Abstract
Main conclusion
Five SEP -like genes were cloned and identified from Platanus acerifolia through the analysis of expression profiles, protein–protein interaction patterns, and transgenic phenotypes, which suggested that they play conservative and diverse functions in floral initiation and development, fruit development, bud growth, and dormancy.
SEPALLATA (SEP) genes have been well characterized in core eudicots and some monocots, and they play important and diverse roles in plant development, including flower meristem initiation, floral organ identity, and fruit development and ripening. However, the knowledge on the function and evolution of SEP-like genes in basal eudicot species is very limited. Here, we cloned and identified five SEP-like genes from London plane (Platanus acerifolia), a basal eudicot tree that is widely used for landscaping in cities. Sequence alignment and phylogenetic analysis indicated that three genes (PlacSEP1.1, PlacSEP1.2, and PlacSEP1.3) belong to the SEP1/2/4 clade, while the other two genes (PlacSEP3.1 and PlacSEP3.2) are grouped into the SEP3 clade. Quantitative real-time PCR (qRT-PCR) analysis showed that all PlacSEPs, except PlacSEP1.1 and PlacSEP1.2, were expressed during the male and female inflorescence initiation, and throughout the flower and fruit development process. PlacSEP1.2 gene expression was only detected clearly in female inflorescence at April. PlacSEP1.3 and PlacSEP3.1 were also expressed, although relatively weak, in vegetative buds of adult trees. No evident PlacSEPs transcripts were detected in various organs of juvenile trees. Overexpression of PlacSEPs in Arabidopsis and tobacco plants resulted in different phenotypic alterations. 35S:PlacSEP1.1, 35S:PlacSEP1.3, and 35S:PlacSEP3.2 transgenic Arabidopsis plants showed evident early flowering, with less rosette leaves but more cauline leaves, while 35S:PlacSEP1.2 and PlacSEP3.1 transgenic plants showed no visible phenotypic changes. 35S:PlacSEP1.1 and 35S:PlacSEP3.2 transgenic Arabidopsis plants also produced smaller and curled leaves. Overexpression of PlacSEP1.1 and PlacSEP3.1 in tobacco resulted in the early flowering and producing more lateral branches. Yeast two-hybrid analysis indicated that PlacSEPs proteins can form homo- or hetero-dimers with the Platanus APETALA1 (AP1)/FRUITFULL (FUL), B-, C-, and D-class MADS-box proteins in different interacting patterns and intensities. Our results suggest that the five PlacSEP genes may play important and divergent roles during floral initiation and development, as well as fruit development, by collaborating with FUL, B-, C-, and D-class MADS-box genes in London plane; PlacSEP1.3 and PlacSEP3.1 genes might also involve in vegetative bud growth and dormancy. The results provide valuable data for us to understand the functional evolution of SEP-like genes in basal eudicot species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Angiosperm flowers are quite diverse in their morphology. Orthologous genes from different species can display divergent functions, which may provide the genetic basis for the floral diversification of flowering plants (Irish and Litt 2005; Theissen and Melzer 2007). In the early 1990s, the ABC model that elucidates how three functionally defined groups of genes (A, B and C) specify the four organ types of a typical eudicot flower was established based on molecular and genetic studies of homeotic mutants in Arabidopsis thaliana and Antirrhinum majus (Coen and Meyerowitz 1991; Bowman et al. 1991). According to this model, the petals are specified by the combination of A and B function genes, the stamens are specified by the combination of B and C function genes, while the sepals and carpels are specified alone by the A and C function genes, respectively; in addition, the A and C genes are mutually antagonistic. It is interesting that all genes involved in the ABC model, except APETALA2 (AP2), encode putative transcriptional regulators and belong to the MADS-box gene family (Weigel and Meyerowitz 1994).
The ABC model has broadly been used as a framework for understanding the flower development of a wide range of angiosperm species (Causier et al. 2010), and later, it was extended into the ABCE and quartet model (Pelaz et al. 2000; Theissen 2001), with the addition of D- and E-class MADS-box genes. The D-class genes were first identified in Petunia hybrida (Angenent et al. 1995), where they specified the ovule identity. In Arabidopsis, the D-function gene, SEEDSTICK (STK), is also required for specifying the identity of ovules, together with AGAMOUS (AG), SHATTERPROOF1 (SHP1), and SHP2, in addition, it plays a role in seed abscission and funiculus development (Pinyopich et al. 2003). The E-function genes are proposed based on a reverse genetic approach showed that four SEPALLATA (SEP) genes (SEP1/2/3/4; formerly AGAMOUS LIKE (AGL)2/4/9/3, respectively) are required for development of sepals, petals, stamens, and carpels in Arabidopsis (Pelaz et al. 2000; Theissen 2001; Ditta et al. 2004). In Arabidopsis, single sep mutants have only subtle phenotype, while sep1 sep2 sep3 triple mutants produce flowers in which all organs develop as sepals (Pelaz et al. 2000), and the sep1 sep2 sep3 sep4 quadruple mutants show an even stronger phenotype than the triple mutants, in which all floral organs are replaced by leaf-like organs (Ditta et al. 2004). Therefore, the different SEP proteins have largely, but not completely redundant functions in determining floral organ identity (Pelaz et al. 2001); they may bind to largely overlapping, but not identical sets of target genes that differ in the arrangement and spacing of the CArG-boxes in their cis-regulatory regions (Jetha et al. 2014).
Besides Arabidopsis, the E-class genes have been isolated from various angiosperms, including eudicots, monocots, and several basal angiosperm species (Li et al. 2005, 2015; Malcomber and Kellogg 2005; Zahn et al. 2005). Phylogenetic analysis of the available SEP genes shows multiple duplications within this subfamily, the first occurring before the origin of extant angiosperms producing the SEP3 and LOFSEP clades (Zahn et al. 2005). Within the SEP3 clade, an early diverging cluster of Asteraceae genes (ASTERACEAE SEP3) hints a second duplication early in angiosperm evolution, and additional duplications occurred at the base of grasses and more recently in some other families. Within the LOFSEP clade, duplications at or near the base of core eudicots produced the SEP1/2, FBP9/23, and SEP4 subclades, and the monocot genes show duplications near the base of grasses, producing the LHS1, OsMADS5, and OsMADS34 subclades (Zahn et al. 2005).
Functional data showed that SEP genes play a crucial role in floral meristem and organ identity. In Arabidopsis, the SEP proteins have been shown to participate in the formation of multimeric complexes with other MADS-box proteins, including the products of A, B, and C genes, to direct flower organ and meristem identity (Honma and Goto 2001; Ditta et al. 2004; Immink et al. 2009). In petunia, the B, C, and D functions require E-class genes (FBP2 and FBP5) to specify petal, stamen, carpel, and ovule development (Vandenbussche et al. 2003; Matsubara et al. 2008). The SEP homologs in tomato, gerbera, birch, poplar, rice, and phalaenopsis have also been shown to be necessary for floral meristem and organ identity, even for inflorescence development (Pnueli et al. 1994; Kotilainen et al. 2000; Lemmetyinen et al. 2004; Uimari et al. 2004; Cseke et al. 2005; Cui et al. 2010; Gao et al. 2010; Pan et al. 2014). In addition, SEP genes have been increasingly proved to play a role in fruit development and ripening, such as LeMADS-RIN and TM29 in tomato (Ampomah-Dwamena et al. 2002; Vrebalov et al. 2002), FaMADS9 in strawberry (Seymour et al. 2011), MdMADS8/9 in apple (Ireland et al. 2013), CaMADS-RIN in pepper (Dong et al. 2014), MgAGL2/9 and MgSEP2 in lotus magnolia (Lovisetto et al. 2015), and MaMADS1/2 in banana (Elitzur et al. 2016). Expression of SEP-like genes in vegetative tissues of some plant species suggests that they may also function in vegetative growth (Ferrario et al. 2003; Tzeng et al. 2003; Cseke et al. 2005; Elitzur et al. 2010; Liu et al. 2010; Li et al. 2014), which remains to be characterized. Recently, the tomato SEP protein SlMBP21 was shown to form protein complexes with JOINTLESS and MACROCALYX as a transcription activator for the development of the flower abscission zone (Liu et al. 2014), suggested that the SEP-like genes may play more diverse roles than what we recognized in plant development via subfunctionalization or even neofunctionalization. So far, SEP genes have been well characterized in some core eudicots and monocots (i.e. Arabidopsis, tomato, and rice); however, we still have very limited knowledge on the evolution and function of SEP genes from basal eudicot and basal angiosperm species (Li et al. 2005, 2015; Liu et al. 2010; Lovisetto et al. 2015).
London plane (Platanus acerifolia) is a basal eudicot tree belonging to the order Proteales that was widely planted in roadside and courtyard for shading. Flowering in trees is often associated with bud dormancy, a physiological state typical for most perennials, where growth is repressed within the buds, usually over the winter. Platanus species are monoecious with unisexual flowers aggregated into compact, spherical inflorescence heads (capitula), and the flowers are unusual in their structure, where the male flowers consist of one whorl of perianth organs followed by an alternating whorl of three-ridged organs and a whorl of stamens, while the female flowers are composed of a whorl of perianth organs followed by a whorl of small club-shaped organs, a whorl of staminodes, and two whorls of carpels (von Balthazar and Schonenberger 2009). Flowering of London plane is spread over two growing seasons. During the first season, lateral buds are formed under the petiole base (namely subpetiolar buds) on developing shoots (April–May), followed by two developmental fates. Most subpetiolar buds of adult trees (frequently located at the middle and upper part of the shoots) differentiate inflorescence and secondary shoot meristems individually in the same bud, hereinafter referred to as mixed flower buds, where floral meristem and floral organ differentiation occur at the early summer (June–July). While some subpetiolar buds located at the bottom part of the shoots or lower shoots of the tree can only differentiate shoot meristems without inflorescence meristems, hereinafter referred to as vegetative subpetiolar buds. By autumn, these lateral buds have ceased growth and become dormant. Flower development continues as the buds resume growth during the spring of the second growing season (Li et al. 2012b). This floral initiation and development process can be divided into inflorescence primordium differentiation phase (late May–early June), flower meristem initiation phase (mid-June), floral organ differentiation phase (late June–late July), first floral organ development phase (August–Oct), bud dormancy stage (Nov–Jan), and second floral organ development phase (Feb–April of next year) followed by anthesis and pollination at April (Li et al. 2012b). The fruits develop from the late April and mature till October.
As an excellent landscaping plant, London plane has an unfavorable characteristic that the abundant pollens shed by mature flowers and the dispersing achenes with numerous stiff hairs released from broken seed balls not only pollute the environment but also may result in serious pollinosis and breathing difficulties if breathed in by people (Lu et al. 2012). To remove this disadvantage, understanding the molecular regulatory mechanisms of flowering and flower development and then breeding non-flowering and fruitless varieties are meaningful and imperative. In this paper, we cloned and characterized five E-class MADS-box genes from P. acerifolia, homologous to the SEP1/2/4 and SEP3 genes in Arabidopsis, respectively. The results indicated that the five genes have divergent expression patterns, and may also possess distinct functions for flower development and bud dormancy, even for fruit and seed development, like the orthologous genes in other plant species.
Materials and methods
Plant materials
Plant samples used in this study were collected from the juvenile (2-year-old) or adult (over 30-year-old) London plane (Platanus acerifolia) trees in the campus of Huazhong Agricultural University on April 2013–April 2014. As described above, Platanus undergo a long period to complete its flower and fruit development process, which span two growing seasons. To catch the comprehensive gene expression profiles during the whole flower and fruit development process, we sampled monthly for different developmental stages according to the previous observation (Li et al. 2012b). Various samples from adult trees include the stems (S), young leaves (YL), mature leaves (ML), shoot apical buds (AB), and lateral subpetiolar buds (SB) in mid-April and mid-May, respectively; vegetative subpetiolar buds (VB), mixed flower buds (MB) containing differentiated inflorescences and vegetative tissues, vegetative tissues in mixed flower buds (MB-V), inflorescences in mixed flower buds (MB-F), and developing fruits (F) during June to September when the male and female inflorescences and flowers begin to differentiate but cannot be distinguished clearly (Li et al. 2012b); different ontogenetic stages of male inflorescences (MF), female inflorescences (FF), and fruits (F) from October to April of second year. The male inflorescences at mid-March and April were sampled by separating into flowers mainly consisting of anthers (MF-A) and the fleshy inflorescence peduncles (MF-P). In addition, roots (JR), stems (JS), young leaves (JYL), mature leaves (JML) and subpetiolar buds (JSB) were sampled from juvenile individuals at June. All samples were collected from three individual trees, respectively, and immediately frozen in liquid nitrogen.
Cloning of Platanus acerifolia SEP-like genes
Total RNA was extracted from London plane tissues using the CTAB method as described by Li et al. (2008). Two micrograms of total RNA were reverse-transcribed using PrimeScript™ RT reagent Kit with gDNA Eraser (Takara, Otsu, Japan) according to the manufacturer’s instructions. RNA and cDNA were stored at −80 °C for long-term use. Primers for PCR of the P. acerifolia SEP-like genes were designed according to the transcriptome sequencing data of London plane (unpublished), and were listed in Table S1. The amplified products were cloned into a pMD18-T vector (Takara), and 4–5 positive clones were randomly selected for sequencing.
Sequence alignment and phylogenetic analysis
Multiple sequence alignment for motif identification was performed using Vector NTI version 11.5 (Invitrogen) Clustal W program with default settings. A total of 75 SEP-like genes (cf. Fig 2, Table S2) were downloaded from the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov) for phylogenetic analysis. The AGL6 and AGL13 genes in AGL6 subfamily of A. thaliana were used as outgroup. Full-length amino-acid sequences were first aligned using the default settings in MUSCLE implemented in MEGA version 6.0 (Kumar et al. 2004), and then adjusted manually with the reference alignment provided by Zahn et al. (2005). Phylogenetic tree was constructed using MEGA v6.0 by the neighbor-joining (NJ) method with 1000 bootstrap replicates.
Gene expression analysis by real-time quantitative RT-PCR
Expression of the PlacSEP1 and PlacSEP3 genes in different tissues and development phases of London plane was investigated by real-time quantitative RT-PCR (qRT-PCR) analysis. Gene-specific primers for qRT-PCR were designed within the non-conservative C-terminal region and 3′ UTR (untranslated region) using the Primer 5.0 software to amplify products between 90 and 300 bp in size (Table S1). The expected size of PCR products was confirmed by agarose gel electrophoresis. Amplification and quantification were carried out using the SYBR Premix Ex Taq (Takara) and the ABI Prism 7500 Sequence Detection System (Applied Biosystems). qRT-PCR products were amplified using 1.0 μl of template from the RT reaction mixture, 5 μl 2 × SYBR Green Master Mix, 0.2 μl forward and reverse primer (10 μmol/μl), and water to a final volume of 10 μl. PCR amplification parameters were as described previously by Zhang et al. (2013). Each PCR was performed in triplicate and data are shown as mean values ± SE (standard error). PCR efficiency for each primer pair was determined by a standard curve generated with serially diluted cDNA. Output data generated by the instrument onboard software Sequence Detector Version 1.3.1 (PE Applied Biosystems) were transferred to a custom-designed Microsoft Excel macro for analysis. Relative expression levels of the target genes were determined using the Relative Expression Software Tool (Multiple Condition Solver REST-MCS v2), normalized to the references genes TPI (triose phosphate isomerase) of P. acerifolia (Zhang et al. 2011, 2013; Lu et al. 2012).
Vector construction
The pMD18-T vectors containing full-length coding sequences (CDS) of PlacSEP1 or PlacSEP3 genes were digested by SalI and KpnI or SalI and SacI restriction enzymes, and the target fragments were ligated into the corresponding sites of vector pMV, modified from the binary vector pBI121 (Zhang et al. 2011, 2013; Lu et al. 2012) containing the CaMV 35S promoter and the Nos 3′ transcriptional terminator, resulting in 35S:PlacSEP1.1, 35S:PlacSEP1.2, 35S:PlacSEP1.3, 35S:PlacSEP3.1, and 35S:PlacSEP3.2 constructions, respectively. All the constructed plasmids were confirmed by PCR and restriction digestions. The resulting plasmids were then transformed into the Agrobacterium tumefaciens strain EHA105 or GV3101 by the electroporation method.
Plant transformation and phenotype analysis
Overexpression of PlacSEP genes in Arabidopsis was carried out in wild-type Columbia (Col-0). Agrobacterium tumefaciens-mediated Arabidopsis transformation was performed by the floral-dipping method (Clough and Bent 1998). The transformed seeds were selected on Murashige and Skoog (MS) agar with 50 μg ml−1 kanamycin and 50 μg ml−1 cefotaxime. Following the segregation tests, 16 kanamycin-resistant transgenic plants of the T2 generation lines which fitted a segregation ratio of 3:1 were chosen to record flowering time and floral phenotype. Seedlings were grown in a growth incubator at 22 °C under a long day (LD) conditions (16/8 h, light/dark).
Agrobacterium tumefaciens EHA105 containing the 35S:PlacSEP1.1 or 35S:PlacSEP3.1 construction was also used to transform tobacco (Nicotiana tabacum) cultivar ‘Xanthi’. Tobacco plants used for transformation were cultured in the plant growth regulator-free MS solid medium at 25 °C under a 16 h photoperiod. Transformation of tobacco plants was performed as described by Horsch et al. (1985). Transgenic lines of T0 and T1 generations were verified by genomic amplification of the transgene using a primer to the 35S promoter (35SF) and a PlacSEP-specific primer (PlacSEP1.1R or PlacSEP3.1R) (Table S1). Phenotype changes of transgenic tobacco plants were investigated in both T0 and T1 generations.
Transgene and downstream target gene expression analysis
To confirm the relationship between PlacSEP transgenes and phenotypic changes of transgenic plants, RT-PCR was performed to analysis the expression of the transgene in Arabidopsis and tobacco, which were carried out on 14-day-old seedlings for Arabidopsis and young leaves and flower buds for tobacco. Total RNA was isolated from wild-type and T1 transgenic lines using the Trizol reagent (Takara), and then reverse-transcribed with the same reagent kit described above. Arabidopsis EF1α (AtEF1α, AT5G60390) and tobacco EF1α (NtEF1α, GenBank accession: D63396.1) were used as the endogenous reference genes to normalize small differences in template amounts. Primers used for the detection of PlacSEPs expression levels in the transgenic plants were listed in Table S1.
To understand the functional conservation of Platanus SEP-like proteins and the underlying mechanism of differential phenotypic changes of 35S::PlacSEPs transgenic Arabidopsis plants, the expression of several flowering time and leaf development-related genes that are regulated directly by SEP3 in Arabidopsis, including FT, SOC1, LFY, AP1, SEP3, AG, GRF1, GRF2, GRF5, TCP3, TCP18, TCP20, and ARF2 (Kaufmann et al. 2009; Pajoro et al. 2014), were investigated using semi-quantitative and quantitative RT-PCR analysis. Total RNA was isolated from 35S::PlacSEPs transgenic (T2) or wild-type Arabidopsis seedlings at 10 days after sowing. Conditions for reverse transcription and RT-PCR are the same as above. Primers used for PCR were listed in Table S1.
The yeast two-hybrid assay
The full-length open reading frame of London plane SEP-like genes, PlacFUL, PlacAP3, PlacPI2a (previously named PaPI2a) (Zhang et al. 2011; Li et al. 2012a), PlacAG, and PlacSTK, were amplified by PCR with primers as listed in Table S1. The PCR products were simultaneously introduced into the pGBKT7 and pGADT7 vector, fused in-frame to the GAL4 binding domain (pBDGAL4, bait) and the GAL4 activation domain (pADGAL4, prey), respectively. The pBDGAL4-PlacPI2a, pADGAL4-PlacAP3, pADGAL4-PlacAG, and pBDGAL4-PlacAG constructs were previously described (Zhang et al. 2011, 2013; Lu et al. 2012). All constructed were confirmed by sequencing analyses. Both of the bait and prey plasmids were transformed into yeast strain AH109 using the Frozen-EZ Yeast Transformation II Kit (Zymo Research Corp, Irvine, CA, USA). The colonies were selected on SD plates lacking Leu and Trp. Interactions between the tested proteins were determined by spotting assay on selective SD media lacking Leu, Trp, His, and Ade, supplemented with X-α-gal.
Results
Identification and phylogenetic analysis of London plane SEPALLATA-like genes
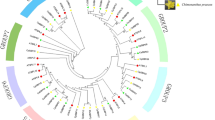
Five London plane SEP-like genes were cloned and identified according to the transcriptome sequencing data of P. acerifolia (GenBank accession nos KT380052–KT380056), amongst them two members show high-sequence identity, over 99% identity at nucleotide sequence and 98.8% identity at amino-acid sequence, respectively, with the PaSEP1 and PaSEP3 isolated previously (Li et al. 2012b), suggesting that they may be the alleles from the same gene locus. Therefore, only the newly isolated genes were investigated further in this study. Sequence alignment and phylogenetic analysis suggested that three of the five P. acerifolia SEPs (designated PlacSEP1.1, PlacSEP1.2, and PlacSEP1.3) belong to the SEP1/2/4 clade, while the other two (designated PlacSEP3.1 and PlacSEP3.2) were grouped into the SEP3 clade (Figs. 1, 2). The full-length CDS of these SEP-like genes encode 244-, 243-, 243-, 240-, and 240 amino-acid proteins, respectively. All predicted amino-acid sequences of PlacSEP proteins possess the conserved MIK domain and a divergent C-terminal domain with the conserved SEP I and SEP II motifs (Fig. 1; Zahn et al. 2005). The three PlacSEP1 sequences share 79.9% nucleotide identity and 74.6% amino-acid identity in their coding regions, and all contain an characteristic SEP1 terminal motif (Malcomber and Kellogg 2005), but PlacSEP1.1 and PlacSEP1.3 are more closely related to each other than either is to PlacSEP1.2 (Fig. 2). The two PlacSEP3 CDS regions share 86.7% nucleotide identity and 83.1% amino-acid identity, and contain an SEP3 motif that is different from the SEP1 terminal motif (Malcomber and Kellogg 2005), whereas the PlacSEP3.2 protein has an incomplete motif with two amino-acid deletion, and the PlacSEP3.1 protein has an amino-acid substitution of G to V in the motif (Fig. 1).
Alignment of Plactanus SEP-like amino-acid sequences with Arabidopsis, Akebia, and Euptelea. The MADS and K domains are marked with black bold lines. The I domain and C-terminal domain are marked with gray lines. SEP I motif and SEP II motif are highlighted by red box, and SEP1 motif and SEP3 motif are marked in dark and blue boxes, respectively
Phylogenetic tree based on the amino-acid alignment of predicted PlacSEP proteins and SEP-like proteins from other plant species. The tree was generated with the MEGA v6.0 software, using the neighbor-joining (NJ) method and 1000 bootstrap replicates, including monocots (green), magnoliids (purple), basal eudicots (pink), rosids (yellow), and asterids (orange). Bootstrap values above 50% are indicated, and five Platanus SEPs are marked with stars
Expression of PlacSEP1 and PlacSEP3 genes in London plane
To investigate the spatio-temporal expression pattern of PlacSEP1 and PlacSEP3 genes in London plane, qRT-PCR was performed for various plant tissues and ontogenetic stages. The results showed that neither PlacSEP1 gene nor PlacSEP3 gene was expressed obviously in juvenile tissues of 2-year-old plants (Fig. 3a). All PlacSEPs except for PlacSEP1.1 and PlacSEP1.2 were expressed during the male and female inflorescence initiation stages and throughout the flower and fruit development process (Fig. 3a). PlacSEP1.1 was not expressed evidently in the mixed flower buds of June (6MB), inflorescences of July to September (7MB-F, 8MB-F and 9MB-F), and the fruits of June–August (6F and 8F), but expressed during subsequent floral and fruit development stages, while PlacSEP1.2 was only detected clearly in female inflorescence at April, absent, or expressed very weakly from all the other tissues during flower initiation and development. PlacSEP1.3 and PlacSEP3.1 were also expressed in vegetative tissues of adult plants, including the shoot apical buds, vegetative subpetiolar buds, and vegetative tissues in mixed flower buds, although the expression levels were lower than that in the reproductive tissues. PlacSEP1.1 and PlacSEP3.2 transcripts were not detectable or only very weakly expressed in the adult vegetative tissues investigated. In addition, two PlacSEP3 genes expressed much higher than PlacSEP1 genes in most tissues and development stages, but all PlacSEPs showed relatively lower expression levels when compared with the housekeeping gene of London plane (Fig. 3a).
Expression profiling of PlacSEP1 and PlacSEP3 in London plane. a Relative expression of PlacSEP1 and PlacSEP3 genes in vegetative tissues of juvenile and adult plants (July mixed flower buds and October inflorescences and fruits were placed here for comparison). PlacSEP1.2 didn’t show here as no expression was detected in all of these tissues. b Relative expression of PlacSEP1 and PlacSEP3 genes in different development stages of inflorescences and fruits. JR roots of juvenile; JS stems of juvenile; JYL young leaves of juvenile; JML mature leaves of juvenile; JSB subpetiolar buds of juvenile; S stems; YL young leaves; ML mature leaves; AB shoot apical buds; SB subpetiolar buds; VB vegetative subpetiolar buds; MB mixed flower buds; MB-V vegetative tissues in mixed flower buds; MB-F inflorescences in mixed flower buds; MF male inflorescences; FF female inflorescences; MF-A male flowers mainly consisting of anthers; MF-P fleshy peduncles of male inflorescences; FF-E female inflorescences at the first day of the month; FF-M female inflorescences at the fifteenth day of the month; F fruits. The numbers indicate the sampling month of the tissues. The level of expression was normalized to London plane TPI (triose phosphate isomerase) gene. Error bars represent SE for three replicates
During the different ontogenetic stages, PlacSEP3.2 started to express in mixed buds at June when the inflorescence and flower meristem began to differentiate (Li et al. 2012b), and then maintained a relative high expression level in both male and female inflorescences till the anthesis stage during March to April in the next year. During the whole annual development cycle, PlacSEP3.2 showed three expression peaks during inflorescence and flower development, at July, October (male) or December (female), and February (male) or March (female), respectively. In the developing fruits, the highest expression level of PlacSEP3.2 was detected at October. PlacSEP3.1 showed a similar expression patterns as that of PlacSEP3.2 during the inflorescence and fruit development processes, but had a relative lower expression levels, especially in the male inflorescences (Fig. 3b; Fig. S1). PlacSEP1.1 increases its expression significantly in the male and female inflorescences at October, later than PlacSEP3.1, and then maintained a relative low expression level till April in the next year, up to the highest expression level in the mature male and female inflorescences, especially in the fleshy inflorescence peduncle. PlacSEP1.3 showed a similar expression tendency as that of PlacSEP1.1 during the male and female flower development processes, but possessed a broader expression patterns and started its expression earlier than PlacSEP1.1 (Fig. 3b; Fig. S1).
Overexpression of PlacSEP genes in Arabidopsis resulted in different phenotypic alterations
To investigate the potential functions of PlacSEP1 and PlacSEP3 genes in flowering transition, floral determination, and organ identity, cauliflower mosaic virus 35S promoter (CaMV35S) was used to ectopically express PlacSEPs in Arabidopsis. Forty-one, eighty-nine, forty-four, twenty-five, and thirty independent T1 transgenic lines were achieved for 35S:PlacSEP1.1, 35S:PlacSEP1.2, 35S:PlacSEP1.3, 35S:PlacSEP3.1, and 35S:PlacSEP3.2, respectively. According to the phenotypic alterations, two to six T1 transgenic lines whose progenies showed a 3:1 segregation ratio for kanamycin resistance, which may indicate a single-copy insertion of transgenes, were chosen for further experiment. Sixteen T2 transgenic plants for each line were used to investigate the flowering time and floral phenotypes.
Compared with the wild-type plants, overexpression of the PlacSEP1 and PlacSEP3 genes in Arabidopsis resulted in different phenotypic alterations. 35S:PlacSEP1.1 and 35S:PlacSEP3.2 transgenic plants showed obvious early flowering with small plant size and curled leaves (the rosette leaves as well as cauline leaves curl inwards and upwards along the axis of the major leaf vein) (Fig. 4a, b, f). Some transgenic lines that exhibit extremely severe phenotypes bolted and flowered after only producing two small rosette leaves, or even immediately after only forming two cotyledons (Fig. 4h, i). In this case, the secondary inflorescence shoots were usually converted into solitary flowers, while the primary inflorescence terminated into an enlarged flower frequently derived from the fusion of several defective flowers, and no progenies can be obtained from these plants (Fig. 4j, k). Most transgenic lines showed moderate phenotypes, which bolted and flowered after production of four to eight rosette leaves and three to six cauline leaves, while the wild-type Arabidopsis plants frequently produced ten to thirteen (11.06 ± 0.97) rosette leaves and two to three (2.13 ± 0.48) cauline leaves before bolting and flowering under the same conditions (Fig. 4a, b; Table S3). Transgenic lines with weak phenotypes, such as PlacSEP3.2-27, showed only smaller curled rosette leaves and more cauline leaves compared with the wild-type plants, without the early flowering (Fig. 4e; Table S3). In addition, it is interesting that two different plant architectures were frequently observed within the same 35S:PlacSEP1.1 or 35S:PlacSEP3.2 transgenic lines, i.e., some individuals produced condensed inflorescences that contained several flowers subtended by sepal-shaped cauline leaves directly within the rosette leaves without evident bolting and peduncle elongation (Fig. 4h, i), while the other individuals bolted extremely early with few rosette leaves and produced elongated inflorescences with more cauline leaves (Fig. 4j, k). The former plants were largely sterile with short empty pods. Most 35S:PlacSEP1.3 transgenic lines also exhibited early flowering with less rosette leaves and more cauline leaves than the wild-type Arabidopsis, but no curled leaves was observed, which was different from that of the 35S:PlacSEP1.1 and 35S:PlacSEP3.2 transgenic plants (Fig. 4c, d). 35S:PlacSEP1.2 and 35S:PlacSEP3.1 transgenic plants did not show severe phenotypic changes with only few lines that expressed high levels of the transgenes (PlacSEP1.2-47, 62 and PlacSEP3.1-17, 20) displaying slightly the early flowering or a little more cauline leaves (Table S3).
Phenotype analysis of transgenic Arabidopsis plants ectopically expressing PlacSEP1 and PlacSEP3 genes. a Wild-type (left) and 35S:PlacSEP1.1 transgenic plants (middle and right) showing early flowering with curled leaves. b Wild-type (left) and 35S:PlacSEP3.2 transgenic plant (right) with early flowering and curled leaves. c, d Wild-type (left) and 35S:PlacSEP1.3 transgenic plants (right) displaying early flowering without curled leaves. e 35S:PlacSEP3.2 transgenic plant that showed curled leaves and more cauline leaves without early flowering. f Phenotypic segregation of T2 generation in 35S:PlacSEP3.2 transgenic line. g Wild-type Col-0 seedling. 35S:PlacSEP1.1 transgenic plants with early flowering, curled leaves (h) and large fused terminal flowers (j). 35S:PlacSEP3.2 transgenic plants with early flowering, curled leaves (i), and fused terminal flower (k). l The numbers of rosette leaves (RL) and cauline leaves (CL) in wild-type and 35S:PlacSEPs lines; asterisks indicate statistically significant differences (P < 0.05) from the wild-type plants. m RT-PCR analysis of transgenes in wild-type Arabidopsis (Col-0) and transgenic lines. Yellow, white, and red arrows indicate curled cotyledon, rosette leaves, and cauline leaf, respectively. Bars 10 mm (a–f), 1 mm (g–k)
RT-PCR analysis indicated that the phenotypic variations of 35S:PlacSEP1.3 and 35S:PlacSEP3.2 transgenic plants are related in a certain extent to the expression levels of the transgenes, namely higher expression levels tend to result in more severe phenotypic changes, such as earlier bolting and flowering time or more cauline leaves, but this correlation seems to be not exist in the 35S:PlacSEP1.1 transgenic lines, such as PlacSEP1.1, which is strongly expressed in lines 3, 17, and 38, moderately expressed in line 40 and weakly expressed in lines 10 and 20; however, the number of rosette leaves in line 38 is even higher than that in lines 10 and 20 (Fig. 4m, l; Table S3). Expression analysis of the SEP3 downstream flowering time and leaf development-related genes in 35S:PlacSEPs transgenic Arabidopsis seedlings indicated that FT, SEP3, AP1, and AG are significantly activated in 10-day-old seedlings of 35S:PlacSEP1.1, 35S:PlacSEP1.3, and 35S:PlacSEP3.2 transgenic lines that display the early flowering when compared with their expression levels in wild-type seedlings (Fig. 5). SEP3 and AP1 genes show the strongest activation in 35S:PlacSEP1.1 and 35S:PlacSEP3.2 transgenic lines with the early flowering and evident leaf curling, followed by AG and FT activation. These genes also showed evidently higher expression levels in transgenic lines displaying both the early flowering and leaf curling (such as PlacSEP1.1-3 and PlacSEP3.2-8) than those in transgenic plants displaying only the early flowering without leaf curling (PlacSEP1.3-9). Expression levels of SOC1 and LFY in these transgenic lines are also significantly higher than that in wild-type seedlings, but they are not activated as strongly as FT, SEP3, AP1, and AG genes. No significantly higher expression levels of FT, SEP3, AP1, and AG were detected in 35S:PlacSEP1.2 and 35S:PlacSEP3.1 transgenic plants relative to wild-type seedlings (Fig. 5). In PlacSEP3.2-27 transgenic line that displays visible leaf curling but no early flowering, only SEP3 and AP1 were upregulated significantly. As for the leaf development-related genes investigated in this study, including GRF1, GRF2, GRF5, TCP3, TCP18, TCP20, and ARF2, only ARF2 showed significantly higher levels of expression in 35S:PlacSEP1.1, 35S:PlacSEP1.2, and 35S:PlacSEP3.1 transgenic lines than in wild-type seedlings (Fig. 5; Fig. S2).
qRT-PCR analysis of endogenous flowering and leaf development-related genes in 10-day-old seedlings of Arabidopsis wild-type and 35S::PlacSEP transgenic lines. 1.1–3, 1.2–47, 1.3–9, 3.1–17, 3.2–8, and 3.2–27 indicate transgenic lines of the five 35S::PlacSEP genes, respectively. Data represent the mean ± SE from three biological replicates, and AtEF1α was used as internal control. WT wild-type seedlings. The asterisks indicate significant differences compared with the WT plants (P < 0.01)
Ectopic expression of the PlacSEP1.1 and PlacSEP3.1 genes in tobacco plants caused early flowering but different axillary shoot branches
The functional roles of the PlacSEP1.1 and PlacSEP3.1, representative of the two clades of SEP gene subfamily in London plane, respectively, were also investigated by ectopic expression in tobacco plants. After selected by kanamycin and confirmed by PCR, thirty-six and thirty-two independent T0 transgenic lines of 35S:PlacSEP1.1 and 35S:PlacSEP3.1 were obtained, respectively. Eighteen of the transgenic lines carrying the 35S:PlacSEP1.1 flowered obviously earlier than the control plants, and exhibited a dwarf phenotype with some plants only approximately 10 cm in height at the time of flowering. RT-PCR analysis was conducted to investigate the expression levels of the transgene in plants with severe (line 33), moderate (line 35), and weak (line 25) phenotypes base on the plant height, internode length, and leaf size, etc. (Table S4). The results revealed that plants showing severe phenotype expressed higher levels of the transgene compared with the transgenic plants exhibiting moderate or weak phenotype (Fig. 6h, i; Table S4), indicating that the transgene is responsible for the phenotypic alterations. Transgenic lines PlacSEP1.1-25, 33, and 35 were selected to examine the heritability of the phenotypes. The results showed that the dwarf and early flowering phenotypes were co-inherited with the 35S:PlacSEP1.1 transgene to the next generation. The T1 transgenic plants displayed early flowering, lower plant height, shorter internode, less leaf number, smaller and greener crumpled leaves, and more lateral branches even before flowering, which was significantly different from those of the wild-type plants (Fig. 6b, e; Table S4).
Phenotype analysis of transgenic tobacco plants constitutively expressing PlacSEP1.1 and PlacSEP3.1 genes. a Wild-type (middle) and 35S:PlacSEP1.1 (left) and 35S:PlacSEP3.1 (right) transgenic tobacco seedlings; white arrow indicates the lateral shoot sprouting from axillary bud. b Wild-type (left) and dwarf 35S:PlacSEP1.1 transgenic plant showing early flowering (right). c Wild-type (left) and 35S:PlacSEP3.1 transgenic plant (right) with early flowering and more lateral branches (white arrow). d 35S:PlacSEP3.1 transgenic plant showing more primary and secondary lateral branches (white arrow) during later development stage. e Close up of the 35S:PlacSEP1.1 transgenic plant in (b). f, g Inflorescence architecture of 35S:PlacSEP3.1 transgenic plant and wild-type plant, respectively; white arrows indicate secondary inflorescences. h Days to flowering of wild-type and 35S:PlacSEPs lines; asterisks indicate statistically significant differences (P < 0.05) from the wild-type plants. i Transgene expression in flowers (left, line number followed by F) and leaves (right, line number followed by L) of T1 transgenic lines with strong, moderate, and weak phenotypes, respectively. WT wild type, W deionized water. Bars 5 cm
In the case of 35S:PlacSEP3.1 transgenic tobacco, 16 out of 32 T0 transgenic plants showed similar phenotypes to that of 35S:PlacSEP1.1 transgenic plants, including early flowering, dwarf plant stature, less leaf number, small and green leaves, and more lateral branches (Fig. 6c, d; Table S4). The inflorescence architecture appears to be simpler in the transgenic plants than in the wild-type plants, with less secondary inflorescences and less flowers for each inflorescence (Fig. 6f, g). It was interesting that the transgenic plants carrying the 35S:PlacSEP3.1 showed more reduced apical dominance and produced more lateral branches which generate secondary branches more easily than the 35S:PlacSEP1.1 transgenic lines, but there was no significantly different internode length were observed between these plants and the wild-type plants (Fig. 6a, d; Table S4). These phenotypic alterations were due to the overexpression of PlacSEP3.1, and the phenotype intensity was related to the expression level of the transgene (Fig. 6h, i). Three transgenic lines, PlacSEP3.1-9, 25 and 7 with severe, moderate, and weak phenotypes, respectively, were selected for further phenotype identification in T1 generation. Unexpectedly, only the PlacSEP3.1-9 T1 plants showed evident early flowering and shorter plant height, while the progenies of PlacSEP3.1-25 and 7 showed no significant difference from the wild-type plants, except for producing more lateral branches (Table S4). No visible changes were observed in flower organs or fruits of both 35S:PlacSEP1.1 and 35S:PlacSEP3.1 transgenic plants. In addition to the overexpression of PlacSEP1.1 and PlacSEP3.1, we also produced transgenic lines constitutively expressing one or both of the antisense transcripts of the two genes, but no plant showed any phenotypic alterations (Table S4).
Interactions between PlacSEPs and other MADS-box proteins from London plane
As overexpression of the three PlacSEP1 and two PlacSEP3 genes in Arabidopsis and tobacco plants resulted in different phenotypes, a yeast two-hybrid analysis was performed to evaluate if it could be caused by their differential ability of protein interactions. The coding sequences of FUL-, APETALA3 (AP3)-, PISTILLATA (PI)-, AGAMOUS (AG)-, SEEDSTICK (STK)-, and SEP-like genes were fused to the AD and BD domains, respectively, and used for comparison. The results indicated that all PlacSEPs interacted with PlacFUL, PlacAP3, PlacAG, and two PlacSTK proteins in different intensity, among which the interactions of PlacSEP3 with PlacAG as well as PlacSEP3.1 with all tested proteins, were weaker than that of the other PlacSEP interactions (Table 1; Fig. S3). All PlacSEPs were also capable of forming homodimers, except for PlacSEP3.1 in which the interaction was weak. Three PlacSEP1 proteins were failed to interact with PlacPI2a, but two PlacSEP3 had a capacity to interact with PlacPI2a, although the interaction between PlacSEP3.1 and PlacPI2a was weak and only in one direction (Table 1; Fig. S3).
Discussion
Evolutionary and functional conservation and diversity of Platanus SEP-like genes
Members of the SEP gene subfamily have been identified and characterized in a wide range of species and have been shown to take on roles in floral meristem identity, floral organ specification, fruit ripening, and plant architecture, and play a major role in the floral evolution of diverse plants (Malcomber and Kellogg 2005). In this study, five SEP-like genes were identified from P. acerifolia, a basal eudicot tree belonging to the family Platanaceae. A phylogenetic tree constructed using amino-acid sequences showed that PlacSEP1.1, PlacSEP1.2, and PlacSEP1.3 belong to the SEP1/2/4 clade, while PlacSEP3.1 and PlacSEP3.2 were grouped into the SEP3 clade (Fig. 2), which were further verified by C-terminal sequence alignment (Fig. 1). Platanaceae together with Proteaceae and Nelumbonaceae form the well-supported order Proteales, which plus Sabiales, Trochodendrales, and Buxales compose the basal eudicot lineages between the first branching eudicot lineage Ranunculales and the core eudicots (Bremer et al. 2009). In accordance with this, the SEP-like genes from P. acerifolia show the highest degree of nucleotide and amino-acid sequence identity with their homologous genes from the species of Nelumbonaceae, Trochodendrales, or Ranunculales, such as Nelumbo nucifera, Trochodendron aralioides, and Euptelea pleiosperma (Table S5). PlacSEP1.1 and PlacSEP3.2 also show high-sequence identity with SEP-like genes of Vitis vinifera. Sequences that have similar structures and features usually have a relatively closer evolutionary relationship, especially if the features appear in a non-conserved region. Several conserved motifs in highly variable C-terminal regions of the SEP gene lineage were found in five P. acerifolia SEP-like genes (Fig. 1), such as the SEP I and SEP II motifs (Zahn et al. 2005); moreover, three PlacSEP1 and two PlacSEP3 genes share the clade-specific SEP1 motif and SEP3 motif in their C-terminal, respectively, suggesting that they are recent duplicated orthologs of SEP1/2/4 and SEP3, respectively.
Based on the expression patterns, protein–protein interactions (PPIs), and phenotypic changes of transgenic Arabidopsis and tobacco plants, we showed that the SEP-like genes of Platanus could have not only conservative but also diverse functions. The previous studies indicated that SEP-like genes from core eudicots and even monocots have relatively conservative expression patterns and/or functions. For instance, almost all SEP3-clade members are expressed in inflorescences and in the inner three floral whorls and fruits; within the LOFSEP clade, all SEP1/2 genes are also expressed in inflorescences, the inner three floral whorls and fruits, most homologs are additionally expressed in sepals and some even in vegetative tissues (Malcomber and Kellogg 2005). In our study, the five Platanus SEP-like genes showed significantly different spatio-temporal expression patterns (Fig. 3). PlacSEP3.2 gene is expressed exclusively in reproductive organs and tissues, including male and female inflorescences, flowers, and fruits, which is similar to that of most SEP homologs in other species (Malcomber and Kellogg 2005). PlacSEP1.1 is also expressed mainly in reproductive tissues. However, the expression profile of the two genes is not identical in both the temporal and quantitative levels. First, the PlacSEP3.2 gene starts its expression at June when the inflorescence began to initiate, then followed by three peaks of expression in male and female inflorescences at the stamen and pistil differentiation stage (July), the first floral organ development stage (October to December), and the second floral organ development stage (February to March), respectively (Fig. 3); while PlacSEP1.1 is activated significantly much later till October when all the floral organ primordia already completed the differentiation, followed by two peaks of expression in male and female inflorescences at the second floral organ development stage (February to March) and the anthesis stage (April), respectively (Fig. 3). In addition, PlacSEP3.2 has a significantly higher expression level than PlacSEP1.1 almost in all the flower and fruit development stages. Another difference between the two genes is that PlacSEP1.1 expression increases gradually during the second floral development period in the spring, while the expression of PlacSEP3.2 declines at the same duration, resulting in the only higher expression level of PlacSEP1.1 than that of PlacSEP3.2 at April (Fig. 3b; Fig S1). In contrast, PlacSEP1.3 and PlacSEP3.1 genes are expressed not only in the reproductive organs like PlacSEP1.1 and PlacSEP3.2 but also in vegetative tissues of adult plants, including the shoot apical buds, vegetative subpetiolar buds, and the vegetative tissues in mixed flower buds (Fig. 3a, b). Likewise, there are evident differences between the expression of PlacSEP1.3 and PlacSEP3.1; for example, PlacSEP3.1 has an evident higher expression level than PlacSEP1.3. Besides, it is notable that the variation tendency of PlacSEP1.3 and PlacSEP3.1 expression is very similar to that of PlacSEP1.1 and PlacSEP3.2, respectively, during the flower and fruit development processes, although they have a relatively lower expression levels, suggesting that PlacSEP1.3 and PlacSEP3.1 may acquire the new expression location in vegetative tissues from PlacSEP1.1 and PlacSEP3.2, respectively, or vice versa, through the cis-regulatory element changes of the genes after their duplication. The most intriguing gene is PlacSEP1.2, which is only expressed at low level in female inflorescence at the anthesis stage and cannot result in visible phenotypic changes when overexpressing in Arabidopsis, indicating that it might have lost the functions, but it still maintains the same interacting capability with other floral MADS-box proteins as the PlacSEP1.1 and PlacSEP1.3 (see below). The differential expression patterns of the SEP-like genes in London plane suggest that they may not only play conservative roles in Platanus flower and fruit development, some members may also play divergent roles in other aspects, such as bud growth and dormancy, after their duplication and subfunctionalization events.
As the E-class MADS-box genes, SEP proteins are involved in the formation of multimeric complexes that contain other floral organ identity proteins. In Arabidopsis, SEP3 was proved to be a ‘glue’ protein to mediate the multimeric complex formation in various development processes (Immink et al. 2009). In other angiosperm species, SEP-like proteins have a conservative tendency of protein–protein interacting behavior with AP1/FUL, B-, C-, and D-function proteins (Malcomber and Kellogg 2005; Ruokolainen et al. 2010; Hu et al. 2015). However, a slightly different situation was also found in various species, especially those that are distantly different from the studied models (Liu et al. 2010; Li et al. 2015). In the previous studies, six PPIs between SEP proteins and other floral MADS-box proteins (A-, B-, C-, and D-class), i.e., AGL2-AP1, AGL9-AP1, AGL2-AG, AGL9-AG, AGL2-STK, and AGL9-STK, was shown to be conservative in angiosperm species, while B-class proteins (AP3 and PI) have gradually lost their interaction capability with SEP proteins, resulting in AGL2-AP3, AGL2-PI, AGL9-AP,3 and AGL9-PI remaining variable, and if any, usually weaker PPIs (Liu et al. 2010; Melzer et al. 2014; Li et al. 2015). Likewise, all five SEP-like proteins in Platanus showed conservative PPIs with AP1/FUL-like (PlacFUL), AG-like (PlacAG), and STK-like (PlacSTK1 and PlacSTK2) proteins in yeast, although in differential intensity and some PPIs merely positive in one-direction assays, confirming the conservative PPIs during angiosperm evolution (Table 1; Fig. S3). However, different from the situation in most angiosperm species, including basal eudicots, where the direct AGL2-AP3 and SEP-PI interactions have lost (Liu et al. 2010), PlacSEP1 and PlacSEP3 (especially PlacSEP3.2) proteins retained their interactions with AP3-like (PlacAP3) and PI-like (PlacPI2a) proteins, respectively, which is similar to the situation in basalmost angiosperm species, Amborella trichopoda (Melzer et al. 2014), suggestive of a slow evolution of PPIs between SEP and B-class proteins in Platanus. Like the situation in most other angiosperms, PlacSEP1 proteins did lost their interacting capability with the PI-like proteins, supporting the quick loss of AGL2-PI interaction during angiosperm evolution (Melzer et al. 2014). Within the SEP subfamily, AGL2-AGL9 has conservative interactions, while AGL2 and AGL9 show variable capability of homodimerization (Liu et al. 2010). In Platanus, all SEP-like proteins can form homodimers varying from weak to strong interaction (Table 1; Fig. S3), which is similar to that in Amborella trichopoda (Melzer et al. 2014), while different from that in the basal eudicot species, Euptelea pleiospermum, where only SEP3-like protein (EUplSEP3) rather than SEP1-like protein (EUplSEP1) can form homodimer (Liu et al. 2010), indicating again the low evolution rate of SEP-related PPIs in London plane. The conservative and differential protein–protein interaction patterns of PlacSEPs with AP1/FUL-like, B-, C-, and D-class MADS-box proteins suggested that they play conservative and divergent functions in Platanus.
The evolutionary conservation and divergence of Platanus SEP-like genes were further suggested by transgenic studies in Arabidopsis and tobacco. Overexpressing three PlacSEP1 and two PlacSEP3 genes in Arabidopsis resulted in different degrees of phenotypic alterations. 35S:PlacSEP1.1 and 35S:PlacSEP3.2 transgenic plants showed most evident phenotypic changes, including the early flowering, reduced plant size, small curled leaves, and usually determinant primary and secondary inflorescences terminating or transforming into solitary flowers (Fig. 4), which is similar to the phenotypic results of ectopic expression of Arabidopsis SEP3 genes or its homologs from some other species, such as petunia FBP2 (Pelaz et al. 2001; Ferrario et al. 2003). 35S:PlacSEP1.3 transgenic plants also bolted and flowered early with small plant and leaf size related to the wild type, but without leaf curling. In contrast, 35S:PlacSEP1.2 and 35S:PlacSEP3.1 transgenic plants did not show any severe phenotypic changes, except few lines with high level of transgene expression flowered slightly early or seemingly produced a little more cauline leaves (Fig. 4l; Table S3). The severity of phenotypes in SEP-like gene overexpressing Arabidopsis plants is frequently reported to be correlated to the levels of transgenes (Zhao et al. 2006; Chang et al. 2009). This relationship was also observed in our experiments for the 35S:PlacSEP1.3 and 35S:PlacSEP3.2 transgenic Arabidopsis plants but not in the 35S:PlacSEP1.1 transgenic lines (Fig. 4m, l; Table S3), which implied that PlacSEP1.1 may have more strong and complicated activity. In addition, differential phenotypes were observed in 35S:PlacSEP1.1 and 35S:PlacSEP3.1 transgenic tobacco plants, with the latter showing easier lateral shoot sprouting (Fig. 6), which may be resulted from their distinct coding sequence and PPI potential (Fig. 1; Table 1). Alternatively, those phenotypic differences just relate to differences in transgene expression could not be excluded. The discrepant phenotypes resulting from the overexpression of PlacSEPs suggested their divergent functions and probably differential capability of activating downstream target genes in Arabidopsis. Genome-wide approaches, such as ChIP-Seq, have identified many growth and development regulators as direct targets of SEP3 protein, including flowering time genes, floral meristem and organ identity genes, and growth regulatory genes, such as FLOWERING LOCUS T (FT), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1), AP1, FUL, AP3, PI, AG, TCPs, ARFs, GRFs, and SEP3 itself (Kaufmann et al. 2009; Pajoro et al. 2014). Moreover, it has been proved that ectopic expression of Arabidopsis SEP3 is sufficient to ectopically activate AP3 and AG in the curled leaves (Castillejo et al. 2005). To understand the molecular basis of differential transgenic phenotypes, the expression levels of a dozen candidate target genes of SEP3 in different transgenic lines were investigated. In accordance with the phenotypic changes, FT, SEP3, AP1, AG, SOC1, and LFY are significantly activated in 10-day-old seedlings of 35S:PlacSEP1.1, 35S:PlacSEP1.3 and 35S:PlacSEP3.2 transgenic lines that display strong early flowering and/or leaf curling, while they are not significantly different in 35S:PlacSEP1.2 and 35S:PlacSEP3.1 transgenic plants showing no visible phenotypic effects related to the expression levels in wild-type seedlings (Fig. 5). The results indicate that PlacSEP1.1, PlacSEP1.3, and PlacSEP3.2 have conservative roles like Arabidopsis SEP3 in activating flowering time and floral development genes, while PlacSEP1.2 and PlacSEP3.1 might lose this capability by subfunctionalization and neofunctionalization. It is worth noting that the expression levels of direct downstream ARF2 gene of SEP3 are significantly higher (although only 3–5 fold) in 35S:PlacSEP1.1, 35S:PlacSEP1.2, and 35S:PlacSEP3.1 transgenic plants, but not in 35S:PlacSEP1.3 and 35S:PlacSEP3.2, than that in the wild-type plants (Fig. 5), which further indicates the divergent functions of PlacSEPs.
The phenotype of 35S:PlacSEP1.1 and 35S:PlacSEP3.2 transgenic plants is also similar to that of 35S:FT transgenic plants (Teper-Bamnolker and Samach 2005) and the curly leaf (clf) or agl15 agl18 agl24 svp mutants in Arabidopsis (Goodrich et al. 1997; Fernandez et al. 2014), including the early flowering and leaf curling. In clf mutants, leaf curling was supposed to be caused by misexpression of the floral homeotic gene AG in leaves (Goodrich et al. 1997); however, AG mRNA and protein were expressed as strongly in clf fpa double mutants as in clf mutants, despite clf fpa mutants lack leaf curling (Lopez-Vernaza et al. 2012), suggesting that AG may be not the only or direct regulator of clf phenotype. Several other genes, including AP3, SEP3, FT, and FLC, have also been found to be misexpressed in clf mutants, among which mutual activation of SEP3 and FT activity are required for the clf phenotype, and SEP3 transcription requires AG activity, while FLC antagonizes the effects of FT/SEP3/AG on flowering and leaf phenotype in clf mutants (Lopez-Vernaza et al. 2012). In agl15 agl18 agl24 svp mutants, the change in leaf morphology is associated with the elevated levels of FT and ectopic expression of SEP3, leading to ectopic expression of floral genes, such as AP1 and AP3 (Fernandez et al. 2014). In 35S:PlacSEPs transgenic Arabidopsis plants, leaf curling was seemingly related to the activated expression levels of SEP3, AG and FT. For example, the expression levels of SEP3, AG and/or FT genes in transgenic lines with leaf curling (35S:PlacSEP1.1-3 and 35S:PlacSEP3.2-8) are evident higher than that in transgenic plants without leaf curling (Fig. 5), suggesting that there may be a threshold of SEP3 activity required for leaf curling and early flowering. Consistent with this, 35S:SEP3 transgenes give variable effects on leaf curling, presumably relating to expression levels (Honma and Goto 2001). However, it is noteworthy that 35S:PlacSEP3.2-27 showed evident leaf curling but with a lower level of SEP3 expression than 35S:PlacSEP1.3-9, indicating that there may be some other regulatory factors involved in the leaf curling phenotype. In addition, the expression of SEP3, AG, and FT is evident higher in lines showing the higher expression of the transgene, such as PlacSEP3.2-8 vs. PlacSEP3.2-27, indicating that the expression levels of SEP3, AG, and FT genes are related to and resulted from the transgene expression (Figs. 4, 5).
Potential roles of London plane SEP-like genes
To exactly understand the functions of a gene usually need to obtain its loss-of-function mutants by gene knockdown or knockout technology, such as transposon or T-DNA insertion, gene inhibition (cosuppression, RNAi, or amiRNA), and targeted genome editing, etc. In woody plants, especially those recalcitrant species for genetic transformation; however, it is difficult and time consuming to obtain the natural or transgenic loss-of-function mutants. In this case, gene expression pattern analysis and ectopically expressing in model species can provide important cues to understand the gene function. Therefore, we investigated the functions of five London plane SEP-like genes in this study by analyzing their expression patterns during growth and flower development processes, the protein–protein interaction manners with other floral MADS-box genes, and transgenic phenotypes in Arabidopsis and tobacco plants.
As discussed above, the five PlacSEP genes showed diverse spatio-temporal expression patterns, with PlacSEP1.1 and PlacSEP3.2 expressed exclusively in reproductive development process, while PlacSEP1.3 and PlacSEP3.1 also transcribed in some vegetative tissues of adult plants (Fig. 3). In favor of the qRT-PCR results, high levels of PlacSEP3.2 transcripts were detected in samples of June other than in samples of the other months by sequencing transcriptomes of London plane subpetiolar buds of April, May, and June, respectively, while PlacSEP1.3 and PlacSEP3.1 transcripts were detected in samples of all the 3 months, although the expression in samples of June is higher (unpublished data). The gene expression patterns indicate that PlacSEP1.1 and PlacSEP3.2 should play crucial but not necessarily identical roles in flower initiation and development as well as fruit development and maturation in Platanus, like most SEP homologs in other species. Consistent with this, transgenic Arabidopsis and tobacco plants ectopically expressing PlacSEP1.1 or PlacSEP3.2 gene showed strong early flowering phenotypes (Figs. 4, 6). PlacSEP1.3 and PlacSEP3.1 genes may not only play roles in flower and fruit development but also have functions in vegetative growth and development, such as bud growth and dormancy. In favor of this, 35S:PlacSEP1.3 transgenic Arabidopsis plants also showed early flowering, but no leaf curling phenotype was observed; 35S:PlacSEP3.1 transgene in Arabidopsis leads to no visible phenotype, but it does result in the early flowering in tobacco plants, and that more lateral branches are produced in transgenic plants (Fig. 6). As for PlacSEP1.2, it is unexpected that no visible early flowering is observed in transgenic Arabidopsis plants, because it maintains similar protein–protein interaction pattern and strength as PlacSEP1.1 and PlacSEP3.2 with other floral MADS-box proteins (Table 1; Fig. S3). Based on its very weak expression levels in all tested tissues and extremely limited expression position during the flower and fruit development processes, PlacSEP1.2 may have lost or changed dramatically its functions, which is supported further by the phylogenetic analysis results that display PlacSEP1.2 is more distantly related to PlacSEP1.1 and PlacSEP1.3.
The SEP genes were first proposed to be the E-function genes that determine all the four whorls of floral organs in the ABCE flower development model (Pelaz et al. 2001; Ditta et al. 2004). Recently, more and more studies showed that SEP-like genes may play complex roles beyond floral organ identity (Seymour et al. 2011; Ireland et al. 2013; Dong et al. 2014; Liu et al. 2014; Elitzur et al. 2016). As in the previous works, our study shows the functions of SEP-like genes are not necessary to correlate with the flower phenotype.
In summary, five SEP-like genes were isolated and characterized from a basal eudicot tree, P. acerifolia. According to the expression profiles, protein–protein interaction patterns, and transgenic phenotype analysis in model species, we suggest that two Platanus SEP-like genes, PlacSEP1.1 and PlacSEP3.2, play crucial and conservative functions in floral initiation and development, as well as in the fruit development; two Platanus SEP-like genes, PlacSEP1.3 and PlacSEP3.1, may also play important roles in vegetative development, such as bud growth and dormancy, besides their functions in flower and fruit development. The results are valuable and informative for us to understand the functional evolution of SEP-class genes in angiosperms and the molecular basis of flower and fruit development in Platanus. However, further studies including in situ hybridization analysis, promoter isolation, and characterization, etc., are needed to uncover the accurate expression location and the underlying mechanism of the divergent expression pattern and functions of these genes.
Author contribution statement
GFL, MZB, and SSZ designed the research. SSZ performed most experiments. SJL, SSY, and LL performed tobacco transformation. HJH performed transcriptome sequencing. JQZ constructed PlacAP3, PlacPI2a, and PlacAG yeast vectors. SSZ, SJL, and GFL analyzed the data and wrote the manuscript. All of the authors approved the final manuscript.
Abbreviations
- AG:
-
AGAMOUS
- AGL:
-
AGAMOUS LIKE
- AP1(3):
-
APETALA1(3)
- ARF:
-
Auxin response factor
- FUL:
-
FRUITFULL
- GRF:
-
Growth-regulating factor
- PPI:
-
Protein–protein interaction
- SEP:
-
SEPALLATA
- STK:
-
SEEDSTICK
References
Ampomah-Dwamena C, Morris BA, Sutherland P, Veit B, Yao JL (2002) Down-regulation of TM29, a tomato SEPALLATA homolog, causes parthenocarpic fruit development and floral reversion. Plant Physiol 130(2):605–617. doi:10.1104/Pp.005223
Angenent GC, Franken J, Busscher M, van Dijken A, van Went JL, Dons HJ, van Tunen AJ (1995) A novel class of MADS box genes is involved in ovule development in petunia. Plant Cell 7(10):1569–1582
Bowman JL, Smyth DR, Meyerowitz EM (1991) Genetic interactions among floral homeotic genes of Arabidopsis. Development 112(1):1–20
Bremer B, Bremer K, Chase MW, Fay MF, Reveal JL, Soltis DE, Soltis PS, Stevens PF, Anderberg AA, Moore MJ, Olmstead RG, Rudall PJ, Sytsma KJ, Tank DC, Wurdack K, Xiang JQY, Zmarzty S, Grp AP (2009) An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc 161(2):105–121
Castillejo C, Romera-Branchat M, Pelaz S (2005) A new role of the Arabidopsis SEPALLATA3 gene revealed by its constitutive expression. Plant J 43(4):586–596. doi:10.1111/j.1365-313X.2005.02476.x
Causier B, Schwarz-Sommer Z, Davies B (2010) Floral organ identity: 20 years of ABCs. Semin Cell Dev Biol 21(1):73–79. doi:10.1016/j.semcdb.2009.10.005
Chang YY, Chiu YF, Wu JW, Yang CH (2009) Four orchid (Oncidium Gower Ramsey) AP1/AGL9-like MADS box genes show novel expression patterns and cause different effects on floral transition and formation in Arabidopsis thaliana. Plant Cell Physiol 50:1425–1438. doi:10.1093/pcp/pcp087
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16(6):735–743. doi:10.1046/j.1365-313x.1998.00343.x
Coen ES, Meyerowitz EM (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353(6339):31–37. doi:10.1038/353031a0
Cseke LJ, Cseke SB, Ravinder N, Taylor LC, Shankar A, Sen B, Thakur R, Karnosky DF, Podila GK (2005) SEP-class genes in Populus tremuloides and their likely role in reproductive survival of poplar trees. Gene 358:1–16. doi:10.1016/j.gene.2005.05.035
Cui R, Han J, Zhao S, Su K, Wu F, Du X, Xu Q, Chong K, Theissen G, Meng Z (2010) Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa). Plant J 61(5):767–781. doi:10.1111/j.1365-313X.2009.04101.x
Ditta G, Pinyopich A, Robles P, Pelaz S, Yanofsky MF (2004) The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr Biol 14(21):1935–1940. doi:10.1016/j.cub.2004.10.028
Dong TT, Chen GP, Tian SB, Xie QL, Yin WC, Zhang YJ, Hu ZL (2014) A non-climacteric fruit gene CaMADS-RIN regulates fruit ripening and ethylene biosynthesis in climacteric fruit. PLoS One 9(4):11. doi:10.1371/journal.pone.0095559
Elitzur T, Vrebalov J, Giovannoni JJ, Goldschmidt EE, Friedman H (2010) The regulation of MADS-box gene expression during ripening of banana and their regulatory interaction with ethylene. J Exp Bot 61(5):1523–1535. doi:10.1093/jxb/erq017
Elitzur T, Yakir E, Quansah L, Zhangjun F, Vrebalov JT, Khayat E, Giovannoni JJ, Friedman H (2016) Banana MaMADS transcription factors are necessary for fruit ripening and molecular tools to promote shelf-life and food security. Plant Physiol 171:380–391. doi:10.1104/pp.15.01866
Fernandez DE, Wang CT, Zheng Y, Adamczyk BJ, Singhal R, Hall PK, Perry SE (2014) The MADS-domain factors AGAMOUS-LIKE15 and AGAMOUS-LIKE18, along with SHORT VEGETATIVE PHASE and AGAMOUS-LIKE24, are necessary to block floral gene expression during the vegetative phase. Plant Physiol 165(4):1591–1603. doi:10.1104/pp.114.242990
Ferrario S, Immink RG, Shchennikova A, Busscher-Lange J, Angenent GC (2003) The MADS box gene FBP2 is required for SEPALLATA function in petunia. Plant Cell 15(4):914–925
Gao X, Liang W, Yin C, Ji S, Wang H, Su X, Guo C, Kong H, Xue H, Zhang D (2010) The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiol 153(2):728–740. doi:10.1104/pp.110.156711
Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G (1997) A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386(6620):44–51. doi:10.1038/386044a0
Honma T, Goto K (2001) Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409(6819):525–529. doi:10.1038/35054083
Horsch RB, Rogers SG, Fraley RT (1985) Transgenic plants. Cold Spring Harb Symp Quant Biol 50:433–437
Hu Y, Liang W, Yin C, Yang X, Ping B, Li A, Jia R, Chen M, Luo Z, Cai Q, Zhao X, Zhang D, Yuan Z (2015) Interactions of OsMADS1 with floral homeotic genes in rice flower development. Mol Plant 8(9):1366–1384. doi:10.1016/j.molp.2015.04.009
Immink RG, Tonaco IA, de Folter S, Shchennikova A, van Dijk AD, Busscher-Lange J, Borst JW, Angenent GC (2009) SEPALLATA3: the ‘glue’ for MADS box transcription factor complex formation. Genome Biol 10(2):R24. doi:10.1186/gb-2009-10-2-r24
Ireland HS, Yao JL, Tomes S, Sutherland PW, Nieuwenhuizen N, Gunaseelan K, Winz RA, David KM, Schaffer RJ (2013) Apple SEPALLATA1/2-like genes control fruit flesh development and ripening. Plant J 73(6):1044–1056. doi:10.1111/tpj.12094
Irish VF, Litt A (2005) Flower development and evolution: gene duplication, diversification and redeployment. Curr Opin Genet Dev 15(4):454–460. doi:10.1016/j.gde.2005.06.001
Jetha K, Theissen G, Melzer R (2014) Arabidopsis SEPALLATA proteins differ in cooperative DNA-binding during the formation of floral quartet-like complexes. Nucleic Acids Res 42(17):10927–10942. doi:10.1093/nar/gku755
Kaufmann K, Muino JM, Jauregui R, Airoldi CA, Smaczniak C, Krajewski P, Angenent GC (2009) Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol 7(4):e1000090. doi:10.1371/journal.pbio.1000090
Kotilainen M, Elomaa P, Uimari A, Albert VA, Yu D, Teeri TH (2000) GRCD1, an AGL2-like MADS box gene, participates in the C function during stamen development in Gerbera hybrida. Plant Cell 12(10):1893–1902
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5(2):150–163. doi:10.1093/bib/5.2.150
Lemmetyinen J, Hassinen M, Elo A, Porali I, Keinonen K, Makela H, Sopanen T (2004) Functional characterization of SEPALLATA3 and AGAMOUS orthologues in silver birch. Physiol Plant 121(1):149–162. doi:10.1111/j.0031-9317.2004.00303.x
Li GS, Meng Z, Kong HZ, Chen ZD, Theissen G, Lu AM (2005) Characterization of candidate class A, B and E floral homeotic genes from the perianthless basal angiosperm Chloranthus spicatus (Chloranthaceae). Dev Genes Evol 215(9):437–449. doi:10.1007/s00427-005-0002-2
Li Z, Liu G, Zhang J, Zhang J, Bao M (2008) Extraction of high-quality tissue-specific RNA from London plane trees (Platanus acerifolia), permitting the construction of a female inflorescence cDNA library. Func Plant Biol 35(2):159–165. doi:10.1071/fp07212
Li Z, Liu G, Zhang J, Lu S, Yi S, Bao M (2012a) Cloning and characterization of paleoAP3-like MADS-box gene in London plane tree. Biol Plant 56(3):585–589. doi:10.1007/s10535-012-0112-4
Li Z, Zhang J, Liu G, Li X, Lu C, Bao M (2012b) Phylogenetic and evolutionary analysis of A-, B-, C- and E-class MADS-box genes in the basal eudicot Platanus acerifolia. J Plant Res 125(3):381–393. doi:10.1007/s10265-011-0456-4
Li XM, Fan T, Song JJ, Sun W, Xia KF, Liao JP, Zhang MY (2014) Functional conservation and divergence of four ginger AP1/AGL9 MADS-box genes revealed by analysis of their expression and protein-protein interaction, and ectopic expression of AhFUL gene in Arabidopsis. PLoS One 9(12):e114134. doi:10.1371/journal.pone.0114134
Li L, Yu XX, Guo CC, Duan XS, Shan HY, Zhang R, Xu GX, Kong HZ (2015) Interactions among proteins of floral MADS-box genes in Nuphar pumila (Nymphaeaceae) and the most recent common ancestor of extant angiosperms help understand the underlying mechanisms of the origin of the flower. J Syst Evol 53(4):285–296. doi:10.1111/jse.12148
Liu CJ, Zhang JA, Zhang N, Shan HY, Su KM, Zhang JS, Meng Z, Kong HZ, Chen ZD (2010) Interactions among proteins of floral MADS-Box genes in basal eudicots: implications for evolution of the regulatory network for flower development. Mol Biol Evol 27(7):1598–1611. doi:10.1093/molbev/msq044
Liu D, Wang D, Qin Z, Zhang D, Yin L, Wu L, Colasanti J, Li A, Mao L (2014) The SEPALLATA MADS-box protein SLMBP21 forms protein complexes with JOINTLESS and MACROCALYX as a transcription activator for development of the tomato flower abscission zone. Plant J 77(2):284–296. doi:10.1111/tpj.12387
Lopez-Vernaza M, Yang S, Muller R, Thorpe F, de Leau E, Goodrich J (2012) Antagonistic roles of SEPALLATA3, FT and FLC genes as targets of the polycomb group gene CURLY LEAF. PLoS One 7(2):e30715. doi:10.1371/journal.pone.0030715
Lovisetto A, Masiero S, Rahim MA, Mendes MAM, Casadoro G (2015) Fleshy seeds form in the basal Angiosperm Magnolia grandiflora and several MADS-box genes are expressed as fleshy seed tissues develop. Evol Dev 17(1):82–91. doi:10.1111/ede.12106
Lu S, Li Z, Zhang J, Yi S, Liu L, Bao M, Liu G (2012) Isolation and expression analysis of a LEAFY/FLORICAULA homolog and its promoter from London plane (Platanus acerifolia Willd.). Plant Cell Rep 31(10):1851–1865. doi:10.1007/s00299-012-1299-8
Malcomber ST, Kellogg EA (2005) SEPALLATA gene diversification: brave new whorls. Trends Plant Sci 10(9):427–435. doi:10.1016/j.tplants.2005.07.008
Matsubara K, Shimamura K, Kodama H, Kokubun H, Watanabe H, Basualdo IL, Ando T (2008) Green corolla segments in a wild Petunia species caused by a mutation in FBP2, a SEPALLATA-like MADS box gene. Planta 228(3):401–409. doi:10.1007/s00425-008-0744-y
Melzer R, Harter A, Rumpler F, Kim S, Soltis PS, Soltis DE, Theissen G (2014) DEF- and GLO-like proteins may have lost most of their interaction partners during angiosperm evolution. Ann Bot 114(7):1431–1443. doi:10.1093/aob/mcu094
Pajoro A, Madrigal P, Muino JM, Matus JT, Jin J, Mecchia MA, Debernardi JM, Palatnik JF, Balazadeh S, Arif M, O’Maoileidigh DS, Wellmer F, Krajewski P, Riechmann JL, Angenent GC, Kaufmann K (2014) Dynamics of chromatin accessibility and gene regulation by MADS-domain transcription factors in flower development. Genome Biol. doi:10.1186/Gb-2014-15-3-R41
Pan ZJ, Chen YY, Du JS, Chen YY, Chung MC, Tsai WC, Wang CN, Chen HH (2014) Flower development of Phalaenopsis orchid involves functionally divergent SEPALLATA-like genes. New Phytol 202(3):1024–1042. doi:10.1111/nph.12723
Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405(6783):200–203. doi:10.1038/35012103
Pelaz S, Gustafson-Brown C, Kohalmi SE, Crosby WL, Yanofsky MF (2001) APETALA1 and SEPALLATA3 interact to promote flower development. Plant J 26(4):385–394
Pinyopich A, Ditta GS, Savidge B, Liljegren SJ, Baumann E, Wisman E, Yanofsky MF (2003) Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424(6944):85–88. doi:10.1038/nature01741
Pnueli L, Hareven D, Broday L, Hurwitz C, Lifschitz E (1994) The TM5 MADS box gene mediates organ differentiation in the three inner whorls of tomato flowers. Plant Cell 6(2):175–186. doi:10.1105/tpc.6.2.175
Ruokolainen S, Ng YP, Albert VA, Elomaa P, Teeri TH (2010) Large scale interaction analysis predicts that the Gerbera hybrida floral E function is provided both by general and specialized proteins. BMC Plant Biol 10:129. doi:10.1186/1471-2229-10-129
Seymour GB, Ryder CD, Cevik V, Hammond JP, Popovich A, King GJ, Vrebalov J, Giovannoni JJ, Manning K (2011) A SEPALLATA gene is involved in the development and ripening of strawberry (Fragaria x ananassa Duch.) fruit, a non-climacteric tissue. J Exp Bot 62(3):1179–1188. doi:10.1093/jxb/erq360
Teper-Bamnolker P, Samach A (2005) The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves. Plant Cell 17(10):2661–2675. doi:10.1105/tpc.105.035766
Theissen G (2001) Development of floral organ identity: stories from the MADS house. Curr Opin Plant Biol 4(1):75–85
Theissen G, Melzer R (2007) Molecular mechanisms underlying origin and diversification of the angiosperm flower. Ann Bot 100(3):603–619. doi:10.1093/aob/mcm143
Tzeng TY, Hsiao CC, Chi PJ, Yang CH (2003) Two lily SEPALLATA-like genes cause different effects on floral formation and floral transition in Arabidopsis. Plant Physiol 133(3):1091–1101. doi:10.1104/pp.103.026997
Uimari A, Kotilainen M, Elomaa P, Yu D, Albert VA, Teeri TH (2004) Integration of reproductive meristem fates by a SEPALLATA-like MADS-box gene. Proc Natl Acad Sci USA 101(44):15817–15822. doi:10.1073/pnas.0406844101
Vandenbussche M, Zethof J, Souer E, Koes R, Tornielli GB, Pezzotti M, Ferrario S, Angenent GC, Gerats T (2003) Toward the analysis of the petunia MADS box gene family by reverse and forward transposon insertion mutagenesis approaches: B, C, and D floral organ identity functions require SEPALLATA-like MADS box genes in petunia. Plant Cell 15(11):2680–2693. doi:10.1105/tpc.017376
von Balthazar M, Schonenberger J (2009) Floral structure and organization in Platanaceae. Int J Plant Sci 170(2):210–225. doi:10.1086/595288
Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J (2002) A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296(5566):343–346. doi:10.1126/science.1068181
Weigel D, Meyerowitz EM (1994) The ABCs of floral homeotic genes. Cell 78(2):203–209. doi:10.1016/0092-8674(94)90291-7
Zahn LM, Kong H, Leebens-Mack JH, Kim S, Soltis PS, Landherr LL, Soltis DE, Depamphilis CW, Ma H (2005) The evolution of the SEPALLATA subfamily of MADS-box genes: a preangiosperm origin with multiple duplications throughout angiosperm history. Genetics 169(4):2209–2223. doi:10.1534/genetics.104.037770
Zhang J, Guo C, Liu G, Li Z, Li X, Bao M (2011) Genetic alteration with variable intron/exon organization amongst five PI-homoeologous genes in Platanus acerifolia. Gene 473(2):82–91. doi:10.1016/j.gene.2010.11.005
Zhang J, Li Z, Guo C, Liu G, Bao M (2013) Isolation and functional analyses of a putative floral homeotic C-function gene in a basal eudicot London plane tree (Platanus acerifolia). PLoS One. doi:10.1371/journal.pone.0063389
Zhao XY, Cheng ZJ, Zhang XS (2006) Overexpression of TaMADS1, a SEPALLATA-like gene in wheat, causes early flowering and the abnormal development of floral organs in Arabidopsis. Planta 223:698–707. doi:10.1007/s00425-005-0123-x
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (30871729, 31272206), the Ministry of Education of China (NCET-12-0867 and IRT13065), the Fundamental Research Funds for the Central Universities (2013PY026), and Huazhong Agricultural University Scientific and Technological Self-innovation Foundation (2012SC29). We thank all colleagues in our laboratory for discussions and technical assistance.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, S., Lu, S., Yi, S. et al. Functional conservation and divergence of five SEPALLATA-like genes from a basal eudicot tree, Platanus acerifolia . Planta 245, 439–457 (2017). https://doi.org/10.1007/s00425-016-2617-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-016-2617-0