® Green qPCR Super Mix (Thermo Fisher Scientific) in a ABI7900 HT (Thermo Fisher Scientific). Each PCR reaction (20 μL) contained 10 μL real-time qRT-PCR Mix, 4 μL of a 1:25 dilution of cDNA and 0.25 μM of each specific primer. The thermal cycling conditions were 50 °C for 2 min, 95 °C for 2 min, followed by 40 cycles of 15 s at 95 °C and 30 s at 60 °C. PCR product melting curves were analysed for the presence of a single peak. All reactions were performed on biological triplicates and technical duplicates and fold change measurements calculated with the 2−ΔΔCT method (Pfaffl 2001, 2004). The ‘Cercola’ ecotype in control condition was used as internal calibrator. Gene expression was normalized on the stably expressed A. donax elongation factor gene (Poli et al. 2017). For primer design, identification of putative A. donax stress-related genes was based on homology searches. Well-known Arabidopsis thaliana stress-responsive genes from the Arabidopsis Stress Responsive Gene Database available at http://srgdb.bicpu.edu.in/ (ASRGDB; Borkotoky et al. 2013) were used to identify the putative homologs and to assess transcript homolog coverage and sequence conservation across species phylogenetically related to A. donax by searching the Phytozome database (Goodstein et al. 2012) with BLASTx with an E value cut-off of 1 × 103 and sequence coverage greater than 50% of the subject length. The retrieved sequences were then used to run TBLASTN homology searches against the A. donax transcriptome available in NCBI at https://www.ncbi.nlm.nih.gov/nuccore/?term=Arundo+donax with an E value cut-off of 1 × 103 and sequence identity greater than 50%. Whenever possible, the giant reed sequences were also verified in other transcriptomic databases (Sablok et al. 2014; Barrero et al. 2015; Fu et al. 2016; Evangelistella et al. 2017). The primers designed on the retrieved A. donax sequences and used for real-time qRT-PCR are provided in Supplementary Table S2, together with the relative homologs of A. thaliana and monocot species.
Proline and free ABA determination
Leaf samples (250 mg) were homogenized in liquid nitrogen and extracted with acidic ninhydrin reagent following a procedure previously described by Claussen (2005). Proline concentration was determined from a standard curve and calculated on a fresh weight basis (μmol proline g−1 FW) using three biological and three technical replicates.
For ABA extraction, freeze-dried samples (150 mg) were homogenized in liquid nitrogen and incubated in 2 mL of water overnight at 4 °C. After sample centrifugation at 9279g for 10 min, quantitative free ABA determination was performed on three biological triplicates and technical duplicates by the competitive ELISA Phytodetek ABA test kit (Agdia-Biofords, Evry, France) following the provider’s protocol. Colour absorbance was read at 405 nm using a plate auto-reader (1420 Multilabel Counter Victor3TM, PerkinElmer) and free ABA content was expressed in nmol g−1 FW.
Determination of total antioxidant activity (TEAC) and lipid peroxidation
Leaf samples were homogenized (250 mg) and extracted twice with water (hydrophilic antioxidants). The residue was re-extracted twice in acetone (lipophilic antioxidants). The TEAC assay was performed as previously described by Butelli et al. (2008). Results were expressed as mmol Trolox kg−1 FW. TEAC was measured on three biological replicates and three technical replicates.
Malondialdehyde content was quantified by the thiobarbituric acid-reactive substance assay (Heath and Packer 1968). Briefly, 100 mg of freeze-dried leaf tissue was homogenized in liquid nitrogen from control and salt-treated plants, and vortexed, for few seconds, in 5 mL of 5% (w/v) trichloroacetic acid. The homogenate was centrifuged at 12,000g for 10 min at room temperature. Supernatant was mixed with an equal volume of thiobarbituric acid [0.5% in 20% (w/v) trichloroacetic acid] and the mixture was then boiled for 25 min at 100 °C. Then, centrifugation for 5 min at 7500g was performed to clarify the solution. Absorbance of the supernatant was measured at 532 nm. MDA equivalents were calculated by the extinction coefficient of 155 mM−1 cm and expressed as nmol MDA g−1 FW.
Methylation-sensitive amplified polymorphism (MSAP) profiling and data analysis
Total DNA was extracted from 50 mg of leaves using the Qiagen Plant DNeasy Kit according to the manufacturer’s instructions (Qiagen). DNA quality and integrity were checked by the Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific) and fluorimeter (Qubit 2.0, Thermo Fisher Scientific). The methylation pattern at the 5′-CCGG sites was analysed using the methylation-sensitive amplification polymorphism (MSAP) technique, which employs the methylation-sensitive isoschizomers, HpaII and MspI. The MSAP protocol developed by Albertini and Marconi 2014 was followed. For selective amplifications, one FAM-labelled EcoRI primer (EcoRI-CCA) and one HEX-labelled EcoRI primer (EcoRI-CAA) were combined with four HpaII/MspI primers for a total of four primer combinations (EcoRI-CCA/HpaII-AAC; EcoRI-CCA/HpaII-ACT; EcoRI-CAA/HpaII-ACC; EcoRI-CCA/HpaII-AGG). The obtained profiles were visualized as band patterns on 2% agarose gel and as electropherograms with the ABI PRISM® 3130 DNA Analyzer system (Thermo Fisher Scientific). Electropherograms’ size calibration was performed with the molecular weight ladder GenScan® 500 ROX™ Size Standard (Thermo Fisher Scientific) and the MSAP peaks were detected using the Peak Scanner® software (Thermo Fisher Scientific). We scored markers as present when peak intensity exceeded 30 and discarded all loci showing a signal in a negative control. To have reproducible and clear banding patterns, each amplification was repeated at least three times, and only peaks showing consistent amplification were considered.
Methylation-sensitive amplification polymorphism profiles were recorded as 1/0 binary matrices, where 1 indicates the presence and 0 the absence of a given fragment. The resultant code describes the presence/absence of each fragment (represented by an amplicon) in the EcoRI/HpaII and EcoRI/MspI digests of a single sample. Four permutations (hereafter called pattern types) are expected, depending on the ability of either enzyme to cut at their recognition sequence. In particular, pattern type I (HpaII = 0/MspI = 0) could be caused either by restriction target absence due to a mutated site when genetically distinct samples are compared, or inhibition of digestion with both enzymes at a fully (both strands) methylated CCGG site when another sample shows the presence of a fragment at that position (Schulz et al. 2013; Fulneček and Kovařík 2014). Therefore, the 0–0 profile is not informative and was excluded from the analysis to avoid the noise produced by confounding the effects of mutation and methylation. Pattern type II (HpaII = 0/MspI = 1) reflects hemi- (only one strand) or full methylation of the internal cytosine, where the band is present in the MspI, but absent in HpaII sample; pattern type III (HpaII = 1/MspI = 0) corresponds to hemi-methylation of the external cytosine sites, in which the amplicon is present only in the HpaII, but not in the MspI sample; pattern type IV (HpaII = 1/MspI = 1) reflects un-methylated sites in which a fragment is detected in both the HpaII- and the MspI-digested samples (Supplementary Fig. S1). In agreement with Karan et al. (2012), 16 banding patterns between control and salinity stress in leaf and root of the three ecotypes were observed to find out the changes in cytosine methylation patterns under salinity stress. In particular, the patterns A–D represented monomorphic class with no methylation changes, the patterns E–J indicated hypomethylation events, while possible hypermethylation induced by salt stress was represented by the patterns K–P. The methylation ratio (MR) parameter was measured to better estimate the predominant event between methylation and de-methylation and calculated as the ratio between the number of fragments showing hyper- and hypo-methylation events according to the following formula: MR = (patterns K–P)/(patterns E–J) (Aversano et al. 2013).
Statistical analysis
The Student’s t test (P ≤ 0.05) was used to compare values of the biometric variables of treated versus control samples within each ecotype. qRT-PCR, biochemical and SPAD data were processed by univariate two-way analysis of variance (ANOVA) (treatment × genotype) and mean separation was performed through the Duncan multiple range test, with critical P value set at 0.05. All analyses were performed using Sigma Plot Software (Systat Software Inc.).
Results
Growth response of A. donax ecotypes under NaCl excess
To understand whether differential response of A. donax ecotypes to saline stress could be associated to distinct morpho-physiological, transcriptional, metabolic and DNA methylation level cues, the three ecotypes ‘Canneto’, ‘Cercola’ and ‘Domitiana’ were selected from a larger collection in reason of their different response to salt stress (De Stefano et al. 2017) and treated for 2 and 21 days with 150 mM NaCl. The 21-day prolonged NaCl treatment did not result in plant mortality or severe stress symptoms, since no leaf senescence was detected by visual observation (Supplementary Fig. S2) or decrease in chlorophyll content, expressed as SPAD value (Supplementary Fig. S3). However, the three ecotypes were separated in terms of shoot and root growth. ‘Canneto’ and ‘Domitiana’ aboveground growth was not affected by the 21-day NaCl treatment, while ‘Cercola’ showed a significant decrease in leaf number, stem height and shoot fresh weight (Table 1). Root biomass under salt stress in ‘Canneto’ and ‘Cercola’ did not differ from the respective control plants, whereas ‘Domitiana’ showed stimulated growth, with significantly higher root FW and length than unstressed plants (Table 1).
Based on the ability to maintain plant growth and aerial biomass production under salinity stress, we calculated the stress susceptibility index (SSI) to discriminate the A. donax ecotypes, with higher SSI indicating lower tolerance to salt stress. Results classified ‘Cercola’ as the less tolerant ecotype (SSI = 1.66), ‘Canneto’ as intermediate (SSI = 1.15) and ‘Domitiana’ as the most tolerant, with the lowest SSI (SSI = 0.48).
Transcriptional response to NaCl in A. donax leaves and roots
Expression profiling of key genes, known to be crucial for maintenance of Na+/H+ homeostasis and regulation of transcription under salinity, indicated that the osmotic stress signal is perceived by leaves already after 2 days of NaCl treatment, before Na+ and Cl− could have accumulated to toxic levels, and highlighted transcriptional differences between the three A. donax ecotypes, indicative of different responses to salt (Fig. 1). However, the largest and most striking transcriptional differences between the ecotypes were observed in control conditions, where intrinsically higher expression levels of most of the analysed genes were detected in ‘Domitiana’ and, to a lower extent, ‘Canneto’. After 21 days of NaCl treatment, in leaves, significant differences between ecotypes were detected almost exclusively in ‘Cercola’, and in roots, the three ecotypes showed strong transcriptional activation of nearly all tested genes (Fig. 1).
Transcriptional profiling of key genes related to Na+/H+ homeostasis and regulation of gene expression in leaves and roots of three A. donax ecotypes after 2 and 21 days of 150 mM NaCl treatment. qRT-PCR results are expressed as fold changes relative to the relevant unstressed control of the ecotype ‘Cercola’. Values are the mean ± SD of three technical replicates from three biological samples. Within each treatment, means followed by the same letter are not statistically different (Duncan’s test; P ≤ 0.05). Within each ecotype, differences between NaCl-treated and control plants indicated by * are significant (Duncan’s test; P ≤ 0.05)
Adjustment to ionic and osmotic stress
After 2 days, the Na+ transporters SOS1, HKT1 and NHX1 showed almost unchanged or diminished transcriptional response to NaCl treatment in leaves of ‘Canneto’, whereas in ‘Cercola’, significant increased expression levels were measured as compared to unstressed leaves (Fig. 1). ‘Domitiana’, had a remarkably, almost eight times, higher transcription of these genes in control leaves respect to the reference (unstressed ‘Cercola’ leaves) and a significant decrease following salt stress (Fig. 1). In roots, ‘Domitiana’, as well as ‘Canneto’, showed higher expression of the three transporter genes under control conditions and down-regulation following salt stress (Fig. 1). Similar to what detected in leaves, ‘Cercola’ unstressed roots had lower expression levels of the three genes, which significantly increased upon salt treatment only for SOS1.
Following prolonged NaCl treatment (21 days), the expression levels of ion transporters showed no significant changes in leaves of ‘Canneto’ and ‘Cercola’ compared to the controls, though in the latter ecotype, they showed a trend towards over-expression (Fig. 1). In unstressed leaves of ‘Domitiana’, expression of the three genes was higher than the other two ecotypes, decreasing under NaCl treatment for SOS1 and HKT1. In roots, a strong induction of these genes (20–40 times increase for SOS1 and 8–10 times increase for HKT1) was detected after 21 days of salt stress in the three ecotypes, except for HKT1 in ‘Domitiana’, which was not significantly different from the control (Fig. 1).
Transcriptional reprogramming
Transcriptional profiling of A. donax at 2 and 21 days showed lower expression levels of DREB2A and WRKY53 TF genes in ‘Canneto’ and ‘Cercola’ than in ‘Domitiana’ unstressed leaves (Fig. 1). In response to the 2-day NaCl treatment, leaf expression of DREB2A increased significantly of almost 3–6 times in ‘Canneto’ and ‘Cercola’, respectively, whereas WRKY53 was not significantly different from the controls. In ‘Domitiana’ leaves, both genes were down-regulated by salt treatment. At 2 days, unstressed ‘Cercola’ roots showed lower expression of both TFs than ‘Canneto’ and ‘Domitiana’, but a significant up-regulation following salt treatment. On the contrary, DREB2A and WRKY53 expression was strongly down-regulated in ‘Canneto’ and ‘Domitiana’ roots at this time point. At 21 days, DREB2A and WRKY53 expression increased in ‘Canneto’ and ‘Cercola’ leaves, though significantly only for ‘Canneto’ WRKY53 (Fig. 1). In ‘Domitiana’, TF genes were down-regulated or did not change significantly. After prolonged NaCl treatment, a strong induction of DREB2A and WRKY53 (almost 20–30 times and 10–80 times, respectively) was recorded in the roots of the three ecotypes, similar to what observed for the ion transporter genes.
Salt stress-induced accumulation of proline and free ABA
Free proline content in unstressed leaves resulted higher in ‘Domitiana’ than in the other two ecotypes both at 2 and 21 days (Table 2). Short exposure to NaCl significantly induced accumulation of this compatible osmolyte in ‘Canneto’ and ‘Cercola’ but not in ‘Domitiana’, whose proline levels in stressed leaves were nonetheless similar to those of the other two ecotypes. After prolonged NaCl treatment, proline accumulated significantly in all the ecotypes (Table 2).
Leaf expression analysis of the proline biosynthetic gene P5CS, coding for Delta-1-pyrroline-5-carboxylate synthetase, showed different transcription levels of this gene in the three ecotypes, that mostly paralleled those observed for the Na+ homeostasis and transcriptional regulators genes. After 2 days, P5CS transcription was significantly lower in control leaves of ‘Cercola’ than in ‘Canneto’ and ‘Domitiana’ and was up-regulated after NaCl treatment only in ‘Canneto’ and ‘Cercola’ (Fig. 2). After 21 days, P5CS expression in control leaves was still significantly higher in ‘Domitiana’ than in ‘Canneto’ and ‘Cercola’, whose P5CS transcription reached the same levels of ‘Domitiana’ only in response to salt treatment.
Expression analysis of the proline biosynthetic gene P5CS in leaves of three A. donax ecotypes after 2 and 21 days of 150 mM NaCl treatment. qRT-PCR results are expressed as fold changes relative to the relevant unstressed control of the ecotype ‘Cercola’. Values are the mean ± SD of three technical replicates from three biological samples. Within each treatment, means followed by the same letter are not statistically different (Duncan’s test; P ≤ 0.05). Within each ecotype, differences between NaCl-treated and control plants indicated by * are significant (Duncan’s test; P ≤ 0.05)
Free ABA quantification in A. donax leaves revealed a strong accumulation of the hormone already after 2 days of NaCl treatment in the three ecotypes, though less so in ‘Canneto’ (73% increase compared to 212 and 232% of ‘Cercola’ and ‘Domitiana’, respectively), suggesting that osmotic stress was inducing stomata closure (Table 2). After 21 days, free ABA content in control leaves was similar between the three ecotypes, and accumulated in response to NaCl with 12, 23 and 88% increase in ‘Cercola’, ‘Canneto’ and ‘Domitiana’, respectively, though significant only in the latter (Table 2).
Ecotype response to NaCl-induced oxidative stress
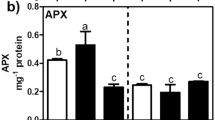
Under control conditions, the levels of hydrophilic and lipophylic antioxidants were lower in leaves of ‘Canneto’ and ‘Cercola’ than in ‘Domitiana’. However, after 21 days of salt treatment, a significant increase was observed for hydrophilic antioxidants in ‘Canneto’ and ‘Domitiana’ and for the lipophilic fraction in ‘Canneto’ and ‘Cercola’, in which it reached the same level found in ‘Domitiana’ (Table 3). Lipid peroxidation, measured as MDA content, was stimulated by salt stress after 21 days in all ecotypes (Table 3), but particularly in ‘Domitiana’, in which it increased by 65%.
Investigation of the redox state following saline stress in the three ecotypes of A. donax through expression analysis of ROS scavenging-related genes at day 21 of NaCl treatment (Fig. 3) revealed that the unstressed leaf transcription rates of glutathione reductase (GR) in ‘Canneto’ and superoxide dismutase (SOD4), catalase (CAT1) and GR in ‘Domitiana’ were higher than in ‘Cercola’ (Fig. 3). NaCl-triggered transcription was significant in leaves of ‘Cercola’ for SOD4 and GR, while in ‘Canneto’ only for GR. In ‘Domitiana’, the only transcriptional change induced by NaCl was the down-regulation of GR. Similarly, to TFs and transporters genes, salt stress strongly induced the expression of SOD4, CAT1 and GR in roots of all the ecotypes, with a higher than 100-fold increase compared to unstressed conditions (Fig. 3).
Expression analysis of ROS detoxification genes in leaves and roots of three A. donax ecotypes after 21 days of 150 mM NaCl treatment. qRT-PCR results are expressed as fold changes relative to the relevant control of the ecotype ‘Cercola’. Values are the mean ± SD of three technical replicates from three biological samples. Within each treatment, means followed by the same letter are not statistically different (Duncan’s test; P ≤ 0.05). Within each ecotype, differences between NaCl-treated and control plants indicated by * are significant (Duncan’s test; P ≤ 0.05)
Changes of the leaf and root methylomes under salt stress
Cytosine methylation status at CCGG sequences in three A. donax ecotypes assayed by the MSAP technique demonstrated DNA methylation changes under salt stress in the leaf and root of all giant reed ecotypes (Table 4). A total of 5010 and 6846 scorable fragments were amplified in leaves and roots, respectively. Under control conditions, in the leaf, total methylated bands averaged 68.4% in ‘Cercola’, 70.5% in ‘Canneto’ and 72.2% in ‘Domitiana’, while in the root, the average ranged from 65.8% (in ‘Domitiana’) to 69.0% (in ‘Cercola’). In this organ, salinity stress increased the percentage of total methylated sites in all ecotypes, whereas in the leaf, a genotype-specific response was observed. ‘Cercola’ showed a higher number of total methylated loci in treated leaf samples (avg 72.0%) compared to its controls, with an average increase of 3.6%. On the counterpart, in ‘Domitiana’, NaCl treatment decreased the total methylated sites (72.2% vs 69.8% in control and stressed leaves, respectively). Total methylation in leaves of ‘Canneto’ remained at a level similar to controls (avg 70.5% vs 70.4%) when exposed to salinity. As for the fully methylated bands, there was an evident increase in ‘Cercola’ samples under salinity in both tissues. Conversely, following NaCl treatment, the percentage of fully methylated sites respective to unstressed control decreased in ‘Domitiana’ leaf (avg 58.8% vs 54.2%) and increased in roots (56.1 vs 63.3%). In ‘Canneto’, salt-triggered changes of hemi-methylated loci resembled those observed in ‘Domitiana’ (Table 4). Regarding the hemi-methylated sites, salinity mainly increased them in all A. donax ecotypes both in the leaf and root. The only exceptions were roots of ‘Domitiana’ and ‘Canneto’, where the level of hemi-methylated bands remained either lower (avg 9.7 vs 7.4 in the former) or similar (avg 10.4 vs 10.6 in the latter). In the leaf, total and fully methylated bands increased in all three ecotype controls when comparing 2 vs 21 days of treatment, while an opposite trend was found in the root. Concerning the hemi-methylated bands, leaf and root samples showed a decrease from 2 to 21 days of treatment in all three ecotypes with percentages of 15.8 vs 8.6 and 10.9 vs 7.9 (avg), respectively. A general trend of higher level of fully and hemi-methylated bands was observed in the leaf and root under both control and high NaCl conditions in the three ecotypes. The fully methylated loci were always more than the hemi-methylated ones (Table 4).
The effects of short (2 days) versus prolonged (21 days) 150 mM NaCl treatment on CCGG sequences were evaluated. Overall, in the leaf, methylated bands decreased with increased exposure time to excess NaCl, except for ‘Domitiana’ (at fully methylated sites), where the 21-day treatment affected the total number of methylated sites (355 in stressed root vs 319 in the control) more than the 2-day stress (461 vs 451). Similarly, in ‘Canneto’, the number of methylated sites was lower in stressed leaves (− 3.3% of average variation). Again, in the root, the trend was towards a reduction of methylated bands from 2 to 21 days of NaCl treatment both in ‘Cercola’ and ‘Canneto’ (with the exceptions of fully methylated sites). However, in ‘Domitiana’, an opposite trend was observed.
The banding patterns between control and NaCl-treated tissues of the three A. donax genotypes were compared to find out all possible changes under salinity stress (Fig. 4, Table S3). The data obtained can be summarized as follows: ‘Cercola’ behaved as an early-responding ecotype, since it showed a higher amount of hypermethylation changes after 2 days compared to the other ecotypes. Regarding long-term stress (21 days) effects, ‘Cercola’ did not show considerable changes respect to 2-day samples. In addition, in the leaf, salt-induced methylation changes were substantially equally distributed between hyper- and hypo-methylation patterns, whereas in the root, hypermethylation was prevalent. Instead, ‘Canneto’ showed greater percentage of hypermethylation changes after 21 than after 2 days. Therefore, ‘Canneto’ behaved as a late-responding ecotype. Finally, ‘Domitiana’ showed an early response in the root (extensive hypomethylation at 2 days) and a late response in both tissues (with a wide hypermethylation at 21 days). To better estimate the predominant event between methylation and de-methylation, a parameter termed methylation ratio (MR) was calculated as the ratio between the hyper- and hypo-methylation frequencies in ‘Cercola’, ‘Canneto’ and ‘Domitiana’ NaCl-treated samples compared to their respective controls (Fig. 5). In ‘Cercola’, the hypermethylation changes were higher in the root than in the leaf at both 2 and 21 days (1.9 and 1.4 root; 1.0 and 0.9 leaf). In ‘Canneto’, a large increment of hypermethylation is registered in leaf and root at 21 days with MR values of 1.3 (root) and 1.7 (leaf). In ‘Domitiana’, we observed a higher hypermethylation at 21 than at 2 days in both tissues.
DNA methylation patterns between control and salt-treated leaf and root of three A. donax ecotypes after 2 and 21 days of 150 mM NaCl exposure. The patterns, expressed as percentage of NaCl-treated over control samples, represent no change, hypomethylation, and hypermethylation events induced by salt stress
DNA methylation ratio (MR) in leaves and roots of three A. donax ecotypes after 2 and 21 days of 150 mM NaCl treatment. MR values were calculated as the percent ratio of the number of MSAP bands revealing hypermethylation over the number of MSAP bands revealing hypomethylation, relative to unstressed leaves and roots
Discussion
Cultivation of the fast-growing invasive species giant reed (A. donax L.) is recently diffusing for ligno-cellulosic biomass production in warm and temperate climates. Due to its peculiar capability of maintaining high growth rates and yield in limiting environments, it appears particularly suitable in marginal lands and especially in saline soils, since it was classified as ‘moderately salt tolerant’ (Nackley and Kim 2015; Pompeiano et al. 2017). Despite the limited genetic variation due to its obligatory agamic reproduction system, recent studies have demonstrated that different varieties and ecotypes of A. donax can have distinct responses to abiotic stresses (Sánchez et al. 2015; De Stefano et al. 2017; Haworth et al. 2017a, b, c). These are of potential use for the selection of genotypes suitable for efficient and sustainable cultivation in specific environments. Therefore, we used three Italian ecotypes to investigate the early and late responses of leaf and root tissues to NaCl at the morpho-physiological, transcriptional, metabolic and DNA methylation level.
A. donax ecotypes differ in their morpho-physiological response to salinity
In our high-salinity experimental conditions (i.e. hydroponic growth in 150 mM NaCl, EC = 1.9 mS cm−1), none of the A. donax ecotypes showed evident phenotypic symptoms of toxicity, such as chlorotic or necrotic leaves, even after prolonged exposure (21 days). Significant reduction in leaf number was detected only in ‘Cercola’, but was associated with decreased shoot height, thus indicating that, if accumulation of Na+ and Cl− in older A. donax leaves was occurring (Pollastri et al. 2017), it did not induce premature leaf senescence in our ecotypes. Similarly, the response of an Italian A. donax ecotype to drought stress did not involve intense leaf shedding, opposite to what observed for a Moroccan ecotype, possibly reflecting different adaptive strategies to the respective environments (Haworth et al. 2017b). Elimination of excess toxic ions through shedding of older leaves seemed not taking place in our experimental conditions and ecotypes, whose salt tolerance might probably rely on the ability to exclude toxic ions and/or partially restrict Na+ translocation and accumulation into the shoots. Indeed, retention of Na+ and Cl− in roots and rhizomes was reported for salt-treated A. donax (Pollastri et al. 2017; Pompeiano et al. 2017). SPAD values, which correlate with chlorophyll A content in A. donax (Spencer 2014), did not change or even slightly increased after salt treatment (Supplementary Fig. S3), in agreement with Sánchez et al. 2015 and Di Mola et al. 2018, indicating that the photosynthetic system was not damaged by excess NaCl. Accordingly, previous results demonstrated that the stress level used in this study did not affect chlorophyll content index or maximum quantum yield of PSII (Fv/Fm) in the three ecotypes (De Stefano et al. 2017). The observed increase in chlorophyll content could be due to increased leaf thickness (Longstreth and Nobel 1979) or to leaf area reduction or shrinkage (Tang and Boyer 2007), though this should not be the case since the fourth leaf width did not change significantly after the 21-day NaCl treatment (Table 1). Nevertheless, lack of stress damage cannot be unequivocally ruled out.
According to growth parameters, salinity negatively affected shoot and leaf biomass only in ‘Cercola’, while in ‘Domitiana’, it significantly stimulated root growth. The adaptive significance of root growth under salt stress remains elusive (Galvan-Ampudia and Testerink 2011). Tolerant Arabidopsis ecotypes reduce main root growth under severe stress to limit excess sodium internalization, whereas in other species, enhanced root growth to avoid locally high salt concentrations is associated to stress tolerance (Bernstein and Kafkafi 2002). Therefore, ‘Domitiana’ may have developed a mechanism of salt avoidance through a root system able to take up water from deep soil layers, as reported for other species (Munns and Tester 2008; Yoshimura 2008; Julkowska and Testerink 2015). From these results, we considered that, in terms of plant productivity (aerial biomass growth), the salt tolerance of the three ecotypes is ‘Cercola’ < ‘Canneto’ < ‘Domitiana’, consistently to what recently reported by (De Stefano et al. 2017).
Transcriptional and metabolic dynamics under short and prolonged salt excess in A. donax are ecotype and tissue dependent
Limitation of Na+ and Cl− accumulation in the more susceptible tissues, such as photosynthetic leaves, and detoxification of excess ions in cells and tissues represent key mechanisms of salt tolerance. Therefore, the study of genes involved in the regulation of Na+ flux as well as salt sensing and signalling is crucial for understanding survival and growth of A. donax in saline soils, especially in light of our results indicating that other mechanisms, such as compartmentalisation in older leaves and elimination through leaf shedding, do not take place in the tested experimental conditions.
In giant reed, the transcriptional behaviour of the plasma membrane SOS1 and the tonoplast NHX1, major players of Na+ exclusion back to the soil and compartmentalization in the vacuole, respectively, as well as of HKT1, an essential contributor to K+/Na+ homeostasis and leaf protection from ion toxicity through unloading of Na+ from the xylem sap (Almeida et al. 2017) demonstrated ecotype-dependent regulation. Salt-induced over-expression of ion transporter genes in ‘Cercola’ after 2 days may indicate that, in response to substrate salinity, ‘Cercola’ regulates ion homeostasis more rapidly than the other two ecotypes, maintaining this trend of regulation after prolonged exposure to NaCl. However, either the putative reduction of ionic stress is not sufficient to protect this ecotype from shoot growth inhibition or shoot biomass reduction is a further mechanism to limit toxic Na+ and Cl− accumulation by decreasing H2O consumption and total evapotranspiration. On the contrary, unchanged or reduced expression suggests that support of plant growth under high salinity in the other two ecotypes may be associated to an already higher expression of ion transporter genes in the absence of stress. Albeit the divergence between ecotypes in the NaCl response may not be associated to genetic variation, the high constitutive level of expression of a set of stress-responsive genes might reflect the adaptation of these ecotypes to the original growing environments. This observation is in agreement with previous studies, which reported that A. donax ecotypes respond in a very different manner to drought stress, depending on their capability to adapt to environmental constraints (Ahrar et al. 2015; Haworth et al. 2017a, b, c).
As far as key TFs contributing to salt tolerance, we evaluated WRKY53 and DREB2A, since they have a synergic role in the improvement of salt stress tolerance (Sakuma et al. 2006; Mallikarjuna et al. 2011; Allu et al. 2014; Van Eck et al. 2014; Song et al. 2016) and have been demonstrated to transduce a wide range of stress-responsive signals (Mallikarjuna et al. 2011) in many plant species. The transcriptional scenario observed for WRKY53 and DREB2A TFs mirrored that of the ion transporters. NaCl-triggered WRKY53 transcription was demonstrated in A. thaliana (Sun and Yu 2015), similar to the trend observed in the A. donax ecotypes ‘Canneto’ and ‘Cercola’, except for ‘Canneto’ roots after short NaCl exposure (Fig. 1). WRKY53 was suggested to be implicated in contrasting NaCl-induced leaf senescence in rice (Khan et al. 2017) and may contribute to the absence of leaf yellowing and decay in our experiments, possibly through activation of the antioxidant machinery. Accordingly, antioxidant accumulation and SOD4 and GR over-expression were more intense in the above two ecotypes (Table 3 and Fig. 3). Indeed, AtWRKY53 could contribute to alleviate oxidative damage, as suggested by decreased H2O2 accumulation during osmotic stress in over-expressing lines (Sun and Yu 2015). The already elevated levels of WRKY53 as well as of antioxidants and antioxidant transcripts in ‘Domitiana’ unstressed leaves (Table 3 and Fig. 3) might allow protection from NaCl-induced oxidative stress and explain the ability of ‘Domitiana’ to tolerate higher levels of lipid peroxidation without compromising plant growth. Rice DREB2A over-expression in soybean up-regulated proline biosynthesis and sugar accumulation, thus showing the involvement of this TF in many stress-related pathways (Zhang et al. 2013). The early (2 days) up-regulation of DREB2A induced by NaCl in ‘Canneto’ and ‘Cercola’ leaves does not unequivocally explain the contribution of this TF to salt tolerance in A. donax. Nevertheless, it is tempting to speculate an interaction with other metabolic partners leading to an increase in leaf proline of ‘Canneto’ and ‘Cercola’ after NaCl treatment, the former reaching the same levels as ‘Domitiana’. Proline accumulation in response to NaCl appears to be complex in A. donax, since in ‘Domitiana’ it does not strictly correlate to the P5CS gene expression. This result suggests that, as hypothesised by Pollastri et al. (2017), de novo synthesis via glutamate or ornithine pathway through the P5CS enzyme might not be the only regulation of proline levels under salt conditions, since reduction of catabolic processes may also contribute to adaptive mechanisms, as demonstrated in Medicago sativa (Miller et al. 2005).
Abscisic acid has a pivotal role in plant adaption to the osmotic component of salinity stress, along with osmolytes, and in the regulation of stomatal closure under long-term adaptation to osmotic stress (Maggio et al. 2007). Therefore, while ‘Cercola’ reacts to salinity with a stronger and earlier activation of the ion homeostasis machinery, the limited effect of salinity on plant biomass in ‘Canneto’ and ‘Domitiana’ may be due, at least in part, to the observed higher increase in free ABA levels following prolonged salt stress, and hence to the higher ability of these two ecotypes to preserve cell turgor and expansion, thus allowing plant growth. However, stomatal closure also limits gas exchange and photosynthesis. Indeed, using the same experimental conditions, we previously demonstrated significant NaCl-dependent inhibition of stomatal conductance and assimilation rate in all the three ecotypes, though in the absence of permanent damage to the photosynthetic apparatus (De Stefano et al. 2017). This suggests dissipation of excess energy through photo-oxidation and production of free radicals in all the three ecotypes, as also indicated by increased leaf accumulation of MDA, a by-product of lipid peroxidation that estimates membrane damage by oxidative stress (Dhindsa et al. 1981). Consequently, the level of leaf antioxidants, hydrophilic and/or lipophilic, increased in all ecotypes, but especially in ‘Canneto’, with ‘Domitiana’ having the highest levels both in control and NaCl-treated leaves. This response corresponds to increased leaf transcription of GR in ‘Canneto’ and ‘Cercola’ and SOD4 in ‘Cercola’. ‘Domitiana’, which accumulated antioxidants already in control conditions, showed decreased or not significantly different leaf expression of antioxidant genes and, consistently, of WRKY53, which is involved in regulation of the antioxidant machinery (Sun and Yu 2015).
Ion transporters and TFs genes, as well as antioxidant genes, were over-expressed at very high levels in roots exposed to prolonged NaCl salinity in all the three ecotypes. Overall, our transcriptional analysis detected the strongest transcriptional response to salinity in late roots, indicating that the stress level in the roots, which are directly in contact with the stressing agent, is high enough at this time point to require the recruitment of all the possible tolerance strategies in all the three ecotypes. Accordingly, higher transcriptional responsivity of roots compared to leaves was also demonstrated in A. donax under water stress (Fu et al. 2016). The root response to 2 days of NaCl treatment is indicative of the different stress tolerance levels of the three ecotypes. ‘Cercola’ is more susceptible and shows over-expression of both TFs, which may hence activate drought-specific transcription control, as well as of the genes responsible for Na+ exclusion and K+ homeostasis, to cope with the stress. Contrastingly, ‘Canneto’ and ‘Domitiana’ appear to have higher constitutive defences that make them tolerate the 2-day NaCl stress intensity without activating transcription of the above genes.
A. donax ecotypes adjust DNA methylation levels differently
To verify whether differences between the ecotypes may extend to the methylome, we profiled the methylation status of the three ecotypes. First, we observed that the amount of DNA methylation varied in all ecotypes from 2 to 21 days of treatment, confirming that plant exposition to salt can induce a reorganization at the cellular level in epigenetic and hence transcriptional terms. Variations of global DNA methylation levels have also been reported in several plant species both for abiotic and biotic constrains (Zhang et al. 2018; De Palma et al. 2019; Ferreira et al. 2019), emphasizing the role of differential methylome flexibility as an important player in stress response. Second, we found that DNA methylation varied between roots and leaves, especially at 2 days after treatment in ‘Canneto’ and ‘Cercola’. This emphasizes that salt-induced DNA methylation changes can be tissue specific, consistently with some authors who argued that tissue-specific biological functions should imply a distinct gene regulation, eventually involving differential DNA methylation (Aceituno et al. 2008). However, how these tissue-specific changes in DNA methylation patterns are triggered by high salinity remains to be elucidated for facilitating breeding programs. Third, we observed that our ecotypes differed in the hyper- and hypo-methylation patterns when grown under high NaCl conditions. In particular, the evident salt-triggered methylation observed in ‘Canneto’ and ‘Domitiana’ (in both leaves and roots) reflects what found in several plant species in response to environmental cues, such as cold, heavy metals and salt (Lízal and Relichová 2001; Aina et al. 2004; Choi and Sano 2007). In contrast, in ‘Cercola’, the length of salinity treatment affected the methylation levels, triggering an overall depletion of mC sites in both root and leaf. These findings agree with other studies demonstrating that closely related genotypes may possess some methylome flexibility. For example, rice cultivars with different salinity tolerance capacities exhibited different DNA methylation patterns in response to NaCl (Ferreira et al. 2015; Garg et al. 2015; Wang et al. 2015). Similarly, salinity inconsistently affected the level of DNA methylation in two different canola genotypes (Marconi et al. 2013). Overall, we found that all giant reed ecotypes were able to rapidly change DNA methylation (either reducing or increasing) upon stress imposition.
Conclusions
This study highlighted ecotype- and tissue-dependent variation in key mechanisms of salt response of A. donax to short- and long-term excess NaCl. The results concur in indicating that, among the ecotypes, the ‘Cercola’ reaction to salt excess was more rapid and intense than the other two ecotypes, though this apparently occurs at the expenses of plant growth. On the contrary, ‘Domitiana’ and, to a lesser extent, ‘Canneto’ may have constitutively elevated defences against high NaCl and, following NaCl treatment, may redirect energy to other tolerance mechanisms, such as stress avoidance through enlarged soil exploration by the root. DNA methylation analyses suggested a link between the plant performance under high salinity and the DNA methylation plasticity, especially in ‘Domitiana’. Our findings demonstrated that tolerant ecotypes possess a wide reservoir of molecular and metabolic resources to face salinity already in the absence of stress, possibly due to adaptation to their original growing environments. This may help in mitigating salinity damage in the early phase of stress, thus allowing the use of a “power saving mode” strategy.
Though the complexity of the salinity tolerance trait in A. donax will require further investigations to elucidate the relative contribution of different mechanisms to adaptation, our results could be useful for design breeding programs aimed at obtaining genotypes more suitable for efficient and sustainable ligno-cellulosic biomass production in limiting environments.
Author contribution statement
TD and MT designed research; TD, RDS, MDP, and EC performed morpho-physiological, transcriptional and metabolic analyses; CV and RA designed and performed MSAP profiling and analysis; TD and MT interpreted data and wrote the paper. All authors read and approved the manuscript.
Abbreviations
- MDA:
-
Malondialdehyde
- MSAP:
-
Methylation-sensitive amplification polymorphism
- TF:
-
Transcription factor
References
Aceituno FF, Moseyko N, Rhee SY, Gutiérrez RA (2008) The rules of gene expression in plants: organ identity and gene body methylation are key factors for regulation of gene expression in Arabidopsis thaliana. BMC Genom 9:438. https://doi.org/10.1186/1471-2164-9-438
Ahmad R, Liow PS, Spencer DF, Jasieniuk M (2008) Molecular evidence for a single genetic clone of invasive Arundo donax in the United States. Aquat Bot 88:113–120. https://doi.org/10.1016/j.aquabot.2007.08.015
Ahrar M, Doneva D, Koleva D et al (2015) Isoprene emission in the monocot Arundineae tribe in relation to functional and structural organization of the photosynthetic apparatus. Environ Exp Bot 119:87–95. https://doi.org/10.1016/j.envexpbot.2015.04.010
Aina R, Sgorbati S, Santagostino A et al (2004) Specific hypomethylation of DNA is induced by heavy metals in white clover and industrial hemp. Physiol Plant 121:472–480. https://doi.org/10.1111/j.1399-3054.2004.00343.x
Albertini E, Marconi G (2014) Methylation-sensitive amplified polymorphism (MSAP) marker to investigate drought-stress response in Montepulciano and Sangiovese grape cultivars. Methods Mol Biol 1112:151–164
Allu AD, Soja AM, Wu A et al (2014) Salt stress and senescence: identification of cross-talk regulatory components. J Exp Bot 65:3993–4008. https://doi.org/10.1093/jxb/eru173
Almeida DM, Margarida Oliveira M, Saibo NJMM et al (2017) Regulation of Na+ and K+ homeostasis in plants: towards improved salt stress tolerance in crop plants. Genet Mol Biol 40:326–345. https://doi.org/10.1590/1678-4685-GMB-2016-0106
Aversano R, Caruso I, Aronne G et al (2013) Stochastic changes affect Solanum wild species following autopolyploidization. J Exp Bot 64:625–635. https://doi.org/10.1093/jxb/ers357
Barrero RA, Guerrero FD, Moolhuijzen P, Goolsby JA, Tidwell J, Bellgard SE, Bellgard MI (2015) Shoot transcriptome of the giant reed, Arundo donax. Data Brief 3:1–6
Bernstein N, Kafkafi U (2002) Root growth under salinity stress. In: Waisel Y, Eshel A, Kafkafi U (eds) Plant roots: the hidden half, 3rd edn. Marcel Dekker, New York, pp 1194–1221
Borkotoky S, Saravanan V, Jaiswal A et al (2013) The Arabidopsis stress responsive gene database. Int J Plant Genom 2013:949564. https://doi.org/10.1155/2013/949564
Butelli E, Titta L, Giorgio M et al (2008) Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotechnol 26:1301–1308. https://doi.org/10.1038/nbt.1506
Choi CS, Sano H (2007) Abiotic-stress induces demethylation and transcriptional activation of a gene encoding a glycerophosphodiesterase-like protein in tobacco plants. Mol Genet Genomics 277:589. https://doi.org/10.1007/s00438-007-0209-1
Claussen W (2005) Proline as a measure of stress in tomato plants. Plant Sci 168:241–248. https://doi.org/10.1016/J.PLANTSCI.2004.07.039
Davenport RJ, Muñoz-Mayor A, Jha D et al (2007) The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant Cell Environ 30:497–507. https://doi.org/10.1111/j.1365-3040.2007.01637.x
De Palma M, Salzano M, Villano C et al (2019) Transcriptome reprogramming, epigenetic modifications and alternative splicing orchestrate the tomato root response to the beneficial fungus Trichoderma harzianum. Hort Res 6:5. https://doi.org/10.1038/s41438-018-0079-1
De Stefano R, Cappetta E, Guida G et al (2017) Screening of giant reed (Arundo donax L.) ecotypes for biomass production under salt stress. Plant Biosystems 152:911–917. https://doi.org/10.1080/11263504.2017.1362059
Deinlein U, Stephan AB, Horie T et al (2014) Plant salt-tolerance mechanisms. Trends Plant Sci 19:371–379. https://doi.org/10.1016/j.tplants.2014.02.001
DeRose-Wilson L, Gaut BS (2011) Mapping salinity tolerance during Arabidopsis thaliana germination and seedling growth. PLoS One 6(8):e22832. https://doi.org/10.1371/journal.pone.0022832
Dhindsa RS, Plumb-Dhindsa P, Thorpe TA (1981) Leaf senescence: correlated with increased levels of membrane permeability and lipid peroxidation, and decreased levels of superoxide dismutase and catalase. J Exp Bot 32(1):93–101
Di Mola I, Guida G, Mistretta C et al (2018) Agronomic and physiological response of giant reed (Arundo donax L.) to soil salinity. Ital J Agron 13:31–39. https://doi.org/10.4081/ija.2018.995
Evangelistella C, Valentini A, Ludovisi R, Firrincieli A, Fabbrini F, Scalabrin S, Cattonaro F, Morgante M, Mugnozza GS, Keurentjes JJB, Harfouche A (2017) De novo assembly, functional annotation, and analysis of the giant reed (Arundo donax L.) leaf transcriptome provide tools for the development of a biofuel feedstock. Biotechnol Biofuels 10(1):138
Ferreira LJ, Azevedo V, Maroco J et al (2015) Salt tolerant and sensitive rice varieties display differential methylome flexibility under salt stress. PLoS One 10(5):e0124060. https://doi.org/10.1371/journal.pone.0124060
Ferreira LJ, Donoghue MT, Barros P, Saibo NJ, Santos AP, Oliveira MM (2019) Uncovering Differentially Methylated Regions (DMRs) in a salt-tolerant rice variety under stress: one step towards new regulatory regions for enhanced salt tolerance. Epigenomes 3:4. https://doi.org/10.3390/epigenomes3010004
Fu Y, Poli M, Sablok G et al (2016) Dissection of early transcriptional responses to water stress in Arundo donax L. by unigene-based RNA-seq. Biotechnol Biofuels 9:54. https://doi.org/10.1186/s13068-016-0471-8
Fulneček J, Kovařík A (2014) How to interpret methylation sensitive amplified polymorphism (MSAP) profiles? BMC Genet 15:2. https://doi.org/10.1186/1471-2156-15-2
Galvan-Ampudia CS, Testerink C (2011) Salt stress signals shape the plant root. Curr Opin Plant Biol 14:296–302. https://doi.org/10.1016/J.PBI.2011.03.019
Garg R, Narayana Chevala V, Shankar R, Jain M (2015) Divergent DNA methylation patterns associated with gene expression in rice cultivars with contrasting drought and salinity stress response. Sci Rep 5:14922. https://doi.org/10.1038/srep14922
Gill S, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48:909–930. https://doi.org/10.1016/j.plaphy.2010.08.016
Golldack D, Li C, Mohan H, Probst N (2014) Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front Plant Sci 5:151. https://doi.org/10.3389/fpls.2014.00151
Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS (2012) Phytozome: a comparative platform for green plant genomics. Nucl Acids Res 40:D1178–D1186. https://doi.org/10.1093/nar/gkr944
Guarino F, Cicatelli A, Brundu G, Improta G, Triassi M, Castiglione S (2019) The use of MSAP reveals epigenetic diversity of the invasive clonal populations of Arundo donax L. PLoS One 14:e0215096. https://doi.org/10.1371/journal.pone.0215096
Hasegawa PM, Bressan RA, Zhu J-K, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51:463–499. https://doi.org/10.1146/annurev.arplant.51.1.463
Haworth M, Catola S, Marino G et al (2017a) Moderate drought stress induces increased foliar dimethylsulphoniopropionate (DMSP) concentration and isoprene emission in two contrasting ecotypes of Arundo donax. Front Plant Sci 8:1016. https://doi.org/10.3389/fpls.2017.01016
Haworth M, Centritto M, Giovannelli A et al (2017b) Xylem morphology determines the drought response of two Arundo donax ecotypes from contrasting habitats. GCB Bioenergy 9:119–131. https://doi.org/10.1111/gcbb.12322
Haworth M, Cosentino SL, Marino G et al (2017c) Physiological responses of Arundo donax ecotypes to drought: a common garden study. GCB Bioenergy 9:132–143. https://doi.org/10.1111/gcbb.12348
Heath RL, Packer L (1968) Photoperoxidation in isolated chloroplasts: I. Kinetics and stoichiometry of fatty acid peroxidation. Arch Biochem Biophys 125:189–198. https://doi.org/10.1016/0003-9861(68)90654-1
Hoagland DR, Arnon DI (1950) The water-culture method for growing plants without soil. Circ Calif Agric Exp Stn 347:32 (citeulike-article-id:9455435)
Ismail A, Takeda S, Nick P (2014) Life and death under salt stress: same players, different timing? J Exp Bot 65:2963–2979. https://doi.org/10.1093/jxb/eru159
Julkowska MM, Testerink C (2015) Tuning plant signaling and growth to survive salt. Trends Plant Sci 20:586–594. https://doi.org/10.1016/j.tplants.2015.06.008
Karan R, DeLeon T, Biradar H, Subudhi PK (2012) Salt stress induced variation in DNA methylation pattern and its influence on gene expression in contrasting rice genotypes. PLoS One 7:e40203. https://doi.org/10.1371/journal.pone.0040203
Khan MS, Akther T, Hemalatha S (2017) Impact of Panchagavya on Oryza sativa L. grown under saline stress. J Plant Growth Regul 36:702–713. https://doi.org/10.1007/s00344-017-9674-x
Kobayashi NI, Yamaji N, Yamamoto H et al (2017) OsHKT1;5 mediates Na+exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. Plant J 91:657–670. https://doi.org/10.1111/tpj.13595
Lewandowski I, Scurlock JMO, Lindvall E, Christou M (2003) The development and current status of perennial rhizomatous grasses as energy crops in the US and Europe. Biomass Bioenergy 25:335–361
Lira-Medeiros CF, Parisod C, Fernandes RA, Mata CS, Cardoso MA, Ferreira PC (2010) Epigenetic variation in mangrove plants occurring in contrasting natural environment. PLoS One 5(4):e10326
Lízal P, Relichová J (2001) The effect of day length, vernalization and DNA demethylation on the flowering time in Arabidopsis thaliana. Physiol Plant 113:121–127. https://doi.org/10.1034/j.1399-3054.2001.1130116.x
Longstreth DJ, Nobel PS (1979) Salinity effects on leaf anatomy consequences for photosynthesis. Plant Physiol 63:700–703. https://doi.org/10.1104/pp.63.4.700
Maggio A, Raimondi G, Martino A, De Pascale S (2007) Salt stress response in tomato beyond the salinity tolerance threshold. Environ Exp Bot 59:276–282. https://doi.org/10.1016/j.envexpbot.2006.02.002
Mallikarjuna G, Mallikarjuna K, Reddy MK, Kaul T (2011) Expression of OsDREB2A transcription factor confers enhanced dehydration and salt stress tolerance in rice (Oryza sativa L.). Biotechnol Lett 33:1689–1697. https://doi.org/10.1007/s10529-011-0620-x
Malone JM, Virtue JG, Williams C, Preston C (2017) Genetic diversity of giant reed (Arundo donax) in Australia. Weed Biol Manag 17:17–28. https://doi.org/10.1111/wbm.12111
Marconi G, Pace R, Traini A et al (2013) Use of MSAP markers to analyse the effects of salt stress on DNA methylation in rapeseed (Brassica napus var. oleifera). PLoS One 8(9):e75597. https://doi.org/10.1371/journal.pone.0075597
Mariani C, Cabrini R, Danin A et al (2010) Origin, diffusion and reproduction of the giant reed (Arundo donax L.): a promising weedy energy crop. Ann Appl Biol 157:191–202. https://doi.org/10.1111/j.1744-7348.2010.00419.x
Miller G, Stein H, Honig A, Kapulnik Y, Zilberstein A (2005) Responsive modes of Medicago sativa proline dehydrogenase genes during salt stress and recovery dictate free proline accumulation. Planta 222(1):70–79
Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7:405–410
Moller IS, Gilliham M, Jha D et al (2009) Shoot Na+ exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na+ transport in Arabidopsis. Plant Cell 21:2163–2178. https://doi.org/10.1105/tpc.108.064568
Moran JF, Becana M, Iturbe-Ormaetxe I et al (1994) Drought induces oxidative stress in pea plants. Planta 194:346–352
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681. https://doi.org/10.1146/annurev.arplant.59.032607.092911
Nackley LL, Kim SH (2015) A salt on the bioenergy and biological invasions debate: salinity tolerance of the invasive biomass feedstock Arundo donax. GCB Bioenergy 7:752–762. https://doi.org/10.1111/gcbb.12184
Negrao S, Schmöckel SM, Tester M et al (2017) Evaluating physiological responses of plants to salinity stress. Ann Bot 119:1–11. https://doi.org/10.1093/aob/mcw191
Papazoglou EG (2007) Arundo donax L. stress tolerance under irrigation with heavy metal aqueous solutions. Desalination 211:304–313. https://doi.org/10.1016/j.desal.2006.03.600
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. https://doi.org/10.1093/nar/29.9.e45
Pfaffl MW (2004) Quantification strategies in real-time PCR. In: Bustin SA (ed) A-Z of quantitative PCR 87–112. International University Line (IUL), La Jolla. https://doi.org/10.1007/s10551-011-0963-1
Pilu R, Cassani E, Landoni M et al (2014) Genetic characterization of an Italian giant reed (Arundo donax L.) clones collection: exploiting clonal selection. Euphytica 196:169–181. https://doi.org/10.1007/s10681-013-1022-z
Poli M, Salvi S, Li M, Varotto C (2017) Selection of reference genes suitable for normalization of qPCR data under abiotic stresses in bioenergy crop Arundo donax L. Sci Rep 7(1):10719. https://doi.org/10.1038/s41598-017-11019-0
Pollastri S, Savvides A, Pesando M et al (2017) Impact of two arbuscular mycorrhizal fungi on Arundo donax L. response to salt stress. Planta 247:573–585. https://doi.org/10.1007/s00425-017-2808-3
Pompeiano A, Landi M, Meloni G et al (2017) Allocation pattern, ion partitioning, and chlorophyll a fluorescence in Arundo donax L. in responses to salinity stress. Plant Biosyst 151(4):613–622. https://doi.org/10.1080/11263504.2016.1187680
Price AH, Atherton NM, Hendry GAF (1989) Plants under drought-stress generate activated oxygen. Free Radic Res 8:61–66. https://doi.org/10.3109/10715768909087973
Qiu Q-S, Guo Y, Dietrich MA et al (2002) Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA 99:8436–8441. https://doi.org/10.1073/pnas.122224699
Sablok G, Fu Y, Bobbio V, Laura M, Rotino GL, Bagnaresi P, Allavena A, Velikova V, Viola R, Loreto F, Li M, Varotto C (2014) Fuelling genetic and metabolic exploration of C3 bioenergy crops through the first reference transcriptome of Arundo donax L. Plant Biotechnol J 12(5):554–567
Sairam RK, Tyagi A (2004) Physiology and molecular biology of salinity stress tolerance in plants. Curr Sci 86:407–421. https://doi.org/10.1016/j.tplants.2005.10.002
Sakuma Y, Maruyama K, Osakabe Y et al (2006) Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18:1292–1309. https://doi.org/10.1105/tpc.105.035881.1
Saltonstall K, Lambert A, Meyerson LA (2010) Genetics and reproduction of common (Phragmites australis) and giant reed (Arundo donax). Invasive Plant Sci Manag 3:495–505. https://doi.org/10.1614/IPSM-09-053.1
Sánchez E, Scordia D, Lino G et al (2015) Salinity and water stress effects on biomass production in different Arundo donax L. clones. BioEneregy Res 8(4):1461–1479. https://doi.org/10.1007/s12155-015-9652-8
Schulz B, Eckstein RL, Durka W (2013) Scoring and analysis of methylation-sensitive amplification polymorphisms for epigenetic population studies. Mol Ecol Resour 13:642–653. https://doi.org/10.1111/1755-0998.12100
Spencer DF (2014) Evaluation of stem injection for managing giant reed (Arundo donax). J Environ Sci Health Part B 49(9):633–638
Song X, Wang J, Ma X et al (2016) Origination, expansion, evolutionary trajectory, and expression bias of AP2/ERF superfamily in Brassica napus. Front Plant Sci 7:1186. https://doi.org/10.3389/fpls.2016.01186
Sun Y, Yu D (2015) Activated expression of AtWRKY53 negatively regulates drought tolerance by mediating stomatal movement. Plant Cell Rep 34:1295–1306. https://doi.org/10.1007/s00299-015-1787-8
Tang AC, Boyer JS (2007) Leaf shrinkage decreases porosity at low water potentials in sunflower. Funct Plant Biol 34:24–30. https://doi.org/10.1071/FP06222
Van Eck L, Davidson RM, Wu S et al (2014) The transcriptional network of WRKY53 in cereals links oxidative responses to biotic and abiotic stress inputs. Funct Integr Genomics 14:351–362. https://doi.org/10.1007/s10142-014-0374-3
Wang W, Huang F, Qin Q et al (2015) Comparative analysis of DNA methylation changes in two rice genotypes under salt stress and subsequent recovery. Biochem Biophys Res Commun 465:790–796. https://doi.org/10.1016/j.bbrc.2015.08.089
Yang Q, Chen ZZ, Zhou XF et al (2009) Overexpression of SOS (salt overly sensitive) genes increases salt tolerance in transgenic Arabidopsis. Mol Plant 2:22–31. https://doi.org/10.1093/mp/ssn058
Yoshimura K (2008) Programmed proteome response for drought avoidance/tolerance in the root of a C3 xerophyte (wild watermelon) under water deficits. Plant Cell Physiol 49:226–241. https://doi.org/10.1093/pcp/pcm180
Zhang XX, Tang YJ, Bin Ma Q et al (2013) OsDREB2A, a rice transcription factor, significantly affects salt tolerance in transgenic soybean. PLoS One 8(12):e83011. https://doi.org/10.1371/journal.pone.0083011
Zhang H, Lang Z, Zhu JK (2018) Dynamics and function of DNA methylation in plants. Nat Rev Mol Cell Biol 19:489–506. https://doi.org/10.1038/s41580-018-0016-z
Zhu J (2016) Abiotic stress signaling and responses in plants. Cell 167:313–324. https://doi.org/10.1016/j.cell.2016.08.029
Acknowledgements
We are thankful to Prof. G. Caruso (Università di Napoli Federico II, Italy) for valuable help with statistical analysis. Assistance of Mr. G. Guarino and Mr. R. Nocerino (CNR-IBBR, Portici, Italy) with artworks and plant growth, respectively, is gratefully acknowledged. This work was partially supported by a research grant from the Italian Ministry of Education, University and Research, Project BioPoliS (PON03PE_00107_1).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Docimo, T., De Stefano, R., De Palma, M. et al. Transcriptional, metabolic and DNA methylation changes underpinning the response of Arundo donax ecotypes to NaCl excess. Planta 251, 34 (2020). https://doi.org/10.1007/s00425-019-03325-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-019-03325-w