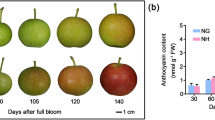
Abstract
Main conclusion
Paper-bagging treatment can transform non-transcribed MdMYB1 - 2 and MdMYB1 - 3 alleles into transcribed alleles through epigenetic regulations, resulting in the red pigmentation of a normally non-red apple cultivar ‘Mutsu.’
Anthocyanin biosynthesis in apples is regulated by MdMYB1/A/10, an R2R3-Type MYB gene. ‘Mutsu,’ a triploid apple cultivar harboring non-transcribed MdMYB1-2 and MdMYB1-3 alleles, retains green skin color under field conditions. However, it can show red/pink pigmentation under natural or artificial ultraviolet-B (UV-B) light exposure after paper-bagging and bag removal treatment. In the present study, we found that in ‘Mutsu,’ paper bagging-induced red pigmentation was due to the activation of non-transcribed MdMYB1-2/-3 alleles, which triggered the expression of downstream anthocyanin biosynthesis genes in a UV-B-dependent manner. By monitoring the epigenetic changes during UV-B-induced pigmentation, no significant differences in DNA methylation and histone modifications in the 5′ upstream region of MdMYB1-2/-3 were recorded between the UV-B-treated fruit skin (red) and the fruit skin treated only by white light (green). In contrast, bag treatment lowered the DNA methylation in this region of MdMYB1-2/-3 alleles. Similarly, higher levels of histone H3 acetylation and trimethylation of H3 tail at lysine 4, and lower level of trimethylation of H3 tail at lysine 27 were observed in the 5′ upstream region of MdMYB1-2/-3 in the skin of the fruit immediately after bag removal. These results suggest that bagging treatment can induce epigenetic changes, facilitating the binding of trans factor(s) to MdMYB1-2/-3 alleles, resulting in the activation of these MYBs after bag removal.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Apple skin color, which is ascribed mainly to anthocyanin pigments, is one of the most important factors determining the market acceptance and value of apples. In general, red cultivars are preferred in Japan, and within a cultivar, better-colored fruits are favored. Two categories of genes affect the biosynthesis of anthocyanin pigments. The first category is structural or biosynthetic genes involving chalcone synthase (CHS), chalcone isomerase (CHI), flavanone-3β-hydroxylase (F3H), dihydroflavonol 4-reductase (DFR), anthocyanin synthase (ANS), and UDP-glucose:flavonoid-3-O-glycosyltransferase (UFGT). The second category includes regulatory genes encoding transcription factors, which influence the intensity and pattern of anthocyanin accumulation through the regulation of downstream genes, including those belonging to the first category. At least three protein families, MYB, bHLH, and WD40, are involved in the regulation of anthocyanin synthesis, but the specific classes and the genes involved in the biosynthesis vary depending on the species (Broun 2005; Hichri et al. 2011).
An apple R2R3 MYB transcription factor, MdMYB1/A/10, reported independently by three research groups (Takos et al. 2006; Ban et al. 2007; Espley et al. 2007), is a key regulator of fruit skin anthocyanin accumulation. MdMYB1, MdMYBA, and MdMYB10 are all located at the same position in the linkage group 9 (Chagné et al. 2007), where only one complete MYB gene is present in the apple genome assembly (Velasco et al. 2010). Therefore, MdMYB1, MdMYBA, and MdMYB10 are considered as alleles (Lin-Wang et al. 2010). As MdMYB1 showed 100 % sequence identity with MdMYBA, MdMYB1 has been used to represent both MdMYB1 and MdMYBA in the present article. Different MdMYB1/10 alleles affect pigmentation of different parts of the fruit. The MdMYB1/MdMYB10 (R1) allele is associated only with fruit skin color, but the MdMYB10 (R6) allele, with six tandem repeats of a minisatellite in its promoter region, leads to red pigmentation in flesh/core and leaves (Espley et al. 2009). MdMYB1 has at least three alleles, namely MdMYB1-1, MdMYB1-2, and MdMYB1-3, among which only MdMYB1-1 co-segregates with red skin color phenotypes, whereas the other two cannot regulate anthocyanin synthesis owing to their poor expression (Takos et al. 2006). The genomic sequences of MdMYB1-1 and MdMYB1-2 are highly similar, showing 100 % sequence identity in the open reading frames and only a few-nucleotide variations in their promoter regions (approximately −2000 bp from the translation initiation codon). Compared with MdMYB1-1, MdMYB1-3 shows a single nucleotide difference in the third exon, resulting in a substitution of arginine with serine in the C-terminal domain. However, this substitution has no effect on the function of the resulting MdMYB1 protein (Takos et al. 2006). Thus, it is interesting that only the MdMYB1-1 allele can confer red pigmentation in apple skins under natural conditions, whereas MdMYB1-2/-3 alleles cannot, despite their high cDNA and amino acid similarities. Presently, variations in skin color of 90 apple cultivars collected in Japan could be explained by different combinations of MdMYB1 alleles, except for the skin color of the cultivar, ‘Granny Smith’ (Hatsuyama, unpublished data). In addition, two types of red flesh have been identified with different core coloration (Chagné et al. 2013). Type 1 red flesh has been attributed to the ectopic expression of MdMYB10, because of the presence of R6 microsatellites in its promoter region (Espley et al. 2009), whereas type 2 red flesh has been attributed to the expression of a duplicated MYB gene (MdMYB110) located on chromosome 17 (Chagné et al. 2013; Umemura et al. 2013).
In Japan, paper bagging of apple fruits during development is a commonly used technique for enhancing the red pigmentation of fruit skin. A commercial paper bag is composed of two layers, an outer layer of light-blocking paper and an inner layer of translucent paper (usually red or blue). The fruits are covered with these bags 1 month after full blossom. The outer bags are removed 2–3 weeks before the expected harvest date, and the inner bags are removed 4–7 days later. This treatment is referred to as the ‘paper-bagging treatment’ in the present report. Paper-bagging treatment can improve pigmentation, but the development of red color after bag removal is retarded in the absence of ultraviolet-B (UV-B) irradiation (Peng et al. 2013), indicating that UV-B might be the key factor for paper bagging-induced red pigmentation. Apart from color improvement in red cultivars, paper-bagging treatment also induces red pigmentation in the skin of yellow/green cultivars, especially ‘Mutsu.’ ‘Mutsu,’ also known as ‘Crispin,’ is a triploid hybrid of non-red cultivars, ‘Golden Delicious’ and ‘Indo’ (that harbors two MdMYB1-2 and one MdMYB1-3 non-transcribed alleles (Peng et al. 2013). ‘Mutsu’ retains green skin color when cultivated without paper-bagging treatment, whereas red/pink color develops after paper-bagging treatment. Although farmers in Japan have routinely applied paper-bagging treatment to ‘Mutsu’ for the production of red/pink colored fruits (Mink 1973), its mechanism has not been well characterized.
Several lines of evidence have suggested the importance of UV-B irradiation in anthocyanin accumulation. Some reports have shown elevated anthocyanin biosynthetic enzyme activities (Arakawa et al. 1986) and induction of their gene expression after UV-B treatment, concomitant with anthocyanin accumulation (Ubi et al. 2006). Peng et al. (2013) showed MdMYBA (=MdMYB1) induction after bag removal in ‘Mutsu.’ The present study was undertaken to clarify this mechanism in apple skins possessing non-transcribed MdMYB1 alleles. The results showed that bagging treatment possibly induced epigenetic changes, facilitating the binding of trans factor(s) to MdMYB1-2/-3 alleles, resulting in the activation of these MYBs after bag removal. Our results provide insight into the mechanisms underlying the regulation of MdMYB1-mediated apple skin coloration.
Materials and methods
Plant materials
The apple cultivar, ‘Mutsu,’ was used in the present study because it clearly showed red pigmentation after bag removal. ‘Mutsu’ apples were cultivated in an orchard in Aomori Prefecture, northeast Japan. Fruits were wrapped with double-layer paper bags (inner red, outer blue) 1 month after full blossom. Paper bags (both inner and outer papers) were removed simultaneously in late September, approximately 1 month before the commercial harvest day. The fruits were exposed to natural sunlight (Fig. 1, bag removal). Some apple fruits were kept in paper bags without removal (Fig. 1, bagged). Apple fruits without paper-bagging treatment were used for comparison with other treatments (Fig. 1, non-bagged). In late October, skin samples from the fruits were collected in a dark room to avoid a light-induced response during sampling; the samples were frozen in liquid nitrogen and immediately stored at −80 °C. To investigate the transcribed MdMYB1 allelic types in red apple skin, ‘Sekaiichi’ and ‘Sensyu’ (Suppl. Fig. S1) cultivated in an orchard in Aomori Prefecture were used. These cultivars possess transcribed MdMYB1-1 and non-transcribed MdMYB1-3 (Hatsuyama, unpublished data), and as mentioned above, these two alleles can be discriminated by the nucleotide 573 in the third exon, which is A for MdMYB1-1 and T for MdMYB1-3.
Illustration of the treatments used in this study. Bag removal ‘Mutsu’ apples were wrapped in early June; the inner and outer paper bags were removed simultaneously in late September, and the fruits were exposed to natural sun light until harvest (late October). Non-bagged ‘Mutsu’ apples were cultivated without paper-bagging treatment. Bagged ‘Mutsu’ apples were wrapped in early June (approximately 1 month after full bloom) and maintained until harvest
For UV-B treatments, paper-bagged fruits (bagged) were harvested on the commercial harvest date. Half the fruits were exposed to UV-B for 96 h at 17 °C after bag removal in a chamber (FLI-301 N, Eyela Co. Ltd., Tokyo Japan) equipped with one white fluorescent lamp (FL; FL15 W, Panasonic, Osaka, Japan) and supplemented with one UV-B lamp (FL/UV-B; G15TBE, Sankyo Denki, Kanagawa, Japan). Some fruits were treated only with FL. Apple skins were carefully harvested at six time points (0, 6, 12, 24, 48, and 96 h). All the skins were immediately frozen in liquid nitrogen and stored at −80 °C until use. Sampling of skin at 0 h was carried out in a dark room, as mentioned earlier.
RNA extraction
Total RNA was extracted from the skins of ‘Mutsu,’ ‘Sekaiichi,’ and ‘Sensyu,’ according to a slightly modified protocol of Gasic et al. (2004). In brief, the ground fruit skins were treated with RNA extraction buffer, and the extracted RNA was subjected to chloroform–isoamyl alcohol (24:1, v/v) extraction twice. RNA was precipitated with isopropanol and washed with 70 % ethanol. After resuspension in 0.1 % SDS, the RNA solution was extracted once with chloroform–isoamyl alcohol (24:1, v/v) and precipitated with 2.5 volumes of 99 % ethanol. Genomic DNA was eliminated from the total RNA preparation with a Turbo DNA-free Kit (Thermo Fisher Scientific Waltham, MA, USA). cDNAs were synthesized from ~500–1000 ng total RNA using a SuperScript VILO cDNA Synthesis Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
Quantitative reverse transcription–PCR (qRT-PCR)
cDNAs obtained from ‘Mutsu’ were used for qRT-PCR. qRT-PCR was performed using an Applied Biosystems 7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA). cDNA corresponding to 10 ng total RNA was used in a 20-μl reaction volume with SYBR Premix Taq II (TaKaRa Bio, Tokyo, Japan) according to the manufacturer’s instructions. Relative expression was determined with the 2−ΔΔT algorithm (Livak and Schmittgen 2001) after normalizing with the SAND gene (Cottage et al. 2001; Imai et al. 2014). The primers used for qRT-PCR are listed in Suppl. Table S1.
Genomic DNA extraction and McrBC-qPCR
The methylation status of the upstream region of MdMYB1-2/-3 was determined by quantitative analysis using real-time qPCR, following the slightly modified procedure of Saito et al. (2013). Briefly, 300 ng genomic DNA from ‘Mutsu’ was incubated for 3 h with 10 U McrBC (New England BioLabs, Tokyo, Japan) at 37 °C in a 30-µl reaction mixture. Mock digestions were performed with 1 µl 50 % glycerol. The reaction mixture (20 µl) for qPCR contained 1.0 µl of the digested sample (equivalent to about 10 ng of the digested or mock-digested genomic DNA), 2.0 µM of each primer and 10 µl SYBR® Green Premix ExTaq II (TaKaRa Bio). Target regions were amplified using the following qPCR conditions: initial denaturation for 3 min at 94 °C, followed by 40 cycles of denaturation at 94 °C for 30 s, annealing at 60 °C for 30 s, and extension for 34 s at 72 °C. The percentage of methylation was determined according to Telias et al. (2011). As DNA methylation is often found in inactive transposons, digestion efficiency was evaluated by a Copia-type retrotransposon, Ppcrt1 (GenBank: AB550651, Kim et al. 2012), for each sample.
Direct sequencing of ‘Mutsu’ MdMYB1 alleles
Regions with single nucleotide polymorphisms (SNPs) were amplified from either DNA or cDNA using primers that could anneal with the three MdMYB1 alleles. To avoid errors in PCR, high fidelity DNA polymerase KOD plus Neo (Toyobo, Tokyo, Japan) was used. PCR products were purified after agarose gel electrophoresis using Wizard SV DNA purification system (Promega, Madison, WI, USA). Ten ng of purified PCR product were used for sequencing with BigDye® Terminator v3.1 (Applied Biosystems) and then purified with X-terminator. Sequencing was carried out using a 3130xl Genetic Analyzer (Applied Biosystems). Reverse transcription was also performed with no reverse transcriptase included in the reaction mixture to confirm the complete elimination of genomic DNA.
Chromatin immunoprecipitation (ChIP) assay
For ChIP, DNA shearing was carried out by a procedure described by Saito et al. (2015) with some modifications. Briefly, apple skins were cross-linked in 1 % formaldehyde, under vacuum, for 10 min; the formaldehyde was quenched by adding glycine to a final concentration of 0.125 M for 5 min. Cross-linked tissues were ground to a powder in liquid nitrogen, and the nuclei were pelleted. Pelleted chromatin was resuspended in a modified micrococcal nuclease (MNase) reaction buffer (20 mM Tris–HCl, pH 8.0, 5 mM NaCl, 2.5 mM CaCl2, 0.1 % NP-40, 1 mM PMSF, and 12.5 % glycerol) and treated with 400 U/ml MNase for 10 min at 37 °C. DNA fragmentation by MNase was stopped by adding EDTA followed by centrifugation at 13,700g for 2 min. After centrifugation, the nuclei pellets were resuspended in a lysis buffer and sonicated briefly to lyse the nuclear membrane, and the resultant clear supernatant was used for immunoprecipitation. Chromatin immunoprecipitation was performed using an EpiQuik Plant ChIP kit (Epigentek Group Inc., Farmingdale, NY, USA) according to the manufacturer’s instructions with minimal modification. Briefly, the chromatin lysate was precleared in Protein G-coated 8-well assay strips for 2 h at 4 °C, and then incubated with antibodies. The antibodies used were anti-trimethyl H3-K4 (Millipore, Billerica, MA, USA, product number 07-473), anti-trimethyl H3-K27 (Millipore, product number 07-449), anti-histone H3 acetylation (Millipore, product number 06-599B), or normal mouse IgG (NM IgG, Millipore, product number 12-371). Primers used for qRT-PCR are listed in Suppl. Table 1.
Sub-cloning and sequencing of the transcribed MdMYB1 alleles from the ‘Sekaiichi’ and ‘Sensyu’ cultivars
cDNAs obtained from the ‘Sekaiichi’ and ‘Sensyu’ cultivars were used for amplification of MdMYB1 alleles. PCR was carried out in a total volume of 50 µl, which contained 125 ng cDNA, 500 µM dNTPs, 375 µM MgCl2, 3.125 µM of each primer (Suppl. Table S1), 0.4 U KOD Plus polymerase and 0.1 × KOD buffer (Toyobo). Followed by a pre-PCR heating at 95 °C for 2 min, a cycling profile of 94 °C for 30 s, 50 °C for 30 s, 72 °C for 30 s was repeated 35 times. A final extension for 10 min at 72 °C was performed. The PCR product was cloned into the pCR® 2.1-TOPO vector (Invitrogen). Sequencing reactions were performed with a BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems), and the products were sequenced on an ABI PRISM 3130xl Genetic Analyzer (Applied Biosystems). After sequencing, the MdMYB1 allelic types were confirmed based on the nucleotides present at position 573.
Results
UV-B in the induction of MdMYB1 expression in the fruit skin of ‘Mutsu’
We analyzed the role of UV-B in the red coloration of ‘Mutsu.’ Bagged fruits were exposed to white fluorescent lamps, with or without UV-B treatment, for up to 96 h. The fruit color of FL/UV-B samples gradually changed to pink at 48 h after incubation and the color intensity increased at 96 h, concomitant with an increase in anthocyanin compounds (Fig. 2a, b). In contrast, the fruit color under the FL treatment remained white (Fig. 2a, b). The transcript levels of the anthocyanin biosynthetic genes MdCHS, MdCHI, MdF3H, MdDFR, MdANS, MdUFGT, and the regulatory genes, MdMYB1 (consensus primers were used to amplify both the MdMYB1-2/-3), MdbHLH3, and MdWD40 (that form the MYB–bHLH–WD40 complex) were compared (Fig. 2c). The transcript levels of the anthocyanin biosynthetic genes were high in FL/UV-B fruits, correlating with the anthocyanin levels. The expression level of these genes remained constant in the FL samples during incubation. Furthermore, the transcription of MdMYB1 in the fruit skin increased substantially after 24 h FL/UV-B treatment, whereas only a slight increase was observed in the FL treatment after 12 and 48 h. In contrast, MdbHLH3 and MdWD40 transcripts showed only 2- to 3-fold changes during the light treatment and the difference was less evident between the FL and FL/UV-B treatment than that of the other tested genes. These results indicate the pivotal role of UV-B on the coloration of fruits in the ‘Mutsu’ cultivar.
UV-B-dependent pigmentation of ‘Mutsu.’ a Bagged fruits were subjected to FL or FL/UV-B treatments at 17 °C for 96 h after bag removal. Note that only the fruits irradiated with UV-B turned red. b Anthocyanin extracted from the fruits of a using the method described in Bai et al. (2014). c Anthocyanin biosynthetic and regulatory gene expression, including MdMYB1 in apple skins subjected to FL or FL/UV-B. Transcription levels were determined by qRT-PCR using SAND as a reference gene. The expression levels in c are values relative to the 0-h sample. Error bars show SE
Bagging treatment and MdMYB1 induction in ‘Mutsu’
We determined the importance of UV-B in the paper bagging-induced red pigmentation (Fig. 2). Although, in the orchard, both non-bagged and bag removal ‘Mutsu’ fruits were subjected to sunlight irradiation, only the bag removal fruits turned red, suggesting that the application of paper-bagging treatment before light irradiation was important for red pigmentation. We compared the expressions of anthocyanin biosynthesis-related genes in bagged and non-bagged fruits (Fig. 1); fruit samples from the bag removal treatment (Fig. 1) were also included in this analysis. As shown in Fig. 3a, skin color did not show red pigmentation in the non-bagged fruits. In contrast, the bag removal fruits showed red pigmentation over the entire skin, with an increase in anthocyanin content. Bagged fruits remained white in color with negligible anthocyanin content (Fig. 3a). High expression of MdMYB1 and the anthocyanin biosynthesis genes, MdCHS, MdCHI, MdF3H, MdDFR, MdANS, and MdUFGT, were observed only in the bag removal fruits, concomitant with the high anthocyanin content. Expression of these genes was negligible but similar in both the non-bagged and bagged fruits (Fig. 3a, b). In addition, these genes showed a dramatic increase only in the bag removal fruits even though non-bagged fruits were exposed to the same environmental conditions, suggesting that bagging treatment can increase the sensitivity of MdMYB1 to UV-B. These results indicate that some events induced MdMYB1 expression during paper bagging. Since MdMYB1 was expressed (Figs. 2c, 3a), we further intended to identify if its transcription was due to the activation of non-transcribed MdMYB1-2 and/or MdMYB1-3.
Effects of bagging treatments on anthocyanin accumulation and expression of MdMYB1 and anthocyanin biosynthetic genes. a ‘Mutsu’ normally shows a green skin color (non-bagged), but the fruits turned red after bag removal (bag removal), concomitant with high anthocyanin accumulation and high MdMYB1 expression. Fruits from bagging treatments showed white color with negligible anthocyanin concentration and MdMYB1 expression. b Expression of different anthocyanin biosynthetic genes. Transcription levels were determined according to the descriptions in Fig. 2. Error bars show SE
Pigmentation of ‘Mutsu’ related to high levels of MdMYB1-2/-3 transcripts
To confirm the MdMYB1 genotype of ‘Mutsu,’ an SNP located at about −1580-bp region (Fig. 4a, b) was used (Bai et al. 2014). Direct sequencing of PCR products amplified from the flanking region of the SNP showed the presence of two alleles, MdMYB1-2 and MdMYB1-3 (Fig. 4c, left), indicating the presence of two copies of MdMYB1-2 in the ‘Mutsu’ genome. Thus, the MdMYB1 genotype of ‘Mutsu’ was re-confirmed to be MdMYB1-2/-2/-3, which was also identified using CAPS markers (Hatsuyama et al. unpublished data). Furthermore, based on the SNP in the third exon of MdMYB1 (Takos et al. 2006) revealed after direct sequencing, we observed that both the alleles were transcribed (Fig. 4c, right), and the peak corresponding to MdMYB1-2 was much higher than that corresponding to MdMYB1-3, which was further confirmed by sequencing of the independent clones (among five independent clones, four clones corresponded to MdMYB1-2 and one to MdMYB1-3). These results indicated that the non-transcribed MdMYB1 alleles, MdMYB1-2/-3, were both actively transcribed in the red/pink fruit skin of ‘Mutsu’ as a result of the paper-bagging treatment. This further raised a question as to why the non-transcribed MdMYB1-2/-3 alleles were transcribed after the paper-bagging treatment.
a Schematic representation of the MdMYB1/A/10 locus on chromosome 9. The three exons are marked with blue bars along with the start codon (ATG). b Alignment of the promoter region around −2000 bp from the translation start codon and a of third the exon sequences of MdMYB1/A/10. Single nucleotide polymorphisms were identified in the 5′ upstream region of the genomic sequence (−1582 and −1574 bp) and in the third exon (573 bp) of the three MdMYB1 alleles, which were used for genotyping in ‘Mutsu.’ c Results for direct sequencing of genomic DNA and RT-PCR products. SNPs marked with an asterisk (−1582 bp) and a triangle (−1574 bp) in the genomic sequence indicate that ‘Mutsu’ harbors MdMYB1-2 and MdMYB1-3. SNP marked with a red circle (573 bp) indicates that both MdMYB1-2 and MdMYB1-3 were present in the transcripts of MdMYB1. RT-PCRs were performed with the no-reverse transcriptase reaction as the negative control to confirm that the RT-PCR products were from the transcripts and not from the genomic DNA
Epigenetic status of the 5′ upstream region of MdMYB1-2/-3 during UV-B treatment after bag removal
In our preliminary study using the ‘Sekaiichi’ and ‘Sensyu’ cultivars, whose MdMYB1 alleles consist of MdMYB1-1 and MdMYB1-3 (Hatsuyama, unpublished data) that can be discriminated by the nucleotide at the position 573 in the third exon (A in the case of MdMYB1-1 and T for MdMYB1-3), we found that only MdMYB1-1 was transcribed under natural conditions (Suppl. Table S2). This result indicates that the functional difference in the inductive ability of MdMYB1 allelic expression could be ascribed to the MdMYB1 genomic structure instead of the trans factor(s). Therefore, we assumed that some events in the transcriptional induction of the non-transcribed MdMYB1-2/-3 alleles would occur in the genomic structure of these alleles after paper bagging. We further assumed that epigenetic changes could be related to the current unexpected induction of MdMYB1-2/-3 expression.
Therefore, we focused on the epigenetic changes in the 5′ upstream region of MdMYB1-2/-3 (conserved sequences were used for both MdMYB1-2/-3) triggered by the paper-bagging treatment. First, we evaluated the cytosine methylation level of MdMYB1 by qPCR. In our system, a highly methylated Copia-type transposable element was used as a positive control to evaluate the efficiency of restriction digestion in each reaction (see the details in ‘Materials and methods’). The nucleotide numbers were calculated based on the 5′ upstream region of MdMYB1-2; this numbering similar in MdMYB1-3. ‘Mutsu’ fruits were used 0, 24, and 48 h after incubation under FL/UV-B or FL because the notable color changes occurred in the skin samples after 48 h under FL/UV-B treatment but not in those under FL treatment (Fig. 2a, b). We expected lower methylation levels, concomitant with red pigmentation in FL/UV-B treatment. Lower methylation levels in the −2025 to −1854-bp region at 24 and 48 h were recorded in the fruit skins under FL/UV-B treatment; these trends were also recorded in the FL treated skin (Fig. 5). Overall, the methylation patterns in the FL/UV-B were similar to those in the FL treatment. These results suggested that the 5′ upstream region of the MdMYB1-2/-3 was epigenetically active, but the results from the two treatments confounded our expectations.
DNA methylation levels in the 5′ upstream region of MdMYB1-2/-3 alleles in FL and FL/UV-B ‘Mutsu’ fruits (see Fig. 2a). Fruit skins were harvested after 0, 24, and 48 h of light treatment. The methylation percentage was estimated using an assay, combining McrBC digestion and qPCR (Telias et al. 2011). The X axis indicates the positions (relative position from the translation start sites) for qPCR amplification. Digestion efficiency was evaluated by a highly methylated Copia-type retrotransposon Ppcrt1 (AB550651, Copia TE). Digestion reactions were performed in triplicate with independently extracted genomic DNA (three biological replications). qPCR was performed with three technical replicates per biological replicate. Error bars show SE
Histone modifications near the transcription initiation and/or translation initiation sites are also a key regulatory mechanism for gene transcription (Guo et al. 2008; Charron et al. 2009; Jang et al. 2011). Therefore, we investigated trimethylation of histone H3 at lysine 4 (H3K4me3), trimethylation of histone H3 at lysine 27 (H3K27me3), and acetylation of histone H3 (H3ac) modifications within the ~700-bp region of MdMYB1-2/-3 in the skin during artificial light treatment (Fig. 2a). We first checked the H3K4me3 status in the 5′ flanking region of ELONGATION FACTOR 1 (EF1) as a positive control; the EF1 locus is under the control of H3K4me3 in petunia (Shibuya et al. 2009). Our results showed that the promoter region of EF1 was highly marked by H3K4me3, as expected (Suppl. Fig. S2), indicating that the chromatin immunoprecipitation protocol performed efficiently. We expected higher occupancy of H3ac and H3K4me3 and/or lower of H3K27me3 in the FL/UV-B samples than the FL samples. No obvious changes were observed in the H3ac and H3K27me3 levels (Fig. 6). Interestingly, H3K4me3 levels in all the tested regions were high at 48 h, but the changes were not significantly different between the FL and FL/UV-B samples (Fig. 6). MdMYB1 expression was elevated for the previous 24 h due to FL/UV-B treatment (Fig. 2c), which was earlier than the histone modifications. Therefore, the increase in H3K4me3 levels at 48 h might not be the key factor responsible for the high transcription of MdMYB1-2/-3 in paper bagging-induced red pigmentation.
Chromatin status in the 5′ upstream region of MdMYB1-2/-3 alleles in FL and FL/UV-B ‘Mutsu’ fruits (see Fig. 2a). Fruit skins were harvested after 0, 24, and 48 h of light treatment. Chromatin status was estimated by ChIP-qPCR using three different modified histone specific antibodies, namely H3ac (active histone mark), H3K4me3 (active histone mark), and H3K27me3 (inactive histone mark). Data are shown as the relative fold change of enriched DNA with specific antibodies compared to the negative control (normal mouse IgG). ChIP analysis was performed in triplicate with independent immunoprecipitation samples (three biological replications). qPCRs were performed with three technical replicates per biological replicate. Error bars show SE
Collectively, these results suggest that UV-B is an essential factor for paper bagging-induced red pigmentation (Fig. 2), but UV-B is not a cause for the induction of non-transcribed MdMYB1-2/-3 expression, which is consistent with the fact that the fruits under natural conditions do not show red pigmentation despite the presence of UV-B (Fig. 3a). Therefore, we could assume that the paper-bagging treatment itself might be involved in the induction of non-transcribed MdMYB1-2/-3 expression.
Epigenetic status of the 5′ upstream region of MdMYB1-2/-3 by paper-bagging treatment
To confirm our assumption, we investigated DNA methylation levels in MdMYB1-2/-3 by comparing the non-bagged and bagged samples (Fig. 1). We collected the bagged samples just after bag removal in a dark room (see ‘Materials and methods’) along with the non-bagged samples at the same time. In both the treatments, high methylation level (more than 90 %) was detected in the 5′ upstream region (−846 to −555 bp upstream from the translation start codon) of MdMYB1, whereas the methylation levels of other regions were around 50 % (Fig. 7). Similar trends have been reported previously in MdMYB10 (Telias et al. 2011). Significant differences in the methylation levels were observed in response to different treatments in some regions, −846 to −651 bp and −704 to −555 bp. We expected lower methylation levels in the bagged fruits than in the non-bagged ones; therefore, the significant low methylation observed in the bagged fruits in the −704 to −555-bp region was in agreement with our expectation, unlike that in the −846 to −651 bp region. Thus, the results were highly valuable, depending on the region. Nevertheless, considering previous reports (Telias et al. 2011; Xu et al. 2012), in which small changes in lower methylation levels had a big impact on red pigmentation, we cannot completely rule out the possible involvement of methylation levels in paper bagging-induced red pigmentation; however, further studies are needed in this regard.
DNA methylation levels in the 5′ upstream region of MdMYB1-2/-3 alleles in non-bagged and bagged ‘Mutsu’ fruits (Fig. 3a). The methylation percentage was estimated using an assay combining McrBC digestion and qPCR. The X axis indicates the position from ATG translation start sites for qPCR amplification. Digestion efficiency was evaluated by a highly methylated Copia-type retrotransposon, Ppcrt1. The statistically different (two-tail Student’s t test) methylation regions are marked with an asterisk (P < 0.05); Error bars show SE. Digestion reactions were performed in triplicate with independently extracted genomic DNA (three biological replications), and qPCR was performed with three technical replicates per biological replicate
Histone modifications were investigated within the ~700-bp region of MdMYB1-2/-3 in the skin of bagged and non-bagged fruits (Fig. 7). Higher levels of H3ac and H3K4me3 mark were recorded in response to the bagging treatment in all the tested regions, although only the H3K4me3 in the −704 to −555-bp region was statistically significant (Fig. 8). In addition, higher levels of inactive modification, H3K27me3, were detected in the −168 to −45-bp region of non-bagged fruits (Fig. 8). These trends were in accordance with our expectation that the higher occupancy of H3ac and H3K4me3, which are related to euchromatin, and lower occupancy of H3K27me3, which is related to heterochromatin, were observed in bagged fruits than in non-bagged ones. Thus, H3ac, H3K4me3, and H3K27me3 in the 5′ upstream region of MdMYB1-2/-3 of the paper-bagging treatment were associated with the paper bagging-induced red pigmentation; however, the results were not statistically significant in all the regions. These results suggest that different H3ac, H3K4me3, and H3K27me3 modifications contribute to the induction of MdMYB1-2/-3 expression.
Chromatin status in the 5′ upstream region of MdMYB1-2/-3 alleles in non-bagged and bagged ‘Mutsu’ fruits (Fig. 3a). Chromatin status was estimated by ChIP-qPCR using three different modified histone specific antibodies, namely H3ac (active histone mark), H3K4me3 (active histone mark), and H3K27me3 (inactive histone mark). Data are shown as the relative value of enriched DNA with specific antibodies compared with the negative control, normal mouse IgG. ChIP analysis was performed in triplicate with independent immunoprecipitation samples (three biological replications). qPCR was performed with three technical replicates per biological replicate. The statistically (two-tail Student’s t test) different methylation regions are marked with an asterisk (P < 0.05); Error bars show SE
Discussion
Paper-bagging treatment positively regulated de novo anthocyanin synthesis in apple skin
Paper bagging-induced red pigmentation in ‘Mutsu’ has been observed for decades and has been reported in other yellow/green cultivars, including ‘Golden Delicious’ and ‘Granny Smith’ (Liu et al. 2013). Increased MdMYB1 levels have been observed in the colored cultivar ‘Granny Smith’ (Wang et al. 2013a). We also observed that the other non-red cultivars, such as ‘Orin’ and ‘Kinsei,’ showed red pigmentation after bag removal, although the anthocyanin accumulation levels varied among the cultivars (data not shown). Paper bagging is also applied to red cultivars to induce more pigmentation in their skin. This suggests that paper bagging-induced red pigmentation is not unique to the cultivar ‘Mutsu,’ and other red and non-red apple cultivars could share the same mechanism.
It has been generally considered that the bright red color of red apple cultivars after paper bagging is due to the low chlorophyll background (green color) and not the higher anthocyanin content (Yonemori 2009). However, this hypothesis cannot explain the red pigmentation in non-red cultivars such as ‘Mutsu,’ because anthocyanin concentrations in the fruits under paper-bagging treatment were significantly higher than those in non-bagged fruits (Fig. 3a), suggesting that anthocyanins were synthesized de novo in such cultivars. In this respect, our finding that non-transcribed MdMYB1 alleles can be transcribed after paper-bagging treatment is fundamental for understanding paper bagging-induced red pigmentation. The photoprotective role of higher anthocyanin concentrations after bag removal was physiologically important. These pigments capture light with short wavelengths and protect the nucleotides and proteins against damage from dramatic changes in light exposure of bagged fruits after bag removal (Merzlyak et al. 2008).
Factors inducing non-transcribed MdMYB1 alleles in paper bagging-induced red pigmentation
Differential expression of MdMYB1 alleles in fruit skins results in different colored apples. However, although non-transcribed alleles such as MdMYB1-2 and MdMYB1-3 have been identified previously, the reasons for their poor expressions have not been well characterized. In blood orange, a retrotransposon element inserted in the promoter region of Ruby, the orthologous gene of MdMYB1, resulted in cold-dependent anthocyanin accumulation (Butelli et al. 2012). Similarly, the increase in anthocyanin accumulation in purple grape cultivars was due to the loss of retrotransposons from the promoter of VvmybA1-1 (Kobayashi et al. 2004). However, to date, no such retrotransposons have been correlated with low expression levels of MdMYB1-2 and MdMYB1-3 in apples. By comparing the promoter regions of different MdMYB1 alleles, we found an SNP that formed an intact light signal responsible G-box only in the MdMYB1-1 allele (Bai et al. 2014), which potentially affected the DNA-binding affinity of transcriptional factors, such as apple ELONGATED HYPOCOTYL 5 (MdHY5) and CONSTANS-like 11 (MdCOL11). However, such an intact G-box may not be the determinant of poor expression, because ‘Granny Smith’ is a non-red cultivar with unknown parents despite the presence of a transcribed MdMYB1-1 (intact G-box) and shows coloration after bagging treatment, similarly to other non-red cultivars (Hatsuyama, unpublished data), such as ‘Mutsu’ and ‘Golden Delicious’ (Liu et al. 2013).
The effects of UV-B on the anthocyanin accumulation in apples have been well characterized. Arakawa et al. (1986) investigated the effects of light of various wavelengths on apple coloration and found that UV-B (~320 nm) was most effective. Ubi et al. (2006) reported that the expression of five anthocyanin biosynthetic genes was enhanced by UV-B and low temperature treatments, concomitantly with the accumulation of anthocyanins. Previous studies on bagging practice of the red cultivar ‘Fuji’ also indicated that paper bagging-induced red pigmentation occurred in a UV-B-dependent manner, while the red color development after bag removal was retarded in the absence of UV-B irradiation (Fan and Mattheis 1998). The present study showed that ‘Mutsu,’ a non-red cultivar, turned red only under light supplemented with UV-B (Fig. 2), confirming that the paper bagging-induced red pigmentation of non-red cultivars was also a UV-B-dependent process, in which the activation of the normally non-transcribed MdMYB1-2/-3 alleles in the non-red cultivars resulted in their red pigmentation (Fig. 2). All these reports suggested that UV-B treatment is indispensable for anthocyanin accumulation, but how MdMYB1/A/10 was activated by UV-B after bag removal, especially in non-red cultivars, such as ‘Mutsu,’ remains unclear. It is worth noting that ‘Mutsu’ fruits in the orchard (without paper-bagging treatment) did not show red pigmentation despite the presence of UV-B in the natural light (Fig. 3). Therefore, we suggest that the paper-bagging treatment itself could be a primary, and UV-B, a secondary factor, for the red pigmentation. We assumed that the paper-bagging treatment elevated the sensitivity of fruit skin to UV-B. To determine the cause of the elevation of this sensitivity, we investigated the possible involvement of epigenetic regulation in MdMYB1-2/-3 expression.
DNA methylation in the 5′ upstream region of MdMYB1-2/-3
DNA methylation has two essential roles in plants and animals—defending the genome against transposons and regulating gene expression. Genome-wide analysis in Arabidopsis indicated that DNA methylation in the promoter regions is often associated with transcriptional gene silencing (Zhang et al. 2006; Zilberman et al. 2007). To date, the pivot roles of DNA methylation on development, stress tolerance, immune response, and local adaptation have been confirmed by the various lines of evidence (Chan et al. 2005; Dowen et al. 2012; Kinoshita and Seki 2014; Yang et al. 2015). In addition, DNA methylation also controls fruit development; a natural epigenetic mutation in an SBP-box gene resulted in its reduced expression, leading to the inhibition of tomato fruit ripening (Manning et al. 2006). In apple, it has been known that methylation levels regulate the expression of MdMYB1/A/10 gene, under certain conditions. Telias et al. (2011) reported that differences in anthocyanin levels between red and green stripes could be attributed to differential expression of MdMYB10, owing to variable methylation levels of MdMYB10 in ‘Honeycrisp’ compared to ‘Royal Gala’ cultivars. DNA methylation in the promoter region of MdMYB1 was higher in the ‘Ralls’ red apple cultivar compared to its blushed sport (Xu et al. 2012). More recent research showed that the alteration of DNA methylation in the MdMYB10 promoter of a red cultivar, ‘Gala,’ resulted in a yellow-skin somatic mutant (El-Sharkawy et al. 2015). These reports suggest that DNA methylation influences the color pattern of the apples through regulation of MdMYB1 expression. Wang et al. (2013b) detected higher methylation levels in the PcMYB10 promoter in the green- than the red-skinned European pear. Moreover, artificial induction of DNA methylation in the PcMYB10 promoter by virus-induced gene silencing in the red cultivar represses PcMYB10 expression, which in turn resulted in less anthocyanin accumulation (Wang et al. 2013b). We also focused on DNA methylation during UV-B treatment after bag removal and in the paper-bagging treatment. The results showed variations depending on the regions tested. The methylation levels in the regions between the current and previous research (Telias et al. 2011; El-Sharkawy et al. 2015) showed variations possibly due to the different allele types of MdMYB, but similar to the DNA methylation patterns of ‘Gala’ and ‘Honeycrisp’ (Telias et al. 2011), relatively high methylations in the regions of −846 to −651 and −704 to −555 were observed, whereas those in the regions of −310 to −178 and −168 to −45 were low (Figs. 5, 7). The changes in methylation levels in the 5′ upstream region of MdMYB1-2/-3 during UV-B treatment might not be related to the regulation of MdMYB1-2/-3 expression because of similar methylation patterns between the FL and FL/UV-B treatments (Fig. 5). In contrast, there were several differences in the methylation levels between the non-bagged and bagged treatments, although they were not significant in several regions (Fig. 7). In the present study, the noticeable differences in methylation levels were recorded in the −2025 to −1854 bp (not statistically significant with an unpaired t test P = 0.07), −846 to −651 bp and −704 to −555 bp (both statistically significant), among which lower methylation levels in the −2025 to −1854 bp and −704 to −555 bp in the bagging treatment were according to our expectation; it could be possible that the paper-bagging treatment itself could be a cause for the induction of MdMYB1-2/-3 transcripts. El-Sharkawy et al. (2015) also reported in two regions (−1246 to −780 bp and −2585 to −2117 bp) the methylation activity in three ‘Gala’ strains were clearly different, with different fruit skin color and methylation levels in both regions negatively correlated with anthocyanin content and MdMYB10 expression. However, all these regions do not include the variable G-box, which is located in the −1580 bp. Thus, we assume that lower cytosine methylation levels caused by the paper-bagging treatment (darkness) could be involved in inducing MdMYB1-2/-3 transcription.
Histone modifications in the 5′ upstream region of MdMYB1
In plants, changes in light condition induce transient alteration of the histone modification patterns (Guo et al. 2008; Charron et al. 2009; Jang et al. 2011). De-etiolation of Arabidopsis during the transition from dark to light has been well characterized, in which activation of ELONGATED HYPOCOTYL 5 (HY5) and HY5 homologue during de-etiolation is accompanied by a significant increase in H3K9ac without detectable differences in the H3K27ac levels (Charron et al. 2009). Dark activation of phyA was also marked by an increase of H3K9ac and H3K27ac (Jang et al. 2011). AtMYB75, the homologous gene of MdMYB1 in Arabidopsis, is marked by H3K27me3 but not by H3K9ac, H3K27ac, or H3K9me3 in both dark conditions and during the transition from dark to light (Guo et al. 2008). These reports indicated the possibility for histone modifications by light treatment including UV-B. Therefore, in the present study, we also investigated the histone modifications both during the UV-B treatment after bag removal and in the paper-bagging treatment. During UV-B treatment, we observed the enrichment of H3K4me3 after 48 h light treatment in both the FL and FL/UV-B treatments (Fig. 6), but such modification was not accompanied by MdMYB1 expression, which peaked 24 h after the light exposure (Fig. 2). Regarding the paper-bagging treatment, the ChIP assay revealed high H3ac and H3K4me3 occupancy in the 5′ upstream region of MdMYB1 in the dark condition, although only H3K4me3 was statistically significant in the −704-bp to −555-bp region (Fig. 8). In contrast, we did not observe general changes in the H3K27me3 level in the 5′ upstream region of MdMYB1, while lower occupancy of H3K27me3 was detected in the −168-bp to −45-bp region in the bagged fruits (Fig. 8). Although the role of H3K4me3 modification during the bagging treatment needs to be studied further, our results suggested the possible involvement of H3K4me3 during the bagging treatment. According to these results, we propose that bagging itself (darkness) could modify histone conformation toward activation with high occupancy of H3K4me3 and low occupancy of H3K27me3, facilitating the binding of trans factor(s) to the MdMYB1-2/-3 alleles, and the consequent increase in the transcripts of MdMYB1-2/-3 could activate the downstream anthocyanin biosynthetic genes. The role of such small epigenetic changes, as observed in the present study, in changing the histone modification status might be subject to skepticism. However, previous reports have shown that small changes in histone modifications of upstream regions of a given gene could have a large impact on endodormancy phase transition in peaches and pears (Leida et al. 2012; Saito et al. 2015). A plausible assumption could be that the observed tendencies of enrichment of both the active H3ac and H3K4me3 marks and a decrease of H3K27me3 in the bagged fruits enhanced the transcriptional activity of MdMYB1, resulting in a higher expression level during the ensuing light exposure.
During the life cycle of plants, continuous darkness only occurs during germination before the seedling protrudes from the soil. Therefore, most of the studies focused on the light responses in early seedling development (Deng et al. 1991; Oyama et al. 1997; Kami et al. 2010; Lau and Deng 2010; Chaves et al. 2011; Rizzini et al. 2011; Wang et al. 2012). In contrast, the effects of continuous darkness on other biological processes remain elusive. In the present study, we provided evidence that continuous darkness (artificial bagging treatment) increased the transcriptional potency of a central anthocyanin biosynthesis regulatory gene, MdMYB1, via epigenetic regulation, which in turn lead to the coloration of the non-red apple cultivar ‘Mutsu.’ However, ‘Mutsu’ fruits were kept in paper bags for more than 4 months and a difference in the degree of DNA methylation levels and some histone modifications between the non-bagged and bagged samples were detected at the end of the treatment. The timing of such epigenetic modifications during the paper-bagging treatment is not clear at present and would need further studies.
In summary, the present study suggests that the paper-bagging treatment is an important factor for inducing red pigmentation, which regulates DNA methylation and histone modifications in the 5′ upstream region of MdMYB1-2/-3 alleles in the ‘Mutsu’ cultivar. Thus, our results indicate novel findings because paper-bagging treatment can transform non-transcribed MdMYB1-2 and MdMYB1-3 alleles to transcribed alleles, possibly through epigenetic regulation. Due to the technical limitation, the genetical confirmation of the effects of chromatin modification is currently impossible in fruit trees. Therefore, it is difficult to clarify the effects of the observed weak significance. However, our work provided a possible explanation for paper bagging-induced red pigmentation of apples, which will open a new avenue for uncovering this interesting phenomenon in the future.
Author contribution statement
TM started the project and conceived the study; SB and TM designed the experiments; SB, PAT and TS carried out the experiments; SB and TM analyzed the experimental data; CH, YH and AI helped in the experiment design and data analysis; SB and TM prepared the manuscript. All authors read and approved the final manuscript.
Abbreviations
- ANS:
-
Anthocyanin synthase
- CHI:
-
Chalcone isomerase
- ChIP:
-
Chromatin immunoprecipitation
- CHS:
-
Chalcone synthase
- DFR:
-
Dihydroflavonol 4-reductase
- F3H:
-
Flavanone-3β-hydroxylase
- FL:
-
Fluorescent lamp
- H3ac:
-
Acetylation of histone H3
- H3K4me3 (H3K27me3):
-
Trimethylation of histone H3 at lysine 4 (at lysine 27)
- SNP:
-
Single nucleotide polymorphism
- UFGT:
-
UDP-glucose:flavonoid-3-O-glycosyltransferase
References
Arakawa O, Hori Y, Ogata R (1986) Characteristics of color development and relationship between anthocyanin synthesis and phenylalanine ammonia-lyase activity in ‘Starking Delicious’, ‘Fuji’ and ‘Mutsu’ apple fruits. J Jpn Soc Hortic Sci 54:424–430
Bai S, Saito T, Honda C, Hatsuyama Y, Ito A, Moriguchi T (2014) An apple B-box protein, MdCOL11, is involved in UV-B- and temperature-induced anthocyanin biosynthesis. Planta 240:1051–1062
Ban Y, Honda C, Hatsuyama Y, Igarashi M, Bessho H, Moriguchi T (2007) Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol 48:958–970
Broun P (2005) Transcriptional control of flavonoid biosynthesis: a complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr Opin in Plant Biol 8:272–279
Butelli E, Licciardello C, Zhang Y, Liu J, Mackay S, Bailey P, Reforgiato-Recupero G, Martin C (2012) Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 24:1242–1255
Chagné D, Carlisle CM, Blond C, Volz RK, Whitworth CJ, Oraguzie NC, Crowhurst RN, Allan AC, Espley RV, Hellens RP, Gardiner SE (2007) Mapping a candidate gene (MdMYB10) for red flesh and foliage colour in apple. BMC Genom 8:212
Chagné D, Lin-Wang K, Espley RV, Volz RK, How NM, Rouse S, Brendolise C, Carlisle CM, Kumar S, De Silva N, Micheletti D, McGhie T, Crowhurst RN, Storey RD, Velasco R, Hellens RP, Gardiner SE, Allan AC (2013) An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiol 161:225–239
Chan SWL, Henderson IR, Jacobsen SE (2005) Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet 6:351–360
Charron JB, He H, Elling AA, Deng XW (2009) Dynamic landscapes of four histone modifications during deetiolation in Arabidopsis. Plant Cell 21:3732–3748
Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen LO, van der Horst GTJ, Batschauer A, Ahmad M (2011) The cryptochromes: blue light photoreceptors in plants and animals. Annu Rev Plant Biol 62:335–364
Cottage A, Edwards YJ, Elgar G (2001) SAND, a new protein family: from nucleic acid to protein structure and function prediction. Comp Funct Genomics 2:226–235
Deng XW, Caspar T, Quail PH (1991) cop1—a regulatory locus involved in light-controlled development and gene-expression in Arabidopsis. Genes Dev 5:1172–1182
Dowen RH, Pelizzola M, Schmitz RJ, Lister R, Dowen JM, Nery JR, Dixon JE, Ecker JR (2012) Widespread dynamic DNA methylation in response to biotic stress. Proc Natl Acad Sci USA 109:E2183–E2191
El-Sharkawy, Liang D, Xu K (2015) Transcriptome analysis of an apple (Malus × domestica) yellow fruit somatic mutation identifies a gene network module highly associated with anthocyanin and epigenetic regulation. J Exp Bot 66:7359–7376
Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC (2007) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J 49:414–427
Espley RV, Brendolise C, Chagné D, Kutty-Amma S, Green S, Volz R, Putterill J, Schouten HJ, Gardiner SE, Hellens RP, Allan AC (2009) Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 21:168–183
Fan X, Mattheis JP (1998) Bagging ‘Fuji’ apples during fruit development affects color development and storage quality. HortScience 33:1235–1238
Gasic K, Hernandez A, Korban S (2004) RNA extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Mol Biol Rep 22:437–438
Guo L, Zhou J, Elling AA, Charron J-B, Deng XW (2008) Histone modifications and expression of light-regulated genes in Arabidopsis are cooperatively influenced by changing light conditions. Plant Physiol 147:2070–2083
Hichri I, Barrieu F, Bogs J, Kappel C, Delrot S, Lauvergeat V (2011) Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J Exp Bot 62:2465–2483
Imai T, Ubi B, Saito T, Moriguchi T (2014) Evaluation of reference genes for accurate normalization of gene expression for real time-quantitative PCR in Pyrus pyrifolia using different tissue samples and seasonal conditions. PLoS ONE 9:e86492
Jang IC, Chung PJ, Hemmes H, Jung C, Chua NH (2011) Rapid and reversible light-mediated chromatin modifications of Arabidopsis phytochrome A locus. Plant Cell 23:459–470
Kami C, Lorrain S, Hornitschek P, Fankhauser C (2010) Light-regulated plant growth and development. Curr Top Dev Biol 91:29–66
Kim H, Terakami S, Nishitani C, Kurita K, Kanamori H, Katayose Y, Sawamura Y, Saito T, Yamamoto T (2012) Development of cultivar-specific DNA markers based on retrotransposon-based insertional polymorphism in Japanese pear. Breed Sci 62:53–62
Kinoshita T, Seki M (2014) Epigenetic memory for stress response and adaptation in plants. Plant Cell Physiol 55:1859–1863
Kobayashi S, Goto-Yamamoto N, Hirochika H (2004) Retrotransposon-induced mutations in grape skin color. Science 304:982
Lau OS, Deng XW (2010) Plant hormone signaling lightens up: integrators of light and hormones. Curr Opin Plant Biol 13:571–577
Leida C, Conesa A, Llácer G, Badenes ML, Ríos G (2012) Histone modifications and expression of DAM6 gene in peach are modulated during bud dormancy release in a cultivar-dependent manner. New Phytol 193:67–80
Lin-Wang K, Bolitho K, Grafton K, Kortstee A, Karunairetnam S, McGhie TK, Espley RV, Hellens RP, Allan AC (2010) An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol 10:50
Liu Y, Che F, Wang L, Meng R, Zhang X, Zhao Z (2013) Fruit coloration and anthocyanin biosynthesis after bag removal in non-red and red apples (Malus × domestica Borkh.). Molecules 18:1549–1563
Livak K, Schmittgen T (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT. Methods 25:402–408
Manning K, Tor M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB (2006) A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet 38:948–952
Merzlyak MN, Melø TB, Naqvi KR (2008) Effect of anthocyanins, carotenoids, and flavonols on chlorophyll fluorescence excitation spectra in apple fruit: signature analysis, assessment, modelling, and relevance to photoprotection. J Exp Bot 59:349–359
Mink G (1973) The apple industry in Japan. HortScience 8:81–86
Oyama T, Shimura Y, Okada K (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11:2983–2995
Peng T, Saito T, Honda C, Ban Y, Kondo S, Liu J, Hatsuyama Y, Moriguchi T (2013) Screening of UV-B-induced genes from apple peels by SSH: possible involvement of MdCOP1-mediated signaling cascade genes in anthocyanin accumulation. Physiol Plant 148:432–444
Rizzini L, Favory JJ, Cloix C, Faggionato D, O’Hara A, Kaiserli E, Baumeister R, Schafer E, Nagy F, Jenkins GI, Ulm R (2011) Perception of UV-B by the Arabidopsis UVR8 protein. Science 332:103–106
Saito T, Bai S, Ito A, Sakamoto D, Ubi B, Imai T, Moriguchi T (2013) Expression and genomic structure of the dormancy-associated MADS box genes MADS13 in Japanese pears (Pyrus pyrifolia Nakai) that differ in their chilling requirement for endodormancy release. Tree Physiol 33:654–667
Saito T, Bai S, Imai T, Ito A, Nakajima I, Moriguchi T (2015) Histone modification and signaling cascade of the dormancy-associated MADS-box gene, PpMADS13-1, in Japanese pear (Pyrus pyrifolia) during endodormancy. Plant Cell Environ 38:1157–1166
Shibuya K, Fukushima S, Takatsuji H (2009) RNA-directed DNA methylation induces transcriptional activation in plants. Proc Natl Acad Sci USA 106:1660–1665
Takos AM, Jaffé FW, Jacob SR, Bogs J, Robinson SP, Walker AR (2006) Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol 142:1216–1232
Telias A, Lin-Wang K, Stevenson DE, Cooney JM, Hellens RP, Allan AC, Hoover EL, Bradeen JM (2011) Apple skin patterning is associated with differential expression of MYB10. BMC Plant Biol 11:93
Ubi BE, Honda C, Bessho H, Kondo S, Wada M, Kobayashi S, Moriguchi T (2006) Expression analysis of anthocyanin biosynthetic genes in apple skin: effect of UV-B and temperature. Plant Sci 170:571–578
Umemura H, Otagaki S, Wada M, Kondo S, Matsumoto S (2013) Expression and functional analysis of a novel MYB gene, MdMYB 110a_ JP, responsible for red flesh, not skin color in apple fruit. Planta 238:65–76
Velasco R, Zharkikh A, Affourtit J et al (2010) The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet 42:833–839
Wang ZY, Bai MY, Oh E, Zhu JY (2012) Brassinosteroid signaling network and regulation of photomorphogenesis. Annu Rev Genet 46:701–724
Wang L, Zhang X, Liu Y, Shi X, Wang Y, Zhang C, Zhao Z (2013a) The effect of fruit bagging on the color, phenolic compounds and expression of the anthocyanin biosynthetic and regulatory genes on the ‘Granny Smith’ apples. Eur Food Res Technol 237:875–885
Wang Z, Meng D, Wang A, Li T, Jiang S, Cong P, Li T (2013b) The methylation of the PcMYB10 promoter is associated with green-skinned sport in max red bartlett pear. Plant Physiol 162:885–896
Xu Y, Feng S, Jiao Q, Liu C, Zhang W, Chen W, Chen X (2012) Comparison of MdMYB1 sequences and expression of anthocyanin biosynthetic and regulatory genes between Malus domestica Borkh. cultivar ‘Ralls’ and its blushed sport. Euphytica 185:157–170
Yang HX, Chang F, You CJ, Cui J, Zhu GF, Wang L, Zheng Y, Qi J, Ma H (2015) Whole-genome DNA methylation patterns and complex associations with gene structure and expression during flower development in Arabidopsis. Plant J 81:268–281
Yonemori K (2009) Japanese pomological magic: producing fruits for gifts. Chron Hortic 49:15–18
Zhang XY, Yazaki J, Sundaresan A, Cokus S, Chan SWL, Chen HM, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, Ecker JR (2006) Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126:1189–1201
Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S (2007) Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet 39:61–69
Acknowledgments
This work was supported by Grants-in-Aid for Scientific Research (No. 24·02214) from the Japanese Society for the Promotion of Science (JSPS). We thank Dr. Ken-ichi Shibuya for technical advice on chromatin immunoprecipitation sample preparation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bai, S., Tuan, P.A., Saito, T. et al. Epigenetic regulation of MdMYB1 is associated with paper bagging-induced red pigmentation of apples. Planta 244, 573–586 (2016). https://doi.org/10.1007/s00425-016-2524-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-016-2524-4