Abstract
Main conclusion
The sulfur-formation ( suf ) genes play important roles in both photosynthesis and respiration of cyanobacteria, but the organism prioritizes Fe–S clusters for respiration at the expense of photosynthesis.
Iron–sulfur (Fe–S) clusters are important to all living organisms, but their assembly mechanism is poorly understood in photosynthetic organisms. Unlike non-photosynthetic bacteria that rely on the iron–sulfur cluster system, Synechocystis sp. PCC 6803 uses the Sulfur-Formation (SUF) system as its major Fe–S cluster assembly pathway. The co-expression of suf genes and the direct interactions among SUF subunits indicate that Fe–S assembly is a complex process in which no suf genes can be knocked out completely. In this study, we developed a condition-controlled SUF Knockdown mutant by inserting the petE promoter, which is regulated by Cu2+ concentration, in front of the suf operon. Limited amount of the SUF system resulted in decreased chlorophyll contents and photosystem activities, and a lower PSI/PSII ratio. Unexpectedly, increased cyclic electron transport and a decreased dark respiration rate were only observed under photoautotrophic growth conditions. No visible effects on the phenotype of SUF Knockdown mutant were observed under heterotrophic culture conditions. The phylogenetic distribution of the SUF system indicates that it has a co-evolutionary relationship with photosynthetic energy storing pathways.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Iron–sulfur (Fe–S) clusters function as the most versatile co-factors and play pivotal roles in various biological processes, such as energy metabolism. There are three known structures of Fe–S clusters, [2Fe-2S], [3Fe-4S], and [4Fe-4S] (Beinert et al. 1997). [2Fe-2S] and [4Fe-4S] are the most common types and occur in free electron transfer proteins such as ferredoxins, or in electron transfer mediators in photosynthetic reaction centers and respiratory complexes. Currently, three main Fe–S cluster assembly pathways are recognized (Fontecave and Ollagnier-de-Choudens 2008; Maio and Rouault 2015). The nitrogen fixation (NIF) system functions in the specialized assembly of Fe–S clusters for nitrogenase and exists mainly in nitrogen-fixing bacteria (Zheng et al. 1993; Frazzon and Dean 2002). The iron–sulfur cluster (ISC) system was proposed to play a housekeeping role in Fe–S cluster assembly and is distributed widely in prokaryotes and the mitochondria of eukaryotes (Zheng et al. 1998). The sulfur formation (SUF) system is the most ancient of the characterized Fe–S cluster assembly pathways and was proposed to play a supporting role in response to oxidative stress and iron starvation but appears to be the major Fe–S assembly system in cyanobacteria and chloroplasts in higher plants (Outten et al. 2004; Jang and Imlay 2010; Outten 2015).
In Escherichia coli, the SUF system is made up of multiple subunits, including SufA–SufE and SufS. SufA is an A-type scaffold protein that functions similarly to IscA and can also transfer [2Fe-2S] to apo-proteins (Ollagnier-de-Choudens et al. 2004; Vinella et al. 2009). The SufB, SufC, and SufD subunits form a protein complex and function as a new type of scaffold for the formation of Fe–S clusters. The model for the function of these proteins is based on the mechanism revealed in E. coli and the containing ratio of SufBCD complex is 1 SufB:2 SufC:1 SufD. SufD is a homolog of SufB but functions in iron trafficking instead of the Fe–S scaffold (Chahal et al. 2009; Saini et al. 2010; Wollers et al. 2010). SufE enhances the activity of SufS, and together, they form an SufS:SufE complex, which carries sulfur atoms to SufB (Outten et al. 2003; Layer et al. 2007). The SUF system is widely distributed from prokaryotes to eukaryotes and the protein sequences show high similarity. SufB and SufC from Arabidopsis can replace SufB and SufC deficiency in E. coli (Xu and Møller 2004; Xu et al. 2005), suggesting that the functions of the SUF system are broadly evolutionarily conserved.
Fe–S cluster containing proteins are involved in photosynthetic electron transport, and nitrogen and sulfur assimilation. The essential role of Fe–S cluster assembly in photosynthetic organisms was proven by lethal knock-out trials (Xu and Møller 2006; Murthy et al. 2007; Nagane et al. 2010; Hu et al. 2017). However, compared with non-photosynthetic organisms, knowledge of Fe–S cluster assembly mechanisms in photosynthetic organisms is rather limited. In higher plants, there are three types of Fe–S cluster assembly systems. The major ISC system is located in mitochondria, and contributes Fe–S clusters to respiratory electron mediators (Frazzon et al. 2007; Balk and Pilon 2011; Balk and Schaedler 2014). The SUF system is the main system in chloroplasts. The cytosolic iron–sulfur cluster assembly (CIA) pathway is a newly identified Fe–S cluster assembly system in the cytosol (Lill and Mühlenhoff 2006; Bych et al. 2008).
Cyanobacteria are the ancestors of chloroplasts in algae and higher plants and were the main players during the Earth’s early environmental development (Dismukes et al. 2001). Cyanobacteria have no physical compartmental differentiation between photosynthetic reactions and respiratory pathways, and share some common electron mediators, such as PQ and cytb 6 f, between the photosynthetic and respiratory electron transfer chains (Scherer 1990). In the genome of Synechocystis sp. PCC 6803, sufB, sufC, sufD, and sufS (sufBCDS) are tightly arranged with the same transcriptional direction, and a transcriptional repressor-encoding gene, sufR, is located upstream of sufB in the opposite transcriptional direction (Seki et al. 2006; Shen et al. 2007) (Fig. 1). However, sufA is not in the sufBDCS cluster of Synechocystis sp. PCC 6803, which differs from the suf operon arrangement in E. coli. It was reported that the knockout of suf genes (any one of sufBCDS and sufE) was lethal in Synechocystis sp. PCC 6803, suggesting that the SUF system might be the major Fe–S cluster assembly pathway (Balasubramanian et al. 2006). In this study, we developed a condition-controlled SUF “knockdown” mutant to investigate the physiological role of the SUF system by decreasing suf gene expression levels. The potential relationship between Fe–S cluster assembly and energy metabolism was analyzed in detail through a series of experiments, including P700+ reduction kinetics and related electron transfer rates.
SUF system in Synechocystis sp. PCC 6803. a Arrangement of the suf operon in the Synechocystis sp. PCC 6803 genome. b Co-transcriptional validation of sufBCDS by PCR. DNA fragments were PCR amplified using primers designed to target pairs of genes in the suf operon (Table S1). Detection of gDNA digestion is conducted, using rnpB primers and taking amplification of 37 cycles with WT gDNA template as a positive control, water as a negative control (up-left). Using rnpB as the internal control, adjust the amount of templates gDNA or cDNA (up-right). Using WT genome as template and taking samples after 24, 28 and 32 cycles and using WT cDNA as template and taking samples after 30 and 35 cycles. Lines 1, 4, 7, 10, and 13 represent sufB-sufC; lines 2, 5, 8, 11 and 14 represent sufC-sufD; lines 3, 6, 9, 12, and 15 represent sufD-sufS. #, cycles. c Yeast two-hybrid experiments using different combinations of SUF subunits
Materials and methods
Cyanobacterial strains, culture conditions, and general methods
A glucose-tolerant wild-type (WT) strain of Synechocystis sp. PCC 6803 was cultured in BG11 medium at 30 °C under continuous illumination of 30 μmol photons m−2 s−1. Heterotrophic growth of Synechocystis sp. PCC 6803 was induced by culture in BG11 medium with 5 mM glucose under dark conditions with exposure to 5 min low light (which did not cause photosynthesis) every day. BG11 plates were prepared by adding 8 mM N-[Tris(hydroxymethyl)methyl]-2-aminopropanesulfonic acid (TES)-NaOH (pH 8.2) and 0.3% Na2S2O3. According to the culture requirements of the mutant, spectinomycin (30 μg ml−1) was added to BG11 medium. Normal BG11 medium (containing 320 nM copper), Cu2+-free BG11 medium and 12 nM Cu2+ BG11 medium, and 25 nM Cu2+ BG11 medium were used for phenotypic assays.
Cell growth was monitored by the optical density recorded at 730 nm (OD730) using a Cary 300 UV–Vis spectrophotometer (Varian Australia Pty Ltd., Australia). For pigment analysis, 1 ml culture was centrifuged and extracted in 95% ethanol. The absorbance of the 95% ethanol extract was recorded at 648.6 and 664.1 nm, and the chlorophyll concentration was calculated following the published formula: Chl a (mg/L) = 13.36*A664.1 − 5.19*A648.6 (Lichtenthaler and Buschmann 2001).
Construction and identification of a sufB knockout mutant
A DNA fragment containing the full-length sufB gene (slr0074) was generated by PCR with the primers slr0074 ko-1 and slr0074 ko-2 (Table S1) using Synechocystis sp. PCC 6803 chromosomal DNA as a template, cloned into the pMD18-T vector (Takara Bio., Japan), and confirmed by sequencing. The kanamycin fragment excised from pRL446 (NCBI GenBank accession No. EU346690) (Elhai and Wolk 1988) by XbaI was inserted into the NheI site of that plasmid, resulting in pHS921, to inactivate sufB in Synechocystis sp. PCC 6803. The plasmid pHS921 was transformed into Synechocystis sp. PCC 6803 to produce a sufB knockout mutant. The segregation of the sufB knockout mutant was assayed by PCR using the primer pair slr0074 ko-1 and slr0074 ko-2 (Table S1) following sequencing confirmation. Genomic DNA from Synechocystis sp. PCC 6803 WT and the plasmid pHS921 were used as positive controls.
Construction of the PpetE-sufB mutant (SUF Knockdown)
For sufB expression under the petE promoter, the plasmid pHS1165 containing PpetE-sufB was constructed following the method described in Ke et al. (2014). sufB expression was controlled by the inserted petE promoter. The plasmid was transformed into Synechocystis sp. PCC 6803 to produce an SUF Knockdown mutant. The petE promoter is regulated by the available Cu2+ concentration and decreases the expression of suf genes under low Cu2+ concentration conditions. The previous studies demonstrated that there is no growth difference between WT and mutant when BG11 media containing >25 nM Cu2+, but the growth will be inhibited when the copper ion content was decreased to 10 nM in the medium (Gao and Xu 2009; Ke et al. 2014). The segregation of the SUF Knockdown mutant was confirmed by PCR using the primer pair slr0074 up-1 and slr0074 down-2 (Table S1) following sequencing confirmation. Genomic DNA from Synechocystis sp. PCC 6803 WT and the plasmid pHS1165 were used as positive controls.
Measurement of chlorophyll fluorescence
The maximal PSII quantum yield (Fv/Fm) was recorded using the saturation pulse method with a WATER-PAM chlorophyll fluorimeter (Walz, Germany). All samples were dark-adapted for 10 min before of measurements (Campbell et al. 1998). The operational PSII quantum yield (Fv′/Fm′) was measured immediately after exposure to the growth light intensity. Photosystem I P700+ reduction kinetics was measured with a Joliot JTS-10 spectrophotometer (BioLogic, France) as described previously in Alric et al. (2010). The samples were adjusted to the same cell density (OD730 = 0.4) to maintain the consistent cell numbers. To determine the cyclic electron flow, 10 µM 3-(3,4-dichlorophenyl)-1, 1-dimethylurea (DCMU) was added to the samples prior to measurements. Methylviologen (MV, 2 mM) was used to block the cyclic electron flow under DCMU conditions. The 77 K fluorescence emission spectra were measured with a Hitachi F-4500 fluorescence spectrophotometer (Hitachi High-Technologies Co., Japan) using an excitation wavelength of 435 nm. All samples were at a concentration of ~3 μg Chl a ml−1. The photosystem stoichiometry (ratio of PSI:PSII) was obtained by comparing the relative fluorescence intensity readings at 720 nm and 685 nm as the methods described in Murakami (1997). The fluorescence spectra were normalized at 685 nm (the fluorescence emission peak of PSII).
Measurement of photosynthetic oxygen evolution and dark respiration
The photosynthetic oxygen evolution and dark respiration of WT and the mutant were monitored using a Clark-type oxygen electrode (Chlorolab 2, Hansatech Instruments, Norfolk, UK) following the method described in Liu et al. (2010). The biomass in the reaction chamber was kept consistent by normalizing the cell numbers.
Extraction of RNA and RT-PCR
About 50 ml of Synechocystis sp. PCC 6803 grown in BG11 or 12 nM Cu2+ BG11 medium (low Cu2+ concentration) was harvested by centrifugation and frozen immediately using liquid nitrogen. Total RNA was extracted using a TRIzol Reagent Kit (Invitrogen, CA, USA), following the manufacturer’s instructions. The extracted RNA was tested for DNA contamination by PCR prior to cDNA synthesis using a digestion reverse kit (Invitrogen). All primers used for RT-PCR are listed in Table S1.
Western blot
WT and SUF Knockdown cells grown in 12 nM Cu2+ BG11 medium were harvested by centrifugation and ruptured by ultrasonication on ice in 40 mM Tris–Cl (pH 8.0). The debris and unbroken cells were removed by centrifugation at 11,900 g and 4 °C for 10 min. Equal amounts of total proteins from the supernatant were loaded after being boiled, separated by 12% SDS-PAGE, transferred to nitrocellulose filters (Millipore), detected with anti-SufB, anti-SufC, and anti-SufD (obtained by inducing SufB, SufC and SufD, immunizing rabbits, and collecting serum), anti-D1 (Agrisera, AS05 084A), anti-PetC (Agrisera, AS08 330), and anti-PsaC (Agrisera, AS10 939) antibodies, and visualized with goat anti-rabbit alkaline phosphatase antibody (Invitrogen) with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate as substrates.
Yeast two-hybrid assays
The protein–protein interactions within the suf gene cluster were detected using the Matchmaker GAL4 Two-Hybrid System 3 (Clontech, Palo Alto, CA, USA). The gene fragments of interest were transformed into pGBKT7 and pGADT7, respectively. The resultant plasmids were co-transformed into Saccharomyces cerevisiae AH109 and cultured on SD/− Trp − Leu − His agar plates for selection. The selected positive transformants were then transferred onto SD/− Trp − Leu − His − Ade plates and incubated at 28 °C for 3 days.
Results
The transcription mode of the suf operon and interactions among SUF subunits in Synechocystis sp. PCC 6803
The suf gene cluster in Synechocystis sp. PCC 6803 contains sufR, sufB, sufC, sufD, and sufS, and shows a similar arrangement to that in E. coli except that sufR was not found in E. coli (Fig. 1a). Both sufA and sufE are absent in the operon and sufR is located upstream of sufB in the reverse direction. Alternative Fe–S assembly system of ISC and NIF is found in the genome of Synechocystis sp. PCC 6803, but they do not form the gene clusters as found in the E. coli genome. Using primers designed to target the non-coding region between the suf genes (open reading frame), the co-transcriptional expression patterns of the sufBCDS gene cluster were investigated by PCR to detect the RNA fragments across the genes. The positive PCR reaction indicated that the co-expression of sufB (slr0074) and sufC (slr0075), sufC and sufD (slr0076), and sufD and sufS (slr0077) (Fig. 1b). The protein interaction between SufB and SufC, SufC and SufD, and SufE and SufS were confirmed using yeast two-hybrid assays (Fig. 1c).
Construction of SUF Knockdown mutant through petE promoter insertion
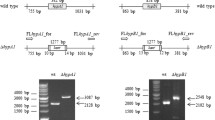
Because of the essential functions of SufBCD, the sufBCD genes cannot be completely knocked out in Synechocystis sp. PCC 6803 (Balasubramanian et al. 2006). Attempts to knock out the sufB gene by inserting the kanamycin resistance gene fragment C.K2 were unsuccessful; the sufB gene was still detected by PCR even after several generations under antibiotic selection pressure conditions (Fig. 2). Attempts to inactivate the sufC and sufD genes produced the same results, i.e., it was impossible to knock out the suf genes (data not shown). This indicated that the suf genes are vital for the survival of Synechocystis sp. PCC 6803 cells.
To overcome these difficulties, an Omega-PpetE fragment was inserted into the upstream region of the suf gene cluster (Fig. 3a). The petE promoter is an inducible promoter controlled by the available copper concentration; therefore, limited expression and partial inactivation of suf genes can be achieved by controlling the copper concentration in the medium (Ke et al. 2014). We named this mutant as “SUF Knockdown”. As shown in Fig. 3b, the petE promoter in the SUF Knockdown mutant was completely segregated. Under a carefully moderated copper concentration (0 nM), cell densities in the SUF Knockdown mutant were decreased in the 4-day-old culture, and eventually, the SUF Knockdown mutant died after 4 days (Fig. 3c). The growth rate of WT showed no differences in BG11 supplemented with different Cu2+ concentrations (Fig. S1), which is consisted with the previous report (Gao and Xu 2009). The SUF Knockdown mutant showed the same growth rate as WT when Cu2+ concentration in BG11 is greater than 25 nM. However, the growth rate of SUF Knockdown mutant in BG11 containing 12 nM Cu2+ was decreased to approximately half of that grown in BG11 containing 25 nM Cu2+ (Fig. S1). SUF Knockdown mutant cannot grow in the BG11 without Cu2+ supplements. Thus, BG11 medium with 12 nM Cu2+ was used for subsequent experiments. The levels of the SufB, SufC and SufD proteins in the SUF Knockdown mutant were decreased when it was cultured in BG11 medium with 12 nM Cu2+ (Fig. 3e).
Construction of SUF Knockdown (PpetE-sufB) mutant. a Schematic of sufB promoter replacement by the petE promoter. The gray area in front of sufB represents a ribosome binding site of about 20 bp. Omega was used as a selectable marker to prevent original promoter activity. b PCR analysis of genomic DNA from the WT, Ppet-sufB plasmid (pHS1165) and SUF Knockdown mutant using the primers slr0074 up-1 and slr0074 down-2, as shown in Table S1. c Photographs of the WT and SUF Knockdown mutant grown in copper-free BG11 medium. The middle and bottom rows show cells at 4 and 8 days after transfer to copper-free BG11 medium. d Loading control for the next western blot; each lane was loaded with 10 μg protein. e Amounts of SufBCD in WT and SUF Knockdown cells cultured in 12 nM Cu2+ BG11
The functions of Synechocystis SUF system in response to photoautotrophic and heterotrophic conditions
Fe–S containing proteins/co-factors play important roles in both photosynthetic and respiratory reactions. The SUF Knockdown mutant showed a significantly decreased growth rate under photoautotrophic (Pa) culture conditions compared with the WT. However, lower but similar growth rates for both strains were noted under heterotrophic (Ht) culture conditions (Fig. 4a). The decreased Chl a content in SUF Knockdown under photoautotrophic culture conditions was consistent with its decreased growth rate (Fig. 4b). The 77 K fluorescence emission spectral comparison showed a relatively decreased PSI content if we assign the fluorescence emission peak of 720 nm from PSI and 685 nm from PSII. A decreased PSI/PSII ratio was observed from SUF Knockdown mutant grown under photoautotrophic conditions, but the same ratio was obtained from the same strain (SUF Knockdown mutant) under heterotrophic conditions (Fig. 4c). The dark respiration rate in the SUF Knockdown mutant was decreased to ~67% of that in the WT strain under photoautotrophic culture conditions (Fig. 4d). In cyanobacteria, the photosynthetic and respiratory chains share the cytochrome b 6 f complex (Scherer 1990). The content of PetC, which is a Fe–S subunit of the Cytb 6 f complex, was decreased in the SUF Knockdown mutant (Fig. 5). Compared with the WT, the content of PsaC (represent PSI) in the SUF Knockdown mutant was decreased (Fig. 5b), which agreed well with the results obtained from 77 K fluorescence emission spectra. The decreased PSII activities (Fv/Fm and Fv′/Fm′) and lower photosynthetic oxygen evolution rates of the SUF Knockdown mutant under photoautotrophic conditions agreed well with the hypothesis that SUF proteins play important roles in photosynthetic reactions (Fig. 5c–e).
Physiological features of the WT and SUF Knockdown mutant under photoautotrophic (Pa) and heterotrophic (Ht) conditions. a Growth determined by the OD730. b Chl a content. c 77 K fluorescence emission spectra using an excitation wavelength of 435 nm. d Dark respiration rate of WT and SUF Knockdown mutant. Black lines and columns represent photoautotrophic (Pa) conditions; gray lines and columns represent heterotrophic (Ht) conditions. All data were from WT and SUF Knockdown mutant cultured in BG11 medium with 12 nM Cu2+ under Pa or Ht conditions
Western blot and photosynthetic activity of the WT and SUF Knockdown mutant. a Loading control for the next western blot, each lane was loaded with 10 μg protein. b Protein levels of D1, PetC, and PsaC detected by anti-DI, anti-PetC, and anti-PsaC. c Fv/Fm. d Fv′/Fm′. e Net photosynthetic rate in the WT and SUF Knockdown mutant under photoautotrophic conditions. f RT-PCR analysis of the relative transcriptional changes of selected genes under photoautotrophic culture conditions. g RT-PCR analysis of the relative transcriptional changes of selected genes under heterotrophic culture conditions. Genes involved in photosynthesis: petE (sll0199), petF1 (sll1382), petF2 (ssl0020), petF3 (slr0150), petH (slr1643) and petJ (sll1796); genes involved in respiration: ndhD (slr0331), ndhD2 (slr1291), ndhI (sll0520), sdhB1 (sll1625); sdhB2 (sll0823); hydrogenase-related genes: hoxE (sll1220), hoxF (sll1221), hoxU (sll1223), hoxY (sll1224), hoxH (sll1226), hypB1 (sll1432), hypA1 (slr1675). *Significant difference in SUF Knockdown mutant compared with WT (t test, p < 0.05)
The transcription changes of selected genes involved in photosynthetic and respiratory electron transport chains under photoautotrophic conditions are shown in Fig. 5f. Comparing with the WT, petJ was downregulated in the SUF Knockdown mutant. Genes encoding Complex I (NADH dehydrogenase) subunits, such as ndhD, ndhD2, and ndhI, were also downregulated. The Complex II encoding gene sdhB2 was significantly upregulated. Interestingly, Hox hydrogenase encoding gene hoxH was also upregulated. The activity of Hox hydrogenase requires the hyp operon (Rosa et al. 2015). Here, hypA1 and hypB1 were upregulated in the SUF Knockdown mutant compared with that in the WT (Fig. 5f). In heterotrophic conditions, almost all selected genes involved in photosynthetic and respiratory electron transport chains in SUF Knockdown mutant were downregulated except petH and sdhB2 (Fig. 5g). Hydrogenase-related genes within hox operon (hoxE, hoxF, and hoxH) and hyp operon (hypA1) from SUF Knockdown mutant showed significant downregulation (Fig. 5g).
P700+ reduction rate constants (s−1) were monitored according to the calculated electron transfer rate. Figure 6 and Table 1 show the P700+ reduction kinetics and rate in the WT and SUF Knockdown mutant under photoautotrophic and heterotrophic culture conditions. There was no significant difference between the WT and SUF Knockdown under either photoautotrophic or heterotrophic conditions initially. Cyclic electron transfer was monitored by adding DCMU prior to P700+ reduction measurement to block the electron transfer between PSII and PSI, i.e., to block the photosynthetic linear electron transfer pathway. The electron transfer rate of 13.37 ± 1.63 s−1 in the SUF Knockdown mutant was higher than that in the WT (6.41 ± 1.73 s−1) under photoautotrophic culture conditions. However, no significant differences were observed between the two strains under heterotrophic culture conditions (t test, P > 0.05). MV functions as an efficient P700+ electron acceptor and can block the cyclic electron flow around PSI. Using MV alone had no effect on P700+ reduction rates (Fig. 6; Table 1). The monitored electron transfers rates in the presence of both DCMU and MV represent the electrons flowing through PSI which are derived from the respiratory electron transport chain. With both DCMU and MV present in the samples, the P700+ reduction rate was higher under heterotrophic than photoautotrophic culture conditions, because the respiratory electron transport pathway functions as the main pathway.
P700+ reduction curves of the WT and SUF Knockdown mutant under various treatments. a P700+ reduction of the WT in photoautotrophic (Pa) conditions. b P700+ reduction of the SUF Knockdown mutant in Pa conditions. c P700+ reduction of the WT in heterotrophic (Ht) conditions. d P700+ reduction of the SUF Knockdown mutant in Ht conditions. Squares, circles, triangles, and diamonds represent no treatment, +DCMU, +DCMU +MV, and +MV treatments, respectively. DCMU, 10 μM DCMU was used to disrupt the linear electron flow; MV, 2 mM MV was used as an efficient PSI electron acceptor to block the cyclic electron flow
The distribution of the suf cluster and its relationship with various photosynthetic types
SUF proteins comprise the most important system for Fe–S formation in cyanobacteria, although some cyanobacteria also have ISC homologous proteins (Fontecave and Ollagnier-de-Choudens 2008). SufBCDSE are highly conserved proteins across all oxygenic photosynthetic organisms (Table 2). Interestingly, Chlorobi and Firmicutes that contain only type I reaction centers (RCIs) use the ISC system to assemble Fe–S clusters instead of the SUF system (Table 2). Notably, all anoxygenic photosynthetic bacteria that have RCII-type reaction centers use the SUF system to assemble Fe–S clusters. The oxygenic photosynthetic organisms use two reaction centers and SUF system was found in all oxygenic photosynthetic organisms.
Discussion
The ISC and SUF systems are two common Fe–S cluster assembly pathways among prokaryotes and eukaryotes and can provide Fe–S clusters to a wide range of apo-proteins. In E. coli, iscRSUA-hscBA-fdx-iscX (ISC system) and sufABCDSE (SUF system) operons co-exist (Mettert and Kiley 2015). The ISC system is the major Fe–S cluster assembly system and functions as housekeeping role. ISC system includes IscS (cysteine desulfurase), IscUA ([Fe–S] assembly scaffold), HscBA (molecular chaperones), and Fdx ([2Fe-2S] ferredoxin) (Schwartz et al. 2000; Ding and Clark 2004; Silberg et al. 2004). IscR is an inhibitory regulator controlling the activity level of the ISC system. Increased iscR activity causes severe growth inhibition due to decreased ISC enzyme activities (Tokumoto and Takahashi 2001). Conversely, the SUF system in E. coli only plays a role under iron starvation or oxidative stress conditions (Outten et al. 2004). However, the SUF system is the main Fe–S cluster assembly system in cyanobacteria and cannot be knocked out completely (Fig. 2) (Balasubramanian et al. 2006; Ayala-Castro et al. 2008). This paper investigated the SUF Fe–S cluster assembly system in Synechocystis sp. PCC 6803 and its functional relationship with photosynthesis and respiration. The transcriptional pattern and assembly mechanism of the suf operon were similar to those in E. coli, although the transcription mechanism relying on SufR is mainly found in cyanobacteria (Shen et al. 2007; Vuorijoki et al. 2017). Our results affirm the regulatory role of SufR in coordinating the expression of the SufBCD proteins suggested by phenotype analysis of knockout (ΔsufR) and overexpression (sufR-OE) mutants. Here, we showed that sufR overexpression (sufR-OE) resulted in a lower growth rate, decreased chlorophyll levels and PSII activities. The noticeably decreased SUF protein levels in the sufR-OE mutant demonstrated that sufR overexpression limited the expression of suf genes (Fig. S2). The decreased SUF protein levels in the SUF Knockdown mutant resulted in the same phenotype—lower growth rate, and decreased PSII activities and total chlorophyll levels—suggesting that SUF proteins are necessary for optimal photosynthetic reactions including the optimal ratio between the two photosystems. The transcriptional level of SUF system was significantly unregulated in WT under heterotrophic conditions except for sufA gene (Fig. S3). However, in SUF Knockdown mutant, the upregulated level of SUF system was limited (Fig. S3).
Iron–sulfur clusters are widely distributed in photosynthetic and respiratory electron transport chains (Frazzon et al. 2007; Balk and Pilon 2011; Balk and Schaedler 2014). The effects of iron–sulfur cluster synthesis on photosynthesis and respiration may be more complicated than results demonstrated (Figs. 4, 5, 6). It is known that Synechocystis sp. PCC 6803 will produce hydrogen in the absence of light and oxygen deprivation (Appel et al. 2000; Dutta and Vermaas 2016). SUF Knockdown mutant showed decreased photosynthetic activities including the decreased oxygen evolution rates under photoautotrophic conditions, which could be the reason for stimulating the activities of hydrogenase-related genes (hox operon and hyp genes) (Fig. 5f). In the contrast, under heterotrophic conditions, the WT cells showed relatively elevated transcriptional expression of hox operon and hyp genes comparing with SUF Knockdown due to the limited iron–sulfur cluster in the mutant (Fig. 5g). In the photoautotrophic conditions, photosynthesis provides the main energy for growth, and the effect of SUF Knockdown on photosynthetic activities is more obvious than its influence on respiration. It suggests that the organism prioritizes Fe–S clusters for respiration at the expense of photosynthesis. Under heterotrophic conditions, the similar growth rates observed indicate the important role of SUF system in heterotrophic growth, in which the respiratory electron transferring chains are the main energy metabolism.
In the current study, we found that suf genes play major roles in photosynthetic organisms that contain type II reaction centers with or without the corresponding isc genes (Table 2), suggesting that the SUF proteins have essential roles in the function of type II reaction centers. Electron acceptor co-factors (Fe–S centers in RCI and pheophytin/quinone complexes in RCII) are the main reason for the evolution of RCs into PSI and PSII (Blankenship 2010), but not for the distribution of the SUF system.
PSI subunits bind several Fe–S containing proteins and the limited activities of suf genes in the SUF Knockdown mutant resulted in a relatively decreased amount of PSI, which is consistent with previous reports (Yabe et al. 2004). However, no significant changes were observed between SUF Knockdown and the WT under heterotrophic culture conditions, suggesting that SUF protein levels have less impact on the respiratory electron transport system (Fig. 4). If we assume that in the presence of DCMU, the electron transfer rate is mainly recorded from cyclic electron transfer around PSI and also electron transfer from respiration, and in the presence of DCMU + MV, electron transfer is mainly recorded from the respiration chain, the cyclic electron transfer rates in the WT were similar between the two types of culture conditions, and similar cyclic electron transfer rates were also noted in the SUF Knockdown mutant grown under heterotrophic conditions, although cyanobacteria grown under heterotrophic conditions demonstrated the highest respiratory electron transfer rates (10.21 ± 0.30 s−1 in the WT and 11.10 ± 0.23 s−1 in SUF Knockdown). Unexpectedly, the highest cyclic electron transfer rate of 13.37 s−1 was found in the SUF Knockdown mutant grown under photoautotrophic conditions (Table 1). Though the relative amount of PSI was decreased in SUF Knockdown, the effects of Fe–S cluster deficiency on energy metabolic pathways in cyanobacteria are not clear at present, which shows the potential challenge in interpreting the phenotypes of the SUF Knockdown mutant. The defects in PSI (or the decreased relative proportion of PSI) coinciding with elevated cyclic electron transfer rates under photoautotrophic culture conditions may compensate for the presence of alternative electron transfer pathways. Further studies are needed to thoroughly investigate these potential electron transfer pathways.
Author contribution statement
SSZ, HBJ, MC, and BSQ designed the experiments, analyzed the data, and wrote the manuscript. SSZ and WYS performed all experiments. All authors read and approved the manuscript.
Abbreviations
- DCMU:
-
3-(3,4-dichlorophenyl)-1, 1-dimethylurea
- Fe–S:
-
Iron–sulfur
- Fv/Fm :
-
Maximal PSII quantum yield
- Fv′/Fm′ :
-
Operational PSII quantum yield
- Ht:
-
Heterotrophic
- ISC:
-
Iron–sulfur cluster
- NIF:
-
Nitrogen fixation
- MV:
-
Methylviologen
- Pa:
-
Photoautotrophic
- PSI:
-
Photosystem I
- PSII:
-
Photosystem II
- RCIs:
-
Type I reaction centers
- RCIIs:
-
Type II reaction centers
- SUF:
-
Sulfur formation
References
Alric J, Lavergne J, Rappaport F (2010) Redox and ATP control of photosynthetic cyclic electron flow in Chlamydomonas reinhardtii (I) aerobic conditions. Biochim Biophys Acta 1797:44–51
Appel J, Phunpruch S, Steinmüller K, Schulz R (2000) The bidirectional hydrogenase of Synechocystis sp. PCC 6803 works as an electron valve during photosynthesis. Arch Microbiol 173:333–338
Ayala-Castro C, Saini A, Outten FW (2008) Fe-S cluster assembly pathways in bacteria. Microbiol Mol Biol Rev 72:110–125
Balasubramanian R, Shen G, Bryant DA, Golbeck JH (2006) Regulatory roles for IscA and SufA in iron homeostasis and redox stress responses in the cyanobacterium Synechococcus sp. strain PCC 7002. J Bacteriol 188:3182–3191
Balk J, Pilon M (2011) Ancient and essential: the assembly of iron–sulfur clusters in plants. Trends Plant Sci 16:218–226
Balk J, Schaedler TA (2014) Iron cofactor assembly in plants. Annu Rev Plant Biol 65:125–153
Beinert H, Holm RH, Münck E (1997) iron-sulfur clusters: nature’s modular, multipurpose structures. Science 277:653–659
Blankenship RE (2010) Early evolution of photosynthesis. Plant Physiol 154:434–438
Bych K, Netz DJA, Vigani G, Bill E, Lill R, Pierik AJ, Balk J (2008) The essential cytosolic iron-sulfur protein Nbp35 acts without Cfd1 partner in the green lineage. J Biol Chem 283:35797–35804
Campbell D, Hurry V, Clarke AK, Gustafsson P, Öquist G (1998) Chlorophyll fluorescence analysis of cyanobacterial photosynthesis and acclimation. Microbiol Mol Biol Rev 62:667–683
Chahal HK, Dai Y, Saini A, Ayala-Castro C, Outten FW (2009) The SufBCD Fe-S scaffold complex interacts with SufA for Fe-S cluster transfer. Biochemistry 48:10644–10653
Ding H, Clark RJ (2004) Characterization of iron binding in IscA, an ancient iron-sulphur cluster assembly protein. Biochem J 379:433–440
Dismukes GC, Klimov VV, Baranov SV, Kozlov YN, DasGupta J, Tyryshkin A (2001) The origin of atmospheric oxygen on Earth: the innovation of oxygenic photosynthesis. Proc Natl Acad Sci USA 98:2170–2175
Dutta I, Vermaas WFJ (2016) The electron transfer pathway upon H2, oxidation by the NiFe bidirectional hydrogenase of Synechocystis sp. PCC 6803 in the light shares components with the photosynthetic electron transfer chain in thylakoid membranes. Int J Hydrogen. Energ 41(28):11949–11959
Elhai J, Wolk CP (1988) Conjugal transfer of DNA to cyanobacteria. Methods Enzymol 167:747–754
Fontecave M, Ollagnier-de-Choudens S (2008) Iron–sulfur cluster biosynthesis in bacteria: mechanisms of cluster assembly and transfer. Arch Biochem Biophys 474:226–237
Frazzon J, Dean DR (2002) Biosynthesis of the nitrogenase iron-molybdenum-cofactor from Azotobacter vinelandii. Met Ions Biol Syst 39:163–186
Frazzon APG, Ramirez MV, Warek U, Balk J, Frazzon J, Dean DR, Winkel BSJ (2007) Functional analysis of Arabidopsis genes involved in mitochondrial iron-sulfur cluster assembly. Plant Mol Biol 64:225–240
Gao H, Xu X (2009) Depletion of Vipp1 in Synechocystis sp. PCC 6803 affects photosynthetic activity before the loss of thylakoid membranes. FEMS Microbiol Lett 292(1):63–70
Hu X, Kato Y, Sumida A, Tanaka A, Tanaka R (2017) The SUFBC2D complex is required for the biogenesis of all major classes of plastid Fe-S proteins. Plant J 90(2):235–248
Jang S, Imlay JA (2010) Hydrogen peroxide inactivates the Escherichia coli Isc iron-sulphur assembly system, and OxyR induces the Suf system to compensate. Mol Microbiol 78:1448–1467
Ke WT, Dai GZ, Jiang HB, Zhang R, Qiu BS (2014) Essential roles of iron superoxide dismutase in photoautotrophic growth of Synechocystis sp. PCC 6803 and heterogeneous expression of marine Synechococcus sp. CC9311 copper/zinc superoxide dismutase within its sodB knockdown mutant. Microbiology-SGM 160:228–241
Layer G, Gaddam SA, Ayala-Castro CN, Ollagnier-de Choudens S, Lascoux D, Fontecave M, Outten FW (2007) SufE transfers sulfur from SufS to SufB for iron-sulfur cluster assembly. J Biol Chem 282:13342–13350
Lichtenthaler HK, Buschmann C (2001) Chlorophylls and carotenoids: measurement and characterization by UV-VIS spectroscopy. In: Wrolstad RE, Acree TE, Decker EA, Penner MH, Reid DS, Schwartz SJ, Shoemaker CF, Smith DM, Sporns P (eds) Current protocols in food analytical chemistry, Supplement 1. Wiley, New York, pp F431–F438
Lill R, Mühlenhoff U (2006) Iron-sulfur protein biogenesis in eukaryotes: components and mechanisms. Annu Rev Cell Dev Biol 22:457–486
Liu Y, Yu L, Ke W, Gao X, Qiu B (2010) Photosynthetic recovery of Nostoc flagelliforme (Cyanophyceae) upon rehydration after 2 years and 8 years dry storage. Phycologia 49:429–437
Maio N, Rouault TA (2015) Iron–sulfur cluster biogenesis in mammalian cells: new insights into the molecular mechanisms of cluster delivery. Biochim Biophys Acta 1853:1493–1512
Mettert EL, Kiley PJ (2015) How is Fe-S cluster formation regulated? Annu Rev Microbiol 69:505–526
Murakami A (1997) Quantitative analysis of 77 K fluorescence emission spectra in Synechocystis sp. PCC 6714 and Chlamydomonas reinhardtii with variable PSI/PSII stoichiometries. Photosynth Res 53:141–148
Nagane T, Tanaka A, Tanaka R (2010) Involvement of AtNAP1 in the regulation of chlorophyll degradation in Arabidopsis thaliana. Planta 231:939–949
Ollagnier-de-Choudens S, Sanakis Y, Fontecave M (2004) SufA/IscA: reactivity studies of a class of scaffold proteins involved in [Fe-S] cluster assembly. J Biol Inorg Chem 9:828–838
Outten FW (2015) Recent advances in the Suf Fe-S cluster biogenesis pathway: beyond the Proteobacteria. Biochim Biophys Acta 1853:1464–1469
Outten FW, Wood MJ, Munoz FM, Storz G (2003) The SufE protein and the SufBCD complex enhance SufS cysteine desulfurase activity as part of a sulfur transfer pathway for Fe-S cluster assembly in Escherichia coli. J Biol Chem 278:45713–45719
Outten FW, Djaman O, Storz G (2004) A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol Microbiol 52:861–872
Rosa ED, Checchetto V, Franchin C, Bergantino E, Berto P, Szabò I, Giacometti GM, Arrigoni G, Costantini P (2015) [NiFe]-hydrogenase is essential for cyanobacterium Synechocystis sp. PCC 6803 aerobic growth in the dark. Sci Rep 5:12424
Saini A, Mapolelo DT, Chahal HK, Johnson MK, Outten FW (2010) SufD and SufC ATPase activity are required for iron acquisition during in vivo Fe-S cluster formation on SufB. Biochemistry 49:9402–9412
Scherer S (1990) Do photosynthetic and respiratory electron transport chains share redox proteins? Trends Biochem Sci 15:458–462
Schwartz CJ, Djaman O, Imlay JA, Kiley PJ (2000) The cysteine desulfurase, IscS, has a major role in in vivo Fe-S cluster formation in Escherichia coli. Proc Natl Acad Sci USA 97:9009–9014
Seki A, Nakano T, Takahashi H, Matsumoto K, Ikeuchi M, Tanaka K (2006) Light-responsive transcriptional regulation of the suf promoters involved in cyanobacterium Synechocystis sp. PCC 6803 Fe-S cluster biogenesis. FEBS Lett 580:5044–5048
Shen G, Balasubramanian R, Wang T, Wu Y, Hoffart LM, Krebs C, Bryant DA, Golbeck JH (2007) SufR coordinates two [4Fe-4S]2+,1+ clusters and functions as a transcriptional repressor of the sufBCDS operon and an autoregulator of sufR in cyanobacteria. J Biol Chem 282:31909–31919
Silberg JJ, Tapley TL, Hoff KG, Vickery LE (2004) Regulation of the HscA ATPase reaction cycle by the co-chaperone HscB and the iron-sulfur cluster assembly protein IscU. J Biol Chem 279:53924–53931
Tokumoto U, Takahashi Y (2001) Genetic analysis of the isc operon in Escherichia coli involved in the biogenesis of cellular iron–sulfur proteins. J Biochem 130:63–71
Vinella D, Brochier-Armanet C, Loiseau L, Talla E, Barras F (2009) Iron-sulfur (Fe/S) protein biogenesis: phylogenomic and genetic studies of A-type carriers. PLoS Genet 5:e1000497
Vuorijoki L, Tiwari A, Kallio P, Aro EM (2017) Inactivation of iron-sulfur cluster biogenesis regulator SufR in Synechocystis sp. PCC 6803 induces unique iron-dependent protein-level responses. Biochim Biophys Acta 1861:1085–1098
Wollers S, Layer G, Garcia-Serres R, Signor L, Clemancey M, Latour JM, Fontecave M, Ollagnier de Choudens S (2010) Iron-sulfur (Fe-S) cluster assembly: the SufBCD complex is a new type of Fe-S scaffold with a flavin redox cofactor. J Biol Chem 285:23331–23341
Xu XM, Møller SG (2004) AtNAP7 is a plastidic SufC-like ATP-binding cassette/ATPase essential for Arabidopsis embryogenesis. Proc Natl Acad Sci USA 101:9143–9148
Xu XM, Møller SG (2006) AtSufE is an essential activator of plastidic and mitochondrial desulfurases in Arabidopsis. EMBO J 25:900–909
Xu XM, Adams S, Chua NH, Møller SG (2005) AtNAP1 represents an atypical SufB protein in Arabidopsis plastids. J Biol Chem 280:6648–6654
Yabe T, Morimoto K, Kikuchi S, Nishio K, Terashima I, Nakai M (2004) The Arabidopsis chloroplastic NifU-like protein CnfU, which can act as an iron-sulfur cluster scaffold protein, is required for biogenesis of ferredoxin and photosystem I. Plant Cell 16:993–1007
Zheng L, White RH, Cash VL, Jack RF, Dean DR (1993) Cysteine desulfurase activity indicates a role for NIFS in metallocluster biosynthesis. Proc Natl Acad Sci USA 90:2754–2758
Zheng L, Cash VL, Flint DH, Dean DR (1998) Assembly of iron–sulfur clusters: identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J Biol Chem 273:13264–13272
Acknowledgements
This study was supported by the National Basic Research Program (973 Program, No. 2008CB418004), the National Natural Science Foundation of China (No. 31470171), and the Fundamental Research Funds for the Central Universities (CCNU16KFY03). M.C holds an Australian Research Council Future Fellowship (FT120100464) and was supported by ARC CE1400015.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zang, SS., Jiang, HB., Song, WY. et al. Characterization of the sulfur-formation (suf) genes in Synechocystis sp. PCC 6803 under photoautotrophic and heterotrophic growth conditions. Planta 246, 927–938 (2017). https://doi.org/10.1007/s00425-017-2738-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-017-2738-0