Abstract
Main conclusion
PP2A catalytic subunit C2 is of special importance for light/dark regulation of nitrate reductase activity. The level of unmethylated PP2A catalytic subunits decreases in darkness.
Protein phosphatase 2A (PP2A) dephosphorylates and activates nitrate reductase (NR) in photosynthetically active tissue when plants are transferred from darkness to light. In the present work, investigation of Arabidopsis thaliana PP2A mutant lines revealed that one of the five PP2A catalytic subunit genes, e.g., C2, was of special importance for NR activation. Impairment of NR activation was, especially pronounced in the c2c4 double mutant. Though weaker, NR activation was also impaired in the c2 single mutant, and c1c2 and c2c5 double mutants. On the other hand, NR activation in the c4c5 double mutant was as efficient as in WT. The c4 single mutant had low PP2A activity, whereas the c2 single mutant possessed WT levels of extractable PP2A activity. PP2A activity was low in both c2c4 and c4c5. Differences in extracted PP2A activity among mutants did not strictly correlate with differences in NR activation, but underpinned that C2 has a special function in NR activation in vivo. The terminal leucine in PP2A catalytic subunits is generally methylated to a high degree, but regulation and impact of methylation/demethylation is barely studied. In WT and PP2A mutants, the level of unmethylated PP2A catalytic subunits decreased during 45 min of darkness, but did not change much when light was switched on. In leucine carboxyl methyl transferase1 (LCMT1) knockout plants, which possess mainly unmethylated PP2A, NR was still activated, although not fully as efficient as in WT.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nitrate reductase (NR) catalyzes the reduction of nitrate to nitrite, a critical step for nitrogen assimilation in plants. NR activity is regulated by light. When plants are exposed to photosynthetic active light, ferredoxin in the photosynthetic electron transport chain reduces a disulphide bridge in chloroplastic thioredoxin. Thioredoxin then reduces, and thereby activates, several enzymes in the Calvin cycle (Lillo 2008; Naranjo et al. 2016). Rubisco activase which is responsible for activation of the CO2 fixation enzyme rubisco is activated through the thioredoxin system (Portis 2003). As for Calvin cycle enzymes and rubisco activase, NR is activated by light and deactivated in darkness. Although CO2 and photosynthetic electron transport is necessary for rapid post-translational activation of NR, the mechanisms linking activation of photosynthesis and regulation of NR is not known (Kaiser and Brendle-Behnisch 1991; Provan and Lillo 1999; Nemie-Feyissa et al. 2013). Notably, NR is localized to the cytosol, but is co-regulated with photosynthesis and nitrite reduction in the chloroplasts.
Phosphorylation of a conserved Ser residue in the hinge one in-between the molybdenum cofactor binding domain and hem binding domain has long been associated with inactivation of NR in darkness. Phosphorylation in itself is, however, not sufficient to inactivate NR, but 14-3-3 proteins and cations, for example Mg2+, are necessary for interacting with the phosphorylated amino acid residue to inactivate NR (Huber et al. 2002). More recently it was shown that the N-terminal acidic domain of NR also binds 14-3-3, and contributes to inactivation of NR (Chi et al. 2015).
Protein phosphatase 2A (PP2A) has long been recognized as a regulator of NR (nitrate reductase), and is important for mediating positive effects of photosynthetic active light on NR (Kaiser and Brendle-Behnisch 1991; MacKintosh 1992; Lillo 2008). PP2A is a trimer made up of a catalytic (C), scaffolding (A) and regulatory (B) subunit. These three types of building blocks are conserved among eukaryotes, but development of isoforms within each group has taken place after evolutionary splitting of plants and animals (Moorhead et al. 2009; Lillo et al. 2014). In Arabidopsis there are five C, three A and 17 B subunits, theoretically enabling up to 255 different combinations in the PP2A complex. The Arabidopsis B subunits are further divided into three groups, two B/B55, nine B′ and six B″ (Farkas et al. 2007). For activation of NR the B/B55 regulatory subunits appear to be important (Heidari et al. 2011). Interestingly, plants have a relatively high number of genes encoding C subunits; whereas Arabidopsis has five, mammals have only two C subunit genes. The reason for high diversity of C subunits in higher plants is not known, but could possibly be linked with special needs for mastering environmental changes. The higher plants C subunits are clustered into subgroups I and II, C1/C2/C5 and C3/C4, respectively, in Arabidopsis. Knocking out all genes within each of these subgroups has serious or detrimental effects on plant growth and development (Ballesteros et al. 2013). Some specific functions of the different C subunits are now starting to be revealed. Knockout of C2 renders Arabidopsis hypersensitive to ABA during germination, indicative of C2 as a negative regulator in ABA signaling (Pernas et al. 2007). The C5 knockout phenotype is indicative of C5 participation in brassinosteroid signaling (Tang et al. 2011) whereas C5 ectopic overexpression confers salt tolerance (Hu et al. 2017). C3 and C4 are important for auxin signaling (Ballesteros et al. 2013). C3 is a positive regulator of the receptor kinase ACR4 and plays an essential role in formative cell division in Arabidopsis (Yue et al. 2016). All single knockouts, except C2, showed less ABA effects during germination, indicative of C1, C3, C4, and C5 being positive components in ABA signaling (Waadt et al. 2015). Changes in PP2A properties may be involved in the light regulation of NR. PP2A itself is subject to post-translational regulation, and from other eukaryotes it is known that methylation of the terminal carboxyl group (of leucine) of the C subunit influences assembly and activity of PP2A (Janssens et al. 2008). The enzymes mediating methylation/demethylation of PP2A, e.g., LCMT1 (leucine carboxyl methyl transferase1) and PME1 (PP2A methyl esterase1) have mainly been studied in animals and yeast but are conserved also in plants (Lillo et al. 2014). In S. cerevisiae, which has only two B subunits, methylation promotes assembly of both B (Cdc55) and B′ (Rts1) with the core AC (scaffolding-catalytic) dimer of PP2A. The methylation is apparently somewhat more important for B than B′ in the assembling process (Wu et al. 2000; Gentry et al. 2005; Janssens et al. 2008). In mammals, assembly of the B (PR55) subunits into the PP2A complex was strongly promoted by C methylation, whereas other regulatory subunits, B′, B″ and B′′′, were only slightly, or not at all influenced (Longin et al. 2007; Janssens et al. 2008). The effects of C methylation on assembly of plant PP2A complexes have not been studied, but if methylation will enhance formation of PP2A-B complexes also in plants, methylation would likely favor NR activation, since B subunits were important for NR activation in Arabidopsis (Heidari et al. 2011). In this work we used a range of knockout mutants to elucidate importance of the different C subunits for light activation of NR, and monitored the degree of terminal methylation of the C subunits.
Materials and methods
Plant growth
Arabidopsis thaliana single mutant T-DNA insert lines lcmt-1 (SALK_079466) (Alonso et al. 2003), and pme1 (GK_804C11) (Kleinboelting et al. 2012) were obtained from the European Arabidopsis Stock Centre in Nottingham, UK. Homozygous mutant selection was done by PCR using primers for T-DNA insertion lines recommended at the SALK institute website. The c2 mutant from CSIC-INIA T-DNA insertion collection in the Wassilewskija background was described by Pernas et al. (2007). This T-DNA mutant was introgressed into the Columbia ecotype after six sequential crosses. Similar phenotype as described by Pernas et al. (2007) were referred to in Ballesteros et al. (2013). Other T-DNA insertion mutants were SALK_102599 (At1g59830, pp2a-c1), SALK_035009 (At3g58500, pp2a-c4), SALK_139822 (At1g69960, pp2a-c5), and the SAIL_182_A02 (At2g42500, pp2a-c3). To obtain the different double mutant combinations, c1 mutant was crossed with c2 (Col-0 background), c3, c4 and c5; c3 was crossed with c4 and c5; c4 was crossed with c5. The effects of these mutations on PP2A-C gene expression was described in Segonzac et al. (2014) indicating that each mutant is indeed a null allele. Seeds were sown directly in regular plant soil and stratified at 4 °C for two days, then grown under artificial light at 22 °C in a 12 h light/12 h dark regimen. Leaves of rosette stage plants were harvested 5–6 weeks after germination, quickly frozen in liquid nitrogen, pulverized, stored at −70 °C, and used for testing NR activity and for immunoblotting to test the methylation state of PP2A catalytic subunits. Since time point accuracy was critical for determining NR activation in plants after transfer from darkness to light, only one or two mutants could be harvested at the same time, and WT was always included as a reference.
Assay of nitrate reductase activity
Rosette leaves (0.5 g) were homogenized in a mortar with 2 mL cold extraction buffer (100 mM Hepes–KOH pH 7.5, 1 mM EDTA, 7 mM cysteine). PVPP was added using 1 g for 30 mL buffer. The extract was filtered through Mira cloth and 50 µL filtered extract was assayed in 700 µL assay volume (50 mM Hepes–KOH pH 7.5, 2 mM KNO3, 200 µM NADH) containing 2 mM EDTA to assay total NR activity (non-phosphorylated plus phosphorylated NR). To measure actual activity (non-phosphorylated NR), NR was tested in the presence of 6 mM MgCl2. The assay was run at 25 °C for 10 min. Nitrite formed was determined by addition of 700 µL 1% sulphanilamide and 0.02% N-(naphtyl)-ethylene-diamine dihydrochloride in 1.2 N HCl. Samples were centrifuged for 30 min at 13,800×g and absorbance was read spectrophotometrically at 540 nm. Absorbance of 1 corresponds with 27 µmol NO2− formed per g fresh weight per h. NR activity state was defined by the ratio of the actual activity (Mg-assay) to the total NR activity (EDTA-assay) multiplied by 100 (Mackintosh et al. 1995; Heidari et al. 2011). Unpaired t test was used to compare NR activation (slope of the line) between mutants and WT.
Immunoblotting
Arabidopsis leaves were crushed in liquid nitrogen, and 50 mg plant powder was homogenized with 100 µL 2× Laemmli buffer (Bio-Rad Laboratories, München, Germany) containing 5 μL β-mercaptoethanol (Sigma-Aldrich, St. Louis, MO, USA). The extract was boiled straight away for 15 min, and centrifuged at 15, 000g for 20 min. The supernatant was used for the immuno-blot analysis. Proteins were separated by SDS PAGE gel electrophoresis using a 12% Mini-PROTEAN® TGX™GEL (Bio-Rad), with Tris/Glycine/SDS as the running buffer (Bio-Rad). Protein transfer was confirmed by Ponceau red (Sigma-Aldrich) staining after electro-blotting of proteins onto a 0.2 µm PVDF membrane (Bio-Rad). Blocking was done overnight at 4 °C with PBS containing 0.1% Tween-20 and 5% dry milk (PBST/MLK). The primary antibody specific for unmethylated PP2A-C (demethylated-PP2A-C (4i57): sc-80990, Santa Cruz Biotechnology Inc., Dallas, TX, USA) was added as a 1:1000 dilution in freshly prepared PBST, followed by 2 h incubation at room temperature. The blot was rinsed 6 × 5 min with PBST and incubated at room temperature for 1.5 h with the secondary antibody (goat anti mouse IgG-HRP, Santa Cruz Biotechnology Inc.) diluted 1:7500 in PBST/MLK. Before developing, the membrane was again washed 6 × 5 min with PBST followed by protein detection using ECL detection kit (Amersham™ECL™Prime Western Blotting Detection Reagent RPN2232, GE healthcare, Buckingshire, UK). The antibody was checked, and found to detect truncated Arabidopsis C2 protein heterologously expressed in Escherichia coli, and detected a peptide corresponding with the nine C-terminal amino acids of C3 and C4, but not peptides corresponding with homologues proteins PP4 or PP6, nor methylated peptide (Supplementary Fig. S1). Quantification of band intensity was performed using Image J software (https://imagej.nih.gov/ij/index.html ). Unpaired t test was used for analysis of statistical significance.
Detection of total PP2A catalytic subunits
The total amount of PP2A-C was determined via alkali-induced de-methylation to cause removal of the methyl group attached to the C-terminal Leu of the PP2A C subunit, and enable detection of total PP2A-C. Alkaline treatment was adapted according to descriptions at Merck-Millipore certificate for analysis following the antibody against methylated PP2A-C (Anti-methyl-PP2A, C subunit, clone 2A10, 04-1479, Merck, Darmstadt, Germany). Alkali treatment was done directly on the PVDF membrane after protein transfer by incubating the blot with NaOH (0.2 M) for 30 min. at 37 °C with gentle shaking. After NaOH treatment, blots were washed 6 × 10 min with PBST followed by blocking and antibody incubation as described.
PP2A activity assay
The Ser/Thr Phosphatase Assay System from Promega (Promega 2009, Madison, WI, USA) was used. Rosette leaves were harvested 3 h into the photoperiod. The frozen plant tissue was homogenized in extraction buffer 50 mM Tris–HCl, pH 7.0, 0.5 mM EDTA, 0.5 mM EGTA, 2 mM DTT and 0.01% w/v Brij 35 (Cohen et al. 1988; McAvoy and Nairn 2010) in a ratio 100 mg of plant tissue to 400 μL of extraction buffer with subsequent centrifugation at 17,000g at 4 °C for 1 h. Extracts were desalted on spin columns, and protein quantified by Bradford assay (Bradford 1×Dye Reagent Quick Start™ from Bio-Rad). Assay mixtures with 0.1 μg/μL proteins and 0.1 mM phosphorylated peptide (RRA(pT)VA) were incubated at 37 °C for 5 min, and terminated by adding 50 μL of the molybdate dye. Assay mixtures terminated at time zero were used as controls. To inhibit the activity of PP2A (Cohen et al. 1989), 2 nM okadaic acid sodium salt (Sigma-Aldrich) was used in the assay buffer. Assay buffer was 50 mM Tris–HCl pH 7.0, 0.2 mM EDTA, 0.02% (v/v) 2-mercaptoethanol, and 0.1 mg/mL bovine serum albumin. PP2A activity was calculated as the difference of phosphate released with and without 2 nM okadaic acid.
Results
Consistent with a large degree of functional overlapping, all single pp2a-c mutants looked normal, and could not be visually distinguished from WT (Supplementary Fig. S2a). The double mutants looked similar to WT, except for the c2c4 mutant that was clearly smaller (Supplementary Fig. S2b). In addition, the c2c5 mutant was slightly smaller than WT (Supplementary Fig. S2c). Moreover, the c2c4 and c2c5 mutants made fewer seeds than WT (not presented). Nitrate reductase activity was tested in PP2A single and double catalytic subunit mutants. Phosphorylation and binding of 14-3-3s form the basis for the differential NR assay; 14-3-3 proteins present in the crude extract will bind to and inactivate NR in the presence of Mg2+, whereas presence of EDTA will release 14-3-3s from NR, and activity then measured represents total NR, e.g., both phosphorylated and non-phosphorylated.
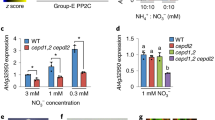
For testing activation of NR, rosette leaves were harvested from plants grown in a 12 h light/12 h darkness regimen. Since NR expression is subjected to diurnal variations, plants were first exposed to 3 h of light to establish a high level of NR protein on the day of harvesting. Thereafter, plants were exposed to 45 min of darkness before light was again switched on. Leaves of single mutant lines of all five catalytic subunits were sampled and assayed after 45 min of darkness and subsequently after 20 min of light. NR was activated in all mutants, where only the c2 knockout line showed significantly slower activation of NR than WT (Fig. 1, left column). Different C subunits may generally be functionally degenerate with respect to their substrates, or be able to replace one another when other subunits are knocked out. To gain further insight into the role of different PP2A subunits, a wide range of PP2A double mutants were tested. Plants were harvested after the 3 h light period, at the end of the 45 min dark period, and after 30 min re-exposure to light (Fig. 2). NR activity state in WT plants declined during darkness and regained during the following 30 min of light. The regain of NR activity was most significantly hampered in the c2c4 mutant. Activation of NR was also hampered in c1c2 and c2c5 double mutants but not in the c4c5 double mutant (Fig. 2, left column). These results underpin that C2 is important for activation of NR. As expected, the total activity of NR (which reflects NR protein level) showed only small variations during the dark/light transitions (Figs. 1, 2, right columns). Interestingly, the total amount of NR was generally higher in those double mutants with impaired NR activation (c2c4, c2c5, c1c2). Several other double mutants were also tested (c1c3, c1c5, c2c3, c2c5), but only C2 knockout in combination with knocked out C4, C5 and C1 impaired NR activation (Fig. 2; Supplementary Fig. S3). Furthermore, c2c4 crossed with b/b55 knockout lines (named pp2a-bα or pp2a-bβ) were tested. However, the triple mutants behaved very similar to the c2c4 mutant (data not shown). This confirmed that presence of one of the two B subunits, Bα or Bβ is sufficient for full activation of NR (Heidari et al. 2011).
Nitrate reductase (NR) activity state and total NR activity in WT and PP2A catalytic single mutant lines (c1-c5) after a dark to light transfer: c1 (a, b), c2 (c, d), c3 (e, f), c4 (g, h), c5 (i, j). Plants were grown in soil in a 12 h light/12 h darkness regimen, and leaves were harvested from 5-week-old rosette stage plants. On the day of harvesting, plants were exposed to 3 h of light, then placed in darkness for 45 min before harvesting (time 0). Thereafter plants were transferred back to light and harvested after 20 min. NR activity in each sample was measured by two different assays, e.g., with Mg2+ to reveal non-phosphorylated NR, and with EDTA for total NR. Activity state (% active NR, e.g., non-phosphorylated NR as percentage of total amount of NR) is shown in left column. Total NR activity, assayed in the presence of EDTA, is given in the right column. Each point is the mean of six biological samples, except for c2 (c, d), which is the mean of nine samples, SE is given. For c2, the rate of NR activation after transfer to light (slope of the line) was significantly lower compared with WT with P = 0.03. For all other mutants (a, e, g, i) the rate was not significantly different from WT, P > 0.2
Nitrate reductase (NR) activity state and total NR activity in WT, PP2A catalytic double mutants, lcmt1, and pme1 after light/dark/light shifts: c1c2 (a, b), c2c4 (c, d), c2c5 (e, f), c4c5 (g, h), lcmt1, pme1 (i, j). Leaves were sampled after 3 h of light (time 0), 45 min of darkness, and 30 min of light, and assayed as described in Fig. 1. NR activity state (% active NR) is presented in the left column. Total NR activity (with EDTA) is given in the right column. Each point is the mean of nine biological samples. SE is given. The rate of NR activation after transfer to light (slope of the line, 45–75 min) was significantly lower compared with WT for c2c4 and lcmt1 at P < 0.01; for c2c1 P = 0.04; for c2c5 P = 0.06. The level of total NR (with EDTA) was always significantly higher compared with WT for c2c4 and c2c5, with P < 0.001. For c1c2 total NR was significantly higher than for WT at the two latter time points with P < 0.01
LCMT1 methylates PP2A catalytic subunits in Arabidopsis (Wu et al. 2011). As in other eukaryotes, methylation is likely to influence PP2A interaction with other proteins, and impaired NR activation was seen in the lcmt1 mutant (Fig. 2i). Knockout of the PME1 gene (At4g10050), which is believed to encode an enzyme that acts antagonistically to LCMT1, did not produce any significant alteration of NR activation (Fig. 2i). However, NR activity was more variable in the pme1 mutant than in other lines, as reflected in the large standard errors (Fig. 2i, j).
A decrease in the level of unmethylated PP2A catalytic subunits (PP2A-C) was detected in WT after 45 min of darkness (Figs. 3c, 4a, b). However, in contrast to the pronounced change in NR activity state (Fig. 3a), no rapid change in PP2A-C methylation status was observed in response to light-on (Fig. 3c). Moreover, the level of total PP2A-C was almost constant during light/dark/light transitions (Fig. 3d). Methylation levels were also tested in the pme1 and lcmt1 mutants and compared with WT after exposure of plants to 3 h of light, 45 min darkness, and 30 min light (Fig. 4a–c). The levels of PP2A-C, unmethylated and total were similar in leaves of pme1 and WT, with indications of somewhat less unmethylated C in pme1 after 3 h light (Fig. 4a, b). Compared with WT or pme1, the level of unmethylated PP2A-C was much higher in lcmt1 under all conditions. PP2A-C levels were also determined for c2c4 and c2c5 mutants and representative blots are shown (Fig. 4d, e). In WT and both double mutants unmethylated PP2A-C decreased by 50–60% after 45 min of darkness, whereas total PP2A-C did not change significantly. In conclusion, a significant decrease of unmethylated PP2A-C was observed in both WT and mutants during darkness. The exception was only lcmt1, which always had very high levels of unmethylated PP2A-C. Extractable PP2A activity from leaves of WT and most relevant mutants was assayed (Table 1). In comparison with WT, c2c4 and c4c5 double mutants possessed low PP2A activity. Although extractable PP2A activity was also significantly lower in the c4c5 mutant, light activation of NR was as for WT (Fig. 2g). Assay of all single mutants showed that only c4 had significantly lower PP2A activity compared with WT (Supplementary Table S1). This gives an explanation why low activity was found for c2c4 and c4c5, but not c2c5. The results underpin that NR activation is not linked to total PP2A in the cell, and gives evidence that the C2 subunit is crucial for controlling NR activity.
Nitrate reductase (NR) activity and levels of unmethylated and total PP2A-C during light/dark/light transitions. WT plants were exposed to light for 3 h before harvesting (time 0), then transferred to darkness and harvested after 45 min, re-exposed to light and harvested after 5, 10, 15, 30 and 45 min. a NR activity state. b Total NR (EDTA assay). Data presented are mean values of at least 6 samples. SE is given. c, d Immunoblots of crude extracts (3 µL). c Upper panel shows the loading control (Ponceau staining). Lower panel shows immunoblot with antibody specific towards unmethylated PP2A-C. d Upper panel shows the loading control (Ponceau staining). Lower panel shows immunoblot with antibody specific towards unmethylated PP2A-C on blot treated with NaOH to demethylated all PP2A-C, giving the total amount of PP2A-C. Immunoblotting was repeated four times with different biological samples. Representative blots are shown. Numbers below blots show quantification of bands according to Image J. Numbers for the depicted blot and mean values of four blots are given. Values significantly different from 3 h light are marked with a star, *P < 0.05
Levels of unmethylated and total PP2A-C in extracts from WT, pme1, lcmt1, c2c4, and c2c5 plants exposed to light/dark shifts. Plants were exposed to 3 h of light before the first harvesting, then transferred to darkness and harvested after 45 min, and in some experiments also harvested after re-exposure to light for 30 min. Upper panels show the loading control (Ponceau staining); lower panels show immunoblots with antibody towards unmethylated-PP2A-C for blots without special treatment (PP2A-C-unmethylated) and blots treated with NaOH to demethylate proteins for visualizing total amount of PP2A-C (PP2A-C-total). a Unmethylated PP2A-C and 5 µL crude extract. b Unmethylated PP2A-C and 10 µL crude extract. c The total amount of PP2A (5 µL crude extract). d Unmethylated PP2A-C (5 µL crude extract). e Total PP2A-C (5 µL crude extract). Representative blots are shown. Numbers below blots show quantification of bands according to Image J for the depicted blots, and (for d, e) also average values from three blots, average values significantly different from WT light are marked with a star, *P < 0.05
Discussion
A special role for the PP2A-C2 subunit
The Arabidopsis PP2A subfamily I contains C1, C2, C5, and subfamily II contains C3 and C4. Subunits within one subfamily share 95% sequence identity and between subfamilies about 80%, which suggests that genes may functionally replace each other. The WT appearance of all pp2a-c single mutants supports this tenet (Yu et al. 2005; Ballesteros et al. 2013). According to expression levels, all C subunits are possible candidates for being involved in NR activation in leaves. All C subunits are expressed in rosette stage leaves with C3 and C4 transcripts at 2–4 times higher levels than the other C transcripts (unpublished RNA-seq data). Except for the c2 mutant, single null mutants of any of the other catalytic subunits were not significantly different from WT plants regarding NR activation. Even in the case of the c2 mutant, the reduction in NR activation was not severe (Fig. 1). This indicates redundancy between the C-subunits regarding NR activation. C2 has previously been implicated in ABA signaling (Pernas et al. 2007), hence appears to have diverse functions in Arabidopsis. Double (c3c4) or triple (c1c2c5) knockouts are extremely dwarfed and misshapen, and do not survive to adult stage in soil (Ballesteros et al. 2013). Noteworthy, the extreme phenotypes are created when all genes within the C subfamilies I or II were knocked out, further indicating that all C subunits are functional. The double mutants used in the present work always had a non-mutated gene from each sub-family present and plants looked normal, except for reduced size and seed set of c2c4 and c2c5. Testing various double mutants revealed that NR activation is strongly impaired when knockout of C2 was accompanied by knockout of C4 and, though less significant also when accompanied with knockout of C1 and C5. The c4c5 double knockout on the other hand, did not show impaired NR activation. This supports the hypothesis of a special role for C2 in NR activation. Extracted PP2A activity was low in c2c4 and c4c5, but high in c2 single mutants, hence differences in total extractable PP2A activity could not explain differences in NR activation, but further underpinned that C2 has a special function in NR activation.
Unmethylated PP2A-C decreases in darkness
Previously, we had shown that during light–dark–light transitions the Arabidopsis PP2A-B subunits were necessary for optimal activation of NR (Heidari et al. 2011). Since methylation of PP2A-C increased the core PP2A-AC dimer affinity towards B subunits in both mammals and yeast (Snabaitis et al. 2006; Longman et al. 2014), it was hypothesized that methylation of Arabidopsis C subunits could increase the level of PP2A complexes containing B subunits and thereby facilitate NR activation; hence PP2A methylation would appear as part of the signal network that activates NR. NR activation after dark to light transition was not paralleled by changes in methylation status of C subunits. Light or photosynthesis did not appear to trigger rapid changes in methylation status, to precede or accompany light-induced NR activation. However, in WT and mutant plants, except for the lcmt1 mutant, the amount of unmethylated C clearly decreased during 45 min of darkness. Since the total amount of C was almost constant during the dark period, this implicates that methylated C likely increased during darkness (Figs. 3, 4). The higher level of methylated C present after 45 min of darkness may be important for the subsequent activation of NR when light is switched on. Consistent with this hypothesis, activation of NR took place less efficiently in the lcmt1 mutant where C subunits remain unmethylated (Figs. 2i, 4a–c) (Wu et al. 2011). This shows that a high methylation degree of C-subunits was not an absolute requirement for NR activation. As shown by Wu et al. (2011), in the lcmt1 knockout mutant unmethylated PP2A-C became localized to cell compartments otherwise occupied only by methylated PP2A-C. This unmehtylated PP2A-C may overtake the function of methylated PP2A when the latter is not present providing an explanation for the NR activation in the mutant. However, the high PP2A-C methylation that occurs in WT plants in darkness likely facilitates binding of B-type PP2A-B subunits upon re-illumination, thus increasing affinity for NR leading to dephosphorylation-mediated enzyme activation.
Author contribution statement
CL, JSS, MTC planned the work. MTC did the main part of the experiments with help from MS, ARAK, BH, DNM and IOA. CL and MTC wrote the manuscript, and all authors discussed and commented on the manuscript.
Abbreviations
- LCMT1:
-
Leucine carboxyl methyl transferase 1
- NR:
-
Nitrate reductase
- PME:
-
PP2A methyl esterase
- PP2A:
-
Protein phosphatase 2A
References
Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301(5633):653–657. doi:10.1126/science.1086391
Ballesteros I, Dominguez T, Sauer M, Paredes P, Duprat A, Rojo E, Sanmartin M, Sanchez-Serrano JJ (2013) Specialized functions of the PP2A subfamily II catalytic subunits PP2A-C3 and PP2A-C4 in the distribution of auxin fluxes and development in Arabidopsis. Plant J 5:862–872. doi:10.1111/tpj.12078
Chi JC, Roeper J, Schwarz G, Fischer-Schrader K (2015) Dual binding of 14-3-3 protein regulates Arabidopsis nitrate reductase activity. J Biol Inorg Chem 20(2):277–286. doi:10.1007/s00775-014-1232-4
Cohen P, Alemany S, Hemmings BA, Resink TJ, Stralfors P, Tung HY (1988) Protein phosphatase-1 and protein phosphatase-2A from rabbit skeletal muscle. Methods Enzymol 159:390–408
Cohen P, Klumpp S, Schelling DL (1989) An improved procedure for identifying and quantitating protein phosphatases in mammalian tissues. FEBS Lett 250(2):596–600
Farkas I, Dombradi V, Miskei M, Szabados L, Koncz C (2007) Arabidopsis PPP family of serine/threonine phosphatases. Trends Plant Sci 12(4):169–176. doi:10.1016/j.tplants.2007.03.003
Gentry MS, Li Y, Wei H, Syed FF, Patel SH, Hallberg RL, Pallas DC (2005) A novel assay for protein phosphatase 2A (PP2A) complexes in vivo reveals differential effects of covalent modifications on different Saccharomyces cerevisiae PP2A heterotrimers. Eukaryot Cell 4(6):1029–1040. doi:10.1128/EC.4.6.1029-1040.2005
Heidari B, Matre P, Nemie-Feyissa D, Meyer C, Rognli OA, Moller SG, Lillo C (2011) Protein phosphatase 2A B55 and A regulatory subunits interact with nitrate reductase and are essential for nitrate reductase activation. Plant Physiol 156(1):165–172. doi:10.1104/pp.111.172734
Hu R, Zhu Y, Wei J, Chen J, Shi H, Shen G, Zhang H (2017) Overexpression of PP2A-C5 that encodes the catalytic subunit 5 of protein phosphatase 2A in Arabidopsis confers better root and shoot development under salt conditions. Plant, Cell Environ 40(1):150–164. doi:10.1111/pce.12837
Huber SC, MacKintosh C, Kaiser WM (2002) Metabolic enzymes as targets for 14-3-3 proteins. Plant Mol Biol 50(6):1053–1063
Janssens V, Longin S, Goris J (2008) PP2A holoenzyme assembly: in cauda venenum (the sting is in the tail). Trends Biochem Sci 33(3):113–121. doi:10.1016/j.tibs.2007.12.004
Kaiser WM, Brendle-Behnisch E (1991) Rapid modulation of spinach leaf nitrate reductase activity by photosynthesis: I. modulation in vivo by CO(2) availability. Plant Physiol 96(2):363–367
Kleinboelting N, Huep G, Kloetgen A, Viehoever P, Weisshaar B (2012) GABI-Kat SimpleSearch: new features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res 40(Database issue):D1211–1215. doi:10.1093/nar/gkr1047
Lillo C (2008) Signalling cascades integrating light-enhanced nitrate metabolism. Biochem J 415(1):11–19. doi:10.1042/BJ20081115
Lillo C, Kataya AR, Heidari B, Creighton MT, Nemie-Feyissa D, Ginbot Z, Jonassen EM (2014) Protein phosphatases PP2A, PP4 and PP6: mediators and regulators in development and responses to environmental cues. Plant, Cell Environ 37(12):2631–2648. doi:10.1111/pce.12364
Longin S, Zwaenepoel K, Louis JV, Dilworth S, Goris J, Janssens V (2007) Selection of protein phosphatase 2A regulatory subunits is mediated by the C terminus of the catalytic subunit. J Biol Chem 282(37):26971–26980. doi:10.1074/jbc.M704059200
Longman MR, Ranieri A, Avkiran M, Snabaitis AK (2014) Regulation of PP2AC carboxylmethylation and cellular localisation by inhibitory class G-protein coupled receptors in cardiomyocytes. PLoS One 9(1):e86234. doi:10.1371/journal.pone.0086234
MacKintosh C (1992) Regulation of spinach-leaf nitrate reductase by reversible phosphorylation. Biochim Biophys Acta 1137(1):121–126
Mackintosh C, Douglas P, Lillo C (1995) Identification of a protein that inhibits the phosphorylated form of nitrate reductase from spinach (Spinacia oleracea) leaves. Plant Physiol 107(2):451–457
McAvoy T, Nairn AC (2010) Serine/threonine protein phosphatase assays. Curr Protoc Mol Biol Chapter 18(Unit18):18. doi:10.1002/0471142727.mb1818s92
Moorhead GB, De Wever V, Templeton G, Kerk D (2009) Evolution of protein phosphatases in plants and animals. Biochem J 417(2):401–409. doi:10.1042/BJ20081986
Naranjo B, Diaz-Espejo A, Lindahl M, Cejudo FJ (2016) Type-f thioredoxins have a role in the short-term activation of carbon metabolism and their loss affects growth under short-day conditions in Arabidopsis thaliana. J Exp Bot 67(6):1951–1964. doi:10.1093/jxb/erw017
Nemie-Feyissa D, Krolicka A, Forland N, Hansen M, Heidari B, Lillo C (2013) Post-translational control of nitrate reductase activity responding to light and photosynthesis evolved already in the early vascular plants. J Plant Physiol 170(7):662–667. doi:10.1016/j.jplph.2012.12.010
Pernas M, Garcia-Casado G, Rojo E, Solano R, Sanchez-Serrano JJ (2007) A protein phosphatase 2A catalytic subunit is a negative regulator of abscisic acid signalling. Plant J 51(5):763–778. doi:10.1111/j.1365-313X.2007.03179.x
Portis AR Jr (2003) Rubisco activase—rubisco’s catalytic chaperone. Photosynth Res 75(1):11–27. doi:10.1023/A:1022458108678
Provan F, Lillo C (1999) Photosynthetic post-translational activation of nitrate reductase. J Plant Physiol 154:605–609
Segonzac C, Macho AP, Sanmartin M, Ntoukakis V, Sanchez-Serrano JJ, Zipfel C (2014) Negative control of BAK1 by protein phosphatase 2A during plant innate immunity. EMBO J 33(18):2069–2079. doi:10.15252/embj.201488698
Snabaitis AK, D’Mello R, Dashnyam S, Avkiran M (2006) A novel role for protein phosphatase 2A in receptor-mediated regulation of the cardiac sarcolemmal Na+/H+ exchanger NHE1. J Biol Chem 281(29):20252–20262. doi:10.1074/jbc.M600268200
Tang W, Yuan M, Wang R, Yang Y, Wang C, Oses-Prieto JA, Kim TW, Zhou HW, Deng Z, Gampala SS et al (2011) PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat Cell Biol 13(2):124–131. doi:10.1038/ncb2151
Waadt R, Manalansan B, Rauniyar N, Munemasa S, Booker MA, Brandt B, Waadt C, Nusinow DA, Kay SA, Kunz HH, Schumacher K, DeLong A, Yates JR 3rd, Schroeder JI (2015) Identification of open stomata1-interacting proteins reveals interactions with sucrose non-fermenting1-related protein kinases2 and with type 2A protein phosphatases that function in abscisic acid responses. Plant Physiol 169(1):760–779. doi:10.1104/pp.15.00575
Wu J, Tolstykh T, Lee J, Boyd K, Stock JB, Broach JR (2000) Carboxyl methylation of the phosphoprotein phosphatase 2A catalytic subunit promotes its functional association with regulatory subunits in vivo. EMBO J 19(21):5672–5681. doi:10.1093/emboj/19.21.5672
Wu G, Wang X, Li X, Kamiya Y, Otegui MS, Chory J (2011) Methylation of a phosphatase specifies dephosphorylation and degradation of activated brassinosteroid receptors. Sci Signal 4(172):ra29. doi:10.1126/scisignal.2001258
Yu RM, Wong MM, Jack RW, Kong RY (2005) Structure, evolution and expression of a second subfamily of protein phosphatase 2A catalytic subunit genes in the rice plant (Oryza sativa L.). Planta 222(5):757–768. doi:10.1007/s00425-005-0018-x
Yue K, Sandal P, Williams EL, Murphy E, Stes E, Nikonorova N, Ramakrishna P, Czyzewicz N, Montero-Morales L, Kumpf R et al (2016) PP2A-3 interacts with ACR4 and regulates formative cell division in the Arabidopsis root. Proc Natl Acad Sci USA 113(5):1447–1452. doi:10.1073/pnas.1525122113
Acknowledgements
This work was supported by a NILS-EEA Grant (019-ABEL-CM-2013) to JJSS and CL, and the Norwegian Research council (NRC) Grant 213853/F20 to CL. BSc students Iren B Helland, Ellen Marie Klinkenberg, Yvonne Sletthaug, and Linn-Kristine Svendsen contributed with NR testing.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Creighton, M.T., Sanmartín, M., Kataya, A.R.A. et al. Light regulation of nitrate reductase by catalytic subunits of protein phosphatase 2A. Planta 246, 701–710 (2017). https://doi.org/10.1007/s00425-017-2726-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-017-2726-4