Abstract
Main conclusion
Comparative and association analyses of the proteome and transcriptome for pear fruit development were conducted for the first time in this study.
Pear fruit development involves complex physiological and biochemical processes, but there is still little knowledge available at proteomic and transcriptomic levels, which would be helpful for understanding the molecular mechanisms of fruit development and quality in pear. In our study, three important stages, including early development (S4-22), middle development (S6-27), and near ripening (S8-30), were investigated in ‘Dangshansuli’ by isobaric tags for relative and absolute quantitation (iTRAQ) labeling technology, identifying a total of 1,810 proteins during pear fruit development. The association analysis of proteins and transcript expression revealed 1,724, 1,722, and 1,718 associated proteins identified in stages S4-22, S6-27, and S8-30, respectively. A total of 237, 318, and 425 unique proteins were identified as differentially expressed during S4-22 vs S6-27, S6-27 vs S8-30, S4-22 vs S8-30, respectively, and the corresponding correlation coefficients of the overall differentially expressed proteins and transcripts data were 0.6336, 0.4113, and 0.7049. The phenylpropanoid biosynthesis pathway, which is related to lignin formation of pear fruit, was identified as a significantly enriched pathway during early stages of fruit development. Finally, a total of 35 important differentially expressed proteins related to fruit quality were identified, including three proteins related to sugar formation, seven proteins related to aroma synthesis, and sixteen proteins related to the formation of lignin. In addition, qRT-PCR verification provided further evidence to support differentially expressed gene selection. This study is the first to reveal protein and associated mRNA variations in pear during fruit development and quality conformation, and identify key genes and proteins helpful for future functional genomics studies, and provides gene resources for improvement of pear quality.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pear, belonging to the subfamily pomoideae in the family Rosaceae, is one of the oldest fruit crops and cultivated worldwide in 85 countries or regions (Wu et al. 2013). Production ranks after just apple and grape within perennial deciduous fruit crops. Pear cultivation can be traced back 3000 years, however, in recent 100 years, breeders and cultivators have been selecting and breeding new varieties with the aim to improve fruit quality and meet consumer demands (Lombard Westwood 1987).
Comprehensive understanding of the genetic elements is very important for understanding the regulation of fruit development regulation. Although the pathways involved in the biological processes of pear fruit development and ripening are complex, usually including comprehensive metabolic changes, there are many studies on related traits of pear, such as pigmentation (Dussi et al. 1995), aroma release (Argenta et al. 2003; Chen et al. 2006), simple sugar and organic acid accumulation (Hudina and Śtampar 2000), the formation of stone cells (Tao et al. 2009; Cai et al. 2010) and morphological changes (Bain 1961). These previous studies have demonstrated the main biological pathways of metabolites, and change patterns related with fruit development. With the adoption of molecular biotechnology in pear research, several genes involved in the biological pathways were identified and successfully cloned. For example, the full-length cDNA of seven anthocyanin biosynthesis-related genes was cloned in the cultivar ‘Early red Doyenne du Comice’ and its green variant strain (Yang et al. 2013), and soluble acid invertase, which plays an important role in accumulating hexoses during fruit enlargement, were cloned (Yamada et al. 2007). 1-aminocyclopropane-1-carboxylic acid synthase genes were identified as controlling ethylene levels during fruit development in Japanese pear (Itai et al. 1999), and Cinnamoyl CoA reductase (CCR) has been demonstrated to be involved in the regulation of lignin biosynthesis and plays an important role in stone cell formation in pear flesh (Lu et al. 2011).
However, it is still difficult to reveal the broader patterns of genetic control and identify key genes responsible for fruit development by a partial biosynthesis procedure, which only encompasses a few known genes. In recent years, many genomic tools have become available to generate a great amount of data rapidly and detect whole gene and protein expression profiles, which provide a good strategy and data platform to compare and determine the important genes and regulation network of fruit development. More and more genome sequences have been released for different fruit tree species (Jaillon et al. 2007; Ming et al. 2008; Velasco et al. 2010; Shulaev et al. 2011; Xu et al. 2013; Verde et al. 2013); these provide a good genomic data platform for further resolving important agronomic traits. For example, reports on transcript level analysis and metabolite profiling reflecting regulation of fruit metabolism during fruit development and ripening have been published in tomato (Mounet et al. 2009; Alba et al. 2005; Osorio et al. 2011) and mRNA expression profiling studies have also been conducted to describe fruit development and ripening regulation in other fruits, such as Chinese bayberry (Feng et al. 2012), citrus (Yu et al. 2012), and apple (Soglio et al. 2009).
In previous studies, proteomics has been shown to be a powerful approach for establishing functional correlations between phenotype and genotype, and characterizing biochemical networks (Palma et al. 2011), such as the studies on developmental fruits that have been done on grape (Giribaldi et al. 2007), cherry tomato (Faurobert et al. 2007), sweet orange (Pan et al. 2012), strawberry (Bianco et al. 2009), and papaya (Nogueira et al. 2012). These researches described tissue-dependent proteome repertoires that show distinctive changes during development. However, for pear, information about protein composition is very limited. iTRAQ labeling technology is a powerful technique with high accuracy and sensitivity, demonstrating a remarkable advantage in simultaneous analysis of multiple samples and subsequently providing relative quantification for hundreds of proteins at one time (Ross et al. 2004). iTRAQ technology can produce high-quality, reproducible results in organelles, whole cell lysates, and enriched complexes. Hence, iTRAQ has been widely used for micro-organisms, fauna, and flora (Zhou et al. 2010; Zhu et al. 2010)
However, functional genomic research has confirmed that information from nucleotide data do not necessarily match the corresponding translated protein complement in an accurate organism physiological manner (Fernie and Stitt 2012). Proteins, as molecules that directly influence biological processes, have their own special activity patterns, and information at the level of gene transcription is not sufficient to reveal the exact function of intracellular processes. In this background, interest in the proteome emerged (Wilkins et al. 1996). Association of transcriptome and proteome data will lead to better knowledge of gene networks. To better clarify potential molecular mechanisms, some studies have been done to correlate the protein and mRNA expression levels of the same samples, attempting to identify reliable genes for amelioration and crop breeding (Maier et al. 2011; Wu et al. 2014a, b)
Pear, as an important fruit species in Rosaceae, has recently had its whole genome sequenced, as well as genome-wide gene annotation and expression related to important fruit quality traits (Wu et al. 2013). However, proteomics analyses during pear fruit development have not been examined until now. Here, iTRAQ labeling technology was used to analyze the proteins dynamic changes during pear fruit development. These changes and overall proteomic association with transcriptome analysis were initiated to explore the potential effect of transcriptome and proteomics on fruit development. The results will facilitate identification of the most variable differentially expressed proteins and genes associated with important traits, such as sugar and organic acid metabolism, aroma synthesis, stone cell and lignin metabolism; and investigation of the dynamic pattern of the protein and gene functional networks in relation to pear fruit development and ripening. They can also lay a foundation for the molecular basis for future improvement of fruit quality.
Materials and methods
Plant materials
Fruit samples of ‘Dangshansuli’ at three stages of fruit development were collected in the pear depository at Pukou District pomology farm of Nanjing Agriculture University. Three time points were sampled depending on the status of pear development in 2010, S4-22 (15 days after full blooming, DAFB) represents the early development stage of fruit; S6-27 (80 DAFB) represents the middle development of fruit, and S8-30 (145 DAFB) represents the near ripening stage of fruit. The phenotypic traits of pear fruit are shown in Fig. 1. About 10–20 pear fruits, depending on the fruit size at different stages, from three different trees were collected. Finally, fruit samples at the same stage were mixed after washing and kernel removing and cut into slices, then snap frozen directly in liquid nitrogen and transported back to the lab. All collected samples were stored at −80 °C until use for proteomic and RNA-seq analysis.
Protein extraction, digestion and iTRAQ labeling LC–MS/MS
The total proteins of the three different developmental stages (S4-22, S6-27, and S8-30) of pear fruit were extracted. Protein extraction was performed according to a previously described protocol (Pedreschi et al. 2008). Finally, Bradford protein quantification and SDS-PAGE were used for accurately quantifying protein concentrations, taking bovine serum albumin as the standard (Bradford 1976). Then 100 μg protein was taken out for treatment from each sample solution, and the protein was digested with Trypsin Gold with the ratio of protein:trypsin = 20:1 at 37 °C for 4 h. Then, Trypsin Gold was added with the ratio of protein:trypsin = 20:1 once more, and digested for 8 h. After this step, peptides were reconstituted in 0.5 M TEAB and processed according to the manufacturer’s instructions for 8-plex iTRAQ reagent (Applied Biosystems). Briefly, one unit of iTRAQ reagent was thawed and reconstituted in 24 μL isopropanol. Each sample was labeled using the iTRAQ tags as follows: S4-22 (114 tag), S6-27 (115 tag), and S8-30 (121 tag). The peptides were labeled with the isobaric tags and incubated at room temperature for 2 h. The labeled peptide mixtures were then dried and pooled by vacuum centrifugation. The pooled mixtures of iTRAQ-labeled peptides were fractionated by strong cationic exchange (SCX) chromatography, using the Shimadzu LC-20AB high pressure liquid chromatography (HPLC) Pump system. The peptides from digestion were reconstituted with 4 mL buffer A (25 mM NaH2PO4 in 25 % ACN, pH 2.7) and loaded directly onto a 4.6 × 250 mm Ultremex SCX column containing 5-μm particles (Phenomenex). The peptides were eluted at a flow rate of 1 mL/min using the following gradient: buffer A for 10 min, and then 5–35 % buffer B (25 mM NaH2PO4, 1 M KCl in 25 % ACN, pH2.7) for 11 min, 35–80 % buffer B for 1 min. The system was then maintained in 80 % buffer B for 3 min before equilibrating with buffer A for 10 min prior to the next injection. Absorbance at 214 nm was used to monitor the eluent, and fractions were collected each minute. The eluted peptides were pooled as ten fractions. Samples were dried using vacuum and desalted using Strata X C18 column (Phenomenex).
LC–ESI–MS/MS analysis by LTQ-Orbitrap HCD
Each SCX fraction was re-suspended with 60 μl of buffer A (2 % ACN, 0.1 % FA) and centrifuged at 20,000g for 10 min. The final concentration of peptide was approximately 0.5 μg/μl, on average in each fraction. A 10 µl sample from each fraction was loaded directly on a Shimadzu LC-20AD nanoHPLC, and then onto a 2 cm C18 capillary trap cartridge (inner diameter 200 μm). Finally, samples were separated on a 10-cm nanoflow C18 column (PepMap 75 μm inner diameter) made in-house. The samples were loaded at 15 μL/min for 4 min, then a linear gradient was run at 400 nL/min starting from 2 to 35 % B (98 % ACN, 0.1 % FA) for 44 min, followed by a linear gradient to 80 % for 2 min, and maintenance 4 min at 80 % B, and finally returned to 2 % in 1 min.
The peptides were subjected to nanoelectrospray ionization, and then analyzed by tandem mass spectrometry (MS/MS) in an LTQ Orbitrap Velos (Thermo) coupled online to the HPLC. Complete peptides were chosen in the Orbitrap at a resolution of 60,000. Peptides were chosen for MS/MS with a normalized collision energy setting of 45 % using high-energy collision dissociation (HCD) operating mode, at last, ion fragments were chosen in the LTQ. A data-dependent program that alternated between one MS scan followed by eight MS/MS scans was applied for the eight most abundant precursor ions above a threshold ion count of 5,000 in the MS survey scan with the following Dynamic Exclusion settings: exclusion duration, 120 s; repeat duration, 30 s; and repeat counts, two. The electrospray voltage of 1.5 kV versus the inlet of the mass spectrometer was used. 1 × 104 ions were accumulated in the ion trap used to produce the HCD spectra; Automatic gain control (AGC) was used to prevent overfilling of the ion trap. For MS scans, the m/z scan was ranged from 350 to 2,000 Da.
Database search and quantification
The 2.3.02 version of the Mascot software (Matrix Science) was used to simultaneously quantify and identify proteins. In this version, only unique peptides used for protein quantification are used, enhancing precision. In this study, the global false discovery rate (FDR) of protein id is 0.07795 %. Searches were made against the database (http://www.peargenome.njau.edu.cn) (Wu et al. 2013). Spectra from the ten fractions were combined into one Mascot Generic Formats file after loading the raw data, and the MGF file was searched. All parameters were set as follows: trypsin was specified as the digestion enzyme; variable modifications of oxidation at Met, fixed modifications of carbamidomethylation at Cys; MS/MS tolerance was set at 0.05 Da, and peptide tolerance was set at 10 ppm. Peptide charge was set as Mr and monoisotopic mass was selected; the iTRAQ 8-plex was selected for quantification during the search simultaneously.
The search results were passed via additional filters before exporting the data. For protein identification, all criteria were set as follows: ion score or expected cutoff less than 0.05 (with 95 % confidence) and significance threshold p < 0.05 (with 95 % confidence). For protein quantitation, all criteria were set as follows: “median” was selected as the protein ratio type (http://www.matrixscience.com/help/quant_config_help.html); minimum peptide was set as two and minimum precursor charge was set as 2+; only unique peptides were used to quantify proteins. Median intensities were set as normalization, and outliers were removed automatically. Finally, the peptide threshold was set as described above for identity.
Proteomic data analysis
The metabolic pathway analysis of the identified proteins was conducted according to Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (Kanehisa and Goto 2000). Gene Ontology (GO) (http://www.geneontology.org) and Cluster of Orthologous Groups of proteins database analyses were conducted according previous research (Tatusov et al. 2001). Proteins with 1.5 fold change between any two stages and p value less than 0.05 were identified as differentially expressed proteins. Metabolic pathway and GO enrichment analysis of the differentially expressed proteins were conducted according to the information from the KEGG Pathway and GO databases, respectively, using the following formula:
N is the number of all identified proteins that can be connected with GO or KEGG Pathway analysis information. n is the number of differential proteins in N. M is the number of proteins that can be connected with a certain GO term or pathway. m is the number of differential proteins with certain GO term or KEGG pathway. If p value ≤ 0.05, we regard this GO term or pathway as a significant enrichment of differential proteins. We can then resolve the main biological function or pathway of differential proteins by GO or KEGG Pathway enrichment analysis during pear fruit development.
Association analysis of proteomics and transcriptomics
RNA sequencing data of three development stages was derived from our previous whole genome-sequencing project (Wu et al. 2013). Cluster analysis of association expression in differentially expressed proteins with corresponding transcript levels data was analyzed by software of cluster 3.0 (Eisen et al. 1998), which was used to identify groups of similarly differentially expressed proteins and transcripts at different stages, and the results output through the software Java Treeview in graphic form. Pearson’s correlations and probabilities of proteins and transcripts were calculated in OriginPro 8.5.
RNA extraction and first-strand cDNA synthesis
In this study, the three time points were sampled depending on the status of pear development in 2013, S4-10 (20 DAFB); S6-4 (75 DAFB), and S8-14 (147 DAFB) for qRT-PCR analysis. Total genomic RNA was extracted from pear fruit according to the CTAB protocol (Gasic et al. 2004), and then total RNA was treated with DNase I to remove genomic DNA contamination (Invitrogen, CA, USA). Finally, approximately 2 µg of total RNA was used for first-strand cDNA synthesis using ReverTra Ace-aFirst Strand cDNA Synthesis Kit (TOYOBO, TOYOBO Biotech Co. Ltd, Japan) base on the manufacturer’s protocol.
Real-time PCR analysis
To validate the expression patterns revealed by transcript abundance measurement results, seven candidate genes were further analyzed by quantitative real-time PCR. Primer sequences for the quantitative real-time PCR assay for each candidate gene and Tubulin were designed by primer5 software and are listed in Supplementary Table 1. All reactions were performed using the LightCycler 480 SYBR GREEN I Master (Roche, USA) according to the manufacturer’s protocol. Reactions were performed in triplicate using 0.4 μl of each primer, 10 µl of LightCycler 480 SYBR GREEN I Master, and 1 µl of diluted cDNA, finally, nuclease-free water was added to a total volume of 20 µl. All reactions were run as duplicates in 96-well plates, and LightCycler 480 (Roche, USA) was used for quantitative real-time PCR analysis and amplification. Each cDNA was analyzed in triplicate, after which the average threshold cycle (Ct) was calculated per sample. The qRT-PCR conditions were as follows: pre-incubation at 95 °C for 10 min; followed by 40 cycles of amplification with 15 s of denaturation at 94 °C, and then 60 °C for 30 s, 72 °C for 30 s, finally, extension at 72 °C for 3 min, and reading the plate for fluorescence data collection at 60 °C. Amplification was followed by a melting curve analysis with continual fluorescence data acquisition during from 60 to 95 °C melt. Finally, the average threshold cycle (Ct) was calculated per sample, and the relative expression levels were calculated with the 2−ΔΔCt method descripted by Livak et al. (Livak and Schmittgen 2001).
Results
Protein identification and quantification
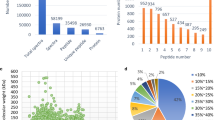
To avoid identification omissions, we confined the peptides matching error of database search strategy to less than 10 ppm. The distribution of errors between the true values and theoretical values of the relative molecular weight of all matching peptides are shown in Fig. 2a. Finally, a total of 56,314 spectra were identified from pear fruit development using iTRAQ technique. After data filtering to eliminate low-scoring spectra, 8,796 unique spectra were matched to 4,131 unique peptides, and a total of 1,810 proteins were identified from pear fruit (Fig. 2b; Supplementary Table 2). Additionally, the bias against lower molecular weight proteins may reflect the fact that smaller proteins, on average, have fewer diagnostic tryptic peptides than large proteins. In terms of protein mass distribution, most identified protein masses were distributed from 10 to 100 kDa with good average coverage, except groups more than 100 kDa (Fig. 2c). These results indicate that the proteomics analyses were reliable.
a Error distribution of spectra match quality of pear fruit. b Proteome identification in pear fruit. Total Spectra represents the total number of secondary spectra, Spectra represents the spectra number that were matched, Unique Spectra represents unique peptides that were matched. Peptide represents the number of identified peptides. Unique Peptide represents the number of unique peptides identified. Protein represents the final number of identified proteins. c Protein relative molecular mass in pear fruit. X-axis is protein molecular weight (Unit: kDa), Y-axis is the percentage of the range of protein mass
Functional classification and annotation
GO analysis provides a dynamically updating controlled vocabulary set to describe genes and gene product attributes in an organism. We conducted GO functional annotation analysis for all identified proteins. The results cover a wide range of molecular functions, biological processes and cellular components, including 38 important functional groups (Fig. 3; Supplementary Table 3). All identified proteins were annotated with categories simultaneously. For biological process category, there were 899 and 1,065 proteins involved in ‘cellular process’ and ‘biological processes’, respectively. In the cellular component category, the proteins associated with ‘cell part’ and ‘cell’ were all 1,329. ‘Binding’ and ‘Catalytic activity’ were the largest categories in molecular function, including 921 and 1,014 protein, respectively. In our study, 1,065 proteins were involved in metabolic processes.
GO categories assigned to pear proteins. Y-axis (left) represents percentages of proteins identified, and Y-axis (right) represent the protein number. The proteins were categorized according to the annotation of GO, and the number of each category is displayed based on biological process, cellular components, and molecular functions
Cluster of Orthologous Groups of proteins is the database for protein orthologous classification. Through comparing identified proteins with COG protein database to predict function of proteins, the results showed that all annotated proteins were classified into 23 clusters (Fig. 4). The highest frequency of occurrence of the functional classification are ‘general function prediction only’, followed by ‘post-translational modification’ and ‘protein turnover, chaperones’. In addition, many identified proteins are involved in ‘energy production and conversion', ‘amino acid transport and metabolism’, ‘carbohydrate transport and metabolism, translation’, ‘ribosomal structure and biogenesis’, indicating these functional classifications were more active than others during pear fruit development.
Functional classification of proteins by COG. COG: Cluster of Orthologous Groups of proteins. a RNA processing and modification. b Chromatin structure and dynamics; c Energy production and conversion. d Cell cycle control, cell division, chromosome. e Amino acid transport and metabolism. f Nucleotide transport and metabolism. g Carbohydrate transport and metabolism. h Coenzyme transport and metabolism. i Lipid transport and metabolism. j Translation, ribosomal structure and biogenesis. k Transcription. l Replication, recombination and repair. m Cell wall/membrane/envelope biogenesis. o Post-translational modification, protein turnover, chaperones. p Inorganic ion transport and metabolism. q Secondary metabolites biosynthesis, transport and catabolism. r General function prediction only. s Function unknown. t Signal transduction mechanisms. u Intracellular trafficking, secretion, and vesicular transport. v Defense mechanisms y Nuclear structure. z Cytoskeleton
In organisms, different proteins coordinate with each other to perform biological functions. According to KEGG pathway database analysis, the main biochemical metabolism and signal transduction pathways that proteins take part in has been described. In the present study, the results indicated that all annotated proteins were mapped onto 244 KEGG pathways. The top 10 % of pathways are shown in Supplementary Table 4. The metabolic pathways were annotated to include the most proteins, followed by biosynthesis of secondary metabolites and microbial metabolism in diverse environments. According to the pathway analysis, we can conclude that more proteins take part in the metabolic pathways, which might affect many aspects of fruit development and quality conformation. As we known, phenylpropanoid biosynthesis is a key component of lignin biosynthesis and one of the most important components for fruit quality. Finally, a total of 47 related proteins participate in phenylpropanoid biosynthesis were identified in this study.
Association and differential expression analysis of proteome and transcriptome data
To obtain an overall concordance of transcript and protein levels during pear fruit development, proteomic data produced in this research and transcriptomic data produced in previous genome research in pear were compared. Approximately, 27,000 transcripts were detected from pear fruit development. The correlations of gene expression at the transcript and protein levels for the three stages were 1,724, 1,722, and 1,718, respectively (Supplementary Fig. 1).
Meanwhile, to confirm differentially expressed proteins between any two stages, in the present study, only proteins that were identified with more than 1.5-fold changes between any two stages were considered as differentially expressed proteins. From this analysis, 115, 153, and 207 differentially expressed proteins were successfully identified as up-regulated, and 122, 165, and 218 differentially expressed proteins were identified as down-regulated during S4-22 vs S6-27, S6-27 vs S8-30, and S4-22 vs S8-30, respectively (Fig. 5a). The results indicated that more differentially expressed proteins were observed during ripening than early stage of fruit development. In addition, to show the differentially expressed proteins, which were up-regulated or down-regulated over the whole course of fruit development, Venn diagrams of identified differentially expressed proteins in any two stages were shown as Fig. 5b, c. Finally, we found that 12 differentially expressed proteins were up-regulated (Fig. 5b) and 18 differentially expressed proteins were down-regulated (Fig. 5c) during pear fruit development.
a The number of differentially expressed proteins in any two different stages. b Venn diagrams representing the overlap of identified differentially expressed proteins, which were up-regulated in any two stages. c Venn diagrams representing the overlap of identified differentially expressed proteins, which were down-regulated in any two stages
To investigate the overall correlation of differential expression and these transcripts, we matched all mRNAs identified from three stages with differentially expressed proteins, and then transformed the volume ratios of all differentially expressed proteins and these transcript ratios between any two stages into log2 forms. Finaly, a good correlation between the protein and mRNA levels at each corresponding developmental stage was observed. The correlation coefficients of the overall transcriptome and proteome data were 0.6336 (S4-22 vs S6-27) and 0.7049 (S4-22 vs S8-30), respectively. However, comparatively low correlation (0.4113) was found in S6-27 vs S8-30 (Fig. 6). In addition, hierarchical cluster analyses were conducted in this study. Figure 7 shows the expression profiles of all differentially expressed proteins and these transcripts during pear fruit development. We found that most proteins differentially expressed during pear fruit development showed similar trends with their corresponding transcripts (Fig. 7). However, although generally good correlation between protein and transcript levels was found, there were also some proteins with a trend opposite to transcript expression trends. A total of 65, 107, and 102 opposite differentially expressed proteins and mRNA were found at stage comparison of S4-22 vs S6-27, S6-27 vs S8-30, and S4-22 vs S8-30, respectively (Supplementary Table 5).
Concordance between changes in the abundance of transcripts and their encoded proteins at stages. The linear association R between log2 form changes on mRNA levels and protein levels of the all differentially expressed proteins during S4-22 vs S6-27 (a), S6-27 vs S8-30 (b) and S4-22 vs S8-30 (c) is 0.6336, 0.4113 and 0.7049, respectively, R is the Pearson correlation coefficient
Clustering of association expression in differentially expressed proteins with corresponding transcripts. S4-22 vs S6-27, S6-27 vs S8-30 and S4-22 vs S8-30 indicate the abundance of mRNA and its encoded differentially expressed protein, respectively. Protein (left) indicates the expression levels of differentially expressed proteins, Transcript (left) indicates the corresponding expression profile for the genes encoding differentially expressed proteins
In addition, the clustering of the intersection of differentially expressed proteins in different periods was analyzed (Fig. 8). Hierarchical cluster analysis showed that most of the intersections of differentially expressed proteins had similar trends. As shown in Fig. 8a, the differentially expressed proteins were down-regulated during pear fruit development, but proteins in Fig. 8b were in opposition, they were up-regulated during pear fruit development. According to the KEGG pathway database annotation, we discovered that these proteins were in functional categories involved in “polygalacturonase”, “alpha-glycosidase”, “UDP-apiose/xylose syntheses”, “large subunit ribosomal protein L36e”, and “large subunit ribosomal protein L19e” (Supplementary Table 6).
GO enrichment analysis for differentially expressed proteins
In the present study, GO enrichment analysis was carried out for differentially expressed proteins to determine the affected biological processes during pear fruit development. A series of biological processes affecting fruit ripening were identified (with p-value less than 0.05, Supplementary Table 7). The result showed that several key cellular components were identified, such as “chloroplast thylakoid”, “chloroplast”, and “chloroplast part”, showing functional changes during pear fruit development from S6-27 to S8-30, suggesting that these components were more active during fruit ripening. On the contrary, some GO terms were found active only during pear fruit development from S4-22 to S6-27, such as proteins related to sugar, including ‘glucan metabolic process’, ‘polysaccharide metabolic process’, ‘cellular glucan metabolic process’; and some proteins related to pigmentation, including ‘pigmentation’, ‘pigment accumulation’, ‘pigment accumulation in response to UV light’, ‘pigment accumulation in tissues in response to UV light’, and ‘pigment accumulation in tissues’, which suggested that those biological processes play a more important role at early stage of fruit development than near ripening stage.
Pathway enrichment analysis for differentially expressed proteins
In this research, pathway enrichment analysis was used to determine the key differentially expressed proteins involved in biochemical metabolic pathways and signal transduction pathways during pear fruit development. A series enrichment of KEGG pathways was identified during pear fruit development, with p value less than 0.05 as threshold. We found that the “Glycosaminoglycan biosynthesis-heparan sulfate”, “Phenylalanine, tyrosine and tryptophan biosynthesis”, “Tight junction”, “Starch and sucrose metabolism”, “Polyketide sugar unit biosynthesis”, and “Glycosaminoglycan biosynthesis-chondroitin sulfate” pathways were only detected during early stages of fruit development, and the enrichment of “Ribosome”, “Cell cycle”, “Regulation of autophagy”, and “Ether lipid metabolism” was only detected during near ripening (Supplementary Table 8). An important pathway, “Ribosome” (KO03010) can only be found during the stage from S6-27 to S8-30. A total of 26 differentially expressed proteins were annotated as ribosome proteins, but only one protein was up-regulated, the others were all down-regulated (Supplementary Fig. 2).
Differentially expressed proteins and transcripts related to fruit quality
According to above analyses, extensive comparison of proteins and mRNA expression changes during pear fruit development in any two stages were carried out. A master table was generated to summarize the physiological and biochemical functions of the changes in major proteins and their transcripts during pear fruit development. However, we are focusing on the processes related to fruit quality, such as sugar and organic acid metabolism, as well as stone cell, chlorophyll, cyanidin, and volatile compound synthetic metabolism pathways. Finally, 35 proteins and their transcripts related to important fruit quality displaying distinctive differential expressions during pear fruit development were detected (Table 1), in addition, we also list all differentially expressed proteins in Supplementary Table 2, in which red indicates up-regulated, and green down-regulated. In this study, three proteins involved in sugar metabolism were identified as differentially expressed during pear fruit development. Proteins belonging to major facilitator super family transporter (MFS, Pbr015095.1, Pbr032130.1) were up-regulated during pear fruit development, while sucrose synthase 1 (Pbr037395.1) was down-regulated during the early stage. For Chlorophyll metabolism, only one Ferritin-like protein (Pbr019418.1) was detected to be up-regulated during the whole pear fruit development. For lignin synthetic metabolism, sixteen differentially expressed proteins were detected. In addition, differentially expressed proteins related to aroma and cyanidin compound synthesis were also detected, as shown in Table 1, including seven differentially expressed proteins related to aroma compound synthesis and eight differentially expressed proteins related to cyanidin compound synthesis.
Verification of differentially expressed genes during pear fruit development and ripening by qRT-PCR
In the present study, to verify the genes and their associated proteins that were actually differentially expressed during pear fruit development, the expression levels of seven randomly selected genes had gene-specific primers designed (Supplementary Table 1), and were analyzed by qRT-PCR (Fig. 9). The results showed that although exact fold changes of seven randomly selected genes between qRT-PCR analysis and RNA-seq varied, the pattern of change in gene expression was consistent with each other. In addition, the qRT-PCR further demonstrated that genes related to stone cells (Pbr000689.1, Pbr022402.1, Pbr035186.1, Pbr040489.1) and aroma (Pbr015376.1, Pbr020361.1 and Pbr039379.1) showed more than twofold changes between any two stages during pear fruit development and ripening.
Discussion
Identification of proteins during pear fruit development
Fruit development and ripening is a highly coordinated and irreversible biological process, including a series of physiological and biochemical changes, finally forming an edible ripe fruit (Seymour et al. 1993), and proteins have their own special activity patterns that directly influence biological processes. In previous studies, proteomics has represented a robust approach to establish functional correlations between phenotype, genotype, and biochemical networks (Palma et al. 2011). iTRAQ-labeling technology is a powerful method that has been successfully used to quantify and characterize changes in protein levels, such as in grape (Lücker et al. 2009; Marsh et al. 2010; Martínez-Esteso et al. 2013) tomato (Pan et al. 2014), mango (Wu et al. 2014a), and other species (Pan et al. 2012; Bianco et al. 2009; Wu et al. 2014b). These studies described tissue-dependent proteome repertoires that show distinctive changes during fruit development. In pear cultivars, although proteomics (Pedreschi et al. 2008; Feng et al. 2011) and transcriptomics have been reported (Xie et al. 2013), there have not been reports about proteomics associated with transcriptome analysis related to pear quality. In the present study, we are the first to use iTRAQ-labeling technology to establish broad proteomics, and the associated transcriptomics, data to identify differentially expressed proteins and transcripts during pear fruit development. The correlation coefficient of the overall differentially expressed proteins and these transcripts indicated a positive correlation between stages from S4-22 to S6-27 and from S4-22 to S8-30 and a poor correlation from S6-27 to S8-30. These discrepancies indicate that some post-translation and post-transcription events might occur in the related protein and RNA expression level processes during pear fruit development (Washburn et al. 2003).
Protein concentrations for the samples S4-22, S6-27, and S8-30 were found to be 2.62, 3.11, and 4.36 μg/μl, respectively (Table 2). However, the same quantity of protein (100 μg) for each sample was used for iTRAQ analysis, in order to not affect the identification of numbers and expression level of proteins at different stages. In the present study, a total of 1,810 proteins were identified during pear fruit development, similar results have also been found in mango, with 1,996 proteins identified during mango fruit development and mapped onto 119 KEGG pathways, with the highest protein representation in ‘metabolic pathways’, followed by ‘biosynthesis of secondary metabolites’ (Wu et al. 2014a). In the present study, 35 proteins and corresponding transcripts were identified as differentially expressed, related to pear quality during fruit development. These important proteins identified can provide an extensive resource for future research on pear quality.
Differentially expressed proteins and transcripts related to sugar transport
The content and composition of soluble sugars are important indicators for pear fruit quality. A previous study reported that the important sugar components in pear are fructose, followed by sucrose and glucose (Chen et al. 2007). In this research, two monosaccharide transporters (MST, Pbr015095.1, Pbr032130.1, Table 1), which play an important role for accumulation of monosaccharides and transport simple sugars, oligosaccharides, and inositols, were identified (Pao et al. 1998). One of the transporters (Pbr032130.1) was up-regulated during all of pear fruit development, while the other (Pbr015095.1) was up-regulated from stage S6-27 to S8-30. The expression level of the two transporters is correlated with the monosaccharide sugar content, which remained low at early stages, and increased sharply from middle stage to near ripening of fruit, indicating that they play an important role for sugar accumulation during pear fruit development and ripening. Since the first monosaccharide transporter cloned from Chlorella (Sauer and Tanner 1989), until now, there are many reports indicating that monosaccharide transporters exist in many plants and play an important role during development and growth (Conde et al. 2007; Cao et al. 2011) Therefore, the two monosaccharide transporters identified in pear deserve further investigation as potentially significant regulators for sugar content.
Differentially expressed proteins and transcripts related to aroma compound synthesis
Aroma is an important trait that can affect the quality of pear fruit; the major components of aroma are derived from metabolism of carbohydrates, amino acids and fatty acids (Schwab et al. 2008). Some major volatile components in fruit are esters, alcohols, hydrocarbons, aldehydes, and ketones (Suwanagul and Richardson 1997). The function of Alcohol dehydrogenases (ADH) is to produce alcohols, precursors of esters and alcohols (Lara et al. 2003); esters are aroma compounds in pear fruit (Chervin et al. 1999). In the present study, we found that the activity of ADH (Pbr039379.1, Table 1) was up-regulated from stage S4-22 to S6-27 and showed no differential expression from S6-27 to S8-30. Meanwhile, expression levels of ADH are up-regulated along with alcohol production, suggesting that the metabolism of alphalinolenic acid is likely to be important for volatile formation during pear fruit development (Wu et al. 2013). In addition, acetaldehyde, which accumulates during fruit development and ripening (Pesis 2005), is converted from pyruvate. The reaction is catalyzed by Pyruvate Decarboxylase (PDC) enzymes. In this study, the two PDC (Pbr016162.1, Pbr019445.1) genes were detected up-regulated from S627 to S830 (Table 1). However, the gene expression patterns were opposite with their protein expression, possibly due to post-translation and post-transcription events (Washburn et al. 2003), which lead to low aroma traits in ‘Dangshansuli’ fruit.
The release of volatiles is another important aspect for perception of flavor and smell. The effect of beta-glucosidase on the enhancement of volatiles in apple fruit (Morales and Duque 2002) and orange juice (Fan et al. 2011) has been reported, but there are few studies reporting the effect of beta-glucosidase on bound volatile compounds in pear fruit. Beta-glucosidase is an enzyme that hydrolyzes the glucose dimer cellobiose to glucose. In the present study, three beta-glucosidases (Pbr042200.2, Pbr028992.1, Pbr020361.1) were identified as differentially expressed proteins, among them, Pbr042200.2 and Pbr020361.1 expression levels were down-regulated during pear fruit ripening (Table 1). This result indicates that low aroma perceived by sensory evaluation at near ripening of the ‘Dangshansuli’ pear cultivar, in difference with European pear, may be due to presence of bound aroma volatiles that are not released (Wu et al. 2013).
Differentially expressed proteins and transcripts related to stone cell metabolism
Stone cells are a crucial and special parameter for the internal quality of pear fruit, and are positively correlated with flesh quality (Choi et al. 2007). Lignin, as a primary component, plays an important role in stone cell formation (Tao et al. 2009), finally influencing pear fruit quality. Over the past few years, the understanding of lignin biosynthesis has made significant progress in both herbaceous and tree crops (Whetten et al. 1998; Boerjan et al. 2003) and enzymes involved in the formation of lignin have been reported (Anterola and Lewis 2002, Baucher et al. 2003). The stone cell formation process (“Phenylpropanoid biosynthesis”; KO00940) is a secondary cell wall thickening process. Lignin, which is a polymer of phenolic compounds, is the main component of the cell wall secondary structure, and is synthesized by the “Phenylpropanoid biosynthesis” pathway. In this study, according to the result of pathway enrichment analysis, “Phenylpropanoid biosynthesis” can be identified as a significantly enriched pathway during early to middle development stages. In addition, at the proteomic and transcriptomic levels, several key enzymes related to “Phenylpropanoid biosynthesis” were identified during pear fruit development (Fig. 10), including PAL (Pbr008387.1), F5H, (Pbr022142.1), 4CL (Pbr030350.1, Pbr012851.1, Pbr001283.1), CCR (Pbr011590.1, Pbr022402.1), CAD (Pbr026289.1), POD (Pbr035186.1, Pbr000691.1, Pbr000689.1, Pbr007903.1, Pbr040489.1), and HCT (Pbr010872.1,). Among these, PAL is an important enzyme in the first step of phenylpropanoid biosynthesis pathway catalyzing the deamination of L-phenylalanine to cinnamic acid (Boerjan et al. 2003). In this research, we identified two PALs (Pbr008387.1; Pbr008363.1). They decrease from S422 to S830 and showed similar trends with lignin content in pear during fruit development. In addition, one F5H (Pbr002142.1), three 4CLs (Pbr030350.1; Pbr012851.1; Pbr001283.1), two 4CL (Pbr012851.1; Pbr001283.1), one HCT (Pbr010872.1), one CCR (Pbr022402.1), and two CAD (Pbr026289.1; Pbr026287.1) were also identified as differentially expressed during pear fruit development. The expression levels were correlated with lignin content (Cai et al. 2010), which indicates the strengthened process of lignin synthesis. PODs, which catalyze the polymerization of monolignol to complete the process of lignification, have been shown to play an important role in the biosynthesis of lignin (Lee et al. 2006). According to the protein and transcript data analysis, we found that three (Pbr035186.1, Pbr000689.1, Pbr040489.1) out of five POD activities showed a similar trend with lignin content (Table 1), indicating that these three POD enzymes might play an important role for stone cell formation during pear development. Similar results were also reported in poplar xylem by Christensen et al. (Christensen et al. 1998), who indicated that POD enzymes were involved in the polymerization of lignin and promoted the development of stone cells. In addition, more structural genes of enzymes were identified in pear fruit (data not shown), which might be involved in different biological processes independently or together, explaining why only some genes and proteins can be identified associated with each other and showing consistent changes with lignin content during pear fruit development.
Regulatory changes in the pathway of Phenylpropanoid biosynthesis. Colors correspond with pear proteins detected with iTRAQ. Red boxes indicate significantly up-regulated proteins in S6-27 compared with S4-22; Green boxes indicate proteins significantly down-regulated in S6-27 compared with S4-22. Phenylalanineammonia-lyase (PAL, EC: 4.3.1.24), Ferulate-5-hydroxylase (F5H), 4-coumarate-CoA ligase (4CL, EC: 6.2.1.12), Cinnamoyl-CoA reductase (CCR, EC: 1.2.1.44), Cinnamyl-alcoholdehydrogenase (CAD, EC:1.1.1.195), Peroxidase (POD, EC:1.11.1.7), and Hydroxycinnamoyl transferase (HCT, EC:23.1.133)
Conclusion
Comparative and association analyses of the proteome and transcriptome for pear fruit development were conducted for the first time in this study. A total of 1,810 proteins were identified and 237, 318, and 415 differentially expressed proteins were screened during S4-22 vs S6-27, S6-27 vs S8-30, and S4-22 vs S8-30 stages. Finally, 35 differentially expressed proteins and their transcripts related to sugar metabolism, lignin metabolism, aroma conformation, and other quality traits were identified. The stone cell-related “Phenylpropanoid biosynthesis” pathway is enriched, and 16 proteins can be detected as differentially expressed in this pathway, suggesting stone cells are actively generated during pear fruit development, especially at the early stages. In addition, other key genes involved in fruit quality were also identified. Overall, the study provides extensive new information on the pear proteome and transcriptome. Compared to separate proteome and transcriptome analysis, it provides more confident results and helps in selecting key candidate genes for fruit development and quality, with both gene and protein expression evidence. This resource can help us to examine mechanisms for protein and transcript level diversification that contributes to understanding the complexity of fruit quality. Information obtained from the ascertained amino acid sequences will be helpful in future molecular cloning and predicting the function of these proteins or genes in other plants, although much more work is still needed to elucidate them.
Author contribution
Jia Ming Li carried out the experimental design, data analysis and drafted the manuscript. Xiao San Huang and Dan Man Zheng contributed to language revision, Lei Ting Li participated in the data analysis and preparation of figures, Cheng Xue contributed to the qRT-PCR experiments analysis, Shaoling Zhang contributed with consultation, Jun Wu managed and designed the research and experiments.
Abbreviations
- DAFB:
-
Day after full blooming,
- iTRAQ:
-
Isobaric tags for relative and absolute quantitation
- SCX:
-
Strong cation exchange
- HPLC:
-
High pressure liquid chromatography
- AGC:
-
Automatic gain control
- HCD:
-
High-energy collision dissociation
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- GO:
-
Gene ontology
- COG:
-
Cluster of orthologous groups of proteins
- PAL:
-
Phenylalanineammonia-lyase
- F5H:
-
Ferulate-5-hydroxylase
- 4CL:
-
4-coumarate-CoA ligase
- CCR:
-
Cinnamoyl-CoA reductase
- CAD:
-
Cinnamyl-alcoholdehydrogenase
- POD:
-
Peroxidase
- HCT:
-
Hydroxycinnamoyl transferase
- MFS:
-
Major facilitator super family transporter
References
Alba R, Payton P, Fei Z, McQuinn R, Debbie P, Martin GB, Tanksley SD, Giovannoni JJ (2005) Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. Plant Cell 17(11):2954–2965
Anterola AM, Lewis NG (2002) Trends in lignin modification: a comprehensive analysis of the effects of genetic manipulations/mutations on lignification and vascular integrity. Phytochemistry 61(3):221–294
Argenta LC, Fan X, Mattheis JP (2003) Influence of 1-methylcyclopropene on ripening, storage life, and volatile production by d’Anjou cv. pear fruit. J Agric Food Chem 51(13):3858–3864
Bain JM (1961) Some morphological, anatomical, and physiological changes in the pear fruit (Pyrus communis var. Williams Bon Chretien) during development and following harvest. Aust J Bot 9(2):99–123
Baucher M, Halpin C, Petit-Conil M, Boerjan W (2003) Lignin: genetic engineering and impact on pulping. Crit Rev Biochem Mol Biol 38(4):305–350
Bianco L, Lopez L, Scalone AG, Di Carli M, Desiderio A, Benvenuto E, Perrotta G (2009) Strawberry proteome characterization and its regulation during fruit ripening and in different genotypes. J Proteomics 72(4):586–607
Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54(1):519–546
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72(1):248–254
Cai Y, Li G, Nie J, Lin Y, Nie F, Zhang J, Xu Y (2010) Study of the structure and biosynthetic pathway of lignin in stone cells of pear. Sci Hortic 125(3):374–379
Cao H, Guo S, Xu Y, Jiang K, Jones AM, Chong K (2011) Reduced expression of a gene encoding a Golgi localized monosaccharide transporter (OsGMST1) confers hypersensitivity to salt in rice (Oryza sativa). J Exp Bot 62(13):4595–4604
Chen JL, Wu JH, Wang Q, Deng H, Hu XS (2006) Changes in the volatile compounds and chemical and physical properties of Kuerle fragrant pear (Pyrus serotina Reld) during storage. J Agric Food Chem 54(23):8842–8847
Chen J, Wang Z, Wu J, Wang Q, Hu X (2007) Chemical compositional characterization of eight pear cultivars grown in China. Food Chem 104(1):268–275
Chervin C, Truett JK, Speirs J (1999) Alcohol dehydrogenase expression and alcohol production during pear ripening. J Am Soc Hortic Sci 124(1):71–75
Choi J-H, Choi J-J, Hong K-H, Kim W-S, Lee S-H (2007) Cultivar differences of stone cells in pear flesh and their effects on fruit quality. Hortic Environ Biotechnol 48(1):27–31
Christensen JH, Bauw G, Welinder KG, Van Montagu M, Boerjan W (1998) Purification and characterization of peroxidases correlated with lignification in poplar xylem. Plant Physiol 118(1):125–135
Conde C, Agasse A, Silva P, Lemoine R, Delrot S, Tavares R, Gerós H (2007) OeMST2 encodes a monosaccharide transporter expressed throughout olive fruit maturation. Plant Cell Physiol 48(9):1299–1308
Dussi MC, Sugar D, Wrolstad RE (1995) Characterizing and quantifying anthocyanins in red pears and the effect of light quality on fruit color. J Am Soc Hortic Sci 120(5):785–789
Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci 95(25):14863–14868
Fan G, Xu Y, Zhang X, Lei S, Yang S, Pan S (2011) Characteristics of immobilised β-glucosidase and its effect on bound volatile compounds in orange juice. Int J Food Sci Technol 46(11):2312–2320
Faurobert M, Mihr C, Bertin N, Pawlowski T, Negroni L, Sommerer N, Causse M (2007) Major proteome variations associated with cherry tomato pericarp development and ripening. Plant Physiol 143(3):1327–1346
Feng S, Chen X, Zhang Y, Wang Y, Song Y, Chen X-L, Li X, Li M, Liu J, Wang Q (2011) Differential expression of proteins in red pear following fruit bagging treatment. Protein J 30(3):194–200
Feng C, Chen M, Xu C-J, Bai L, Yin X-R, Li X, Allan AC, Ferguson IB, Chen K-S (2012) Transcriptomic analysis of Chinese bayberry (Myrica rubra) fruit development and ripening using RNA-Seq. BMC Genom 13(1):19
Fernie AR, Stitt M (2012) On the discordance of metabolomics with proteomics and transcriptomics: coping with increasing complexity in logic, chemistry, and network interactions scientific correspondence. Plant Physiol 158(3):1139–1145
Gasic K, Hernandez A, Korban SS (2004) RNA extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Mol Biol Rep 22(4):437–438
Giribaldi M, Perugini I, Sauvage FX, Schubert A (2007) Analysis of protein changes during grape berry ripening by 2-DE and MALDI-TOF. Proteomics 7(17):3154–3170
Hudina M, Śtampar F (2000) Sugars and organic acids contents of European (Pyrus communis L.) and Asian (Pyrus serotina Rehd.) pear cultivars. Acta Alimentaria 29(3):217–230
Itai A, Kawata T, Tanabe K, Tamura F, Uchiyama M, Tomomitsu M, Shiraiwa N (1999) Identification of 1-aminocyclopropane-1-carboxylic acid synthase genes controlling the ethylene level of ripening fruit in Japanese pear (Pyrus pyrifolia Nakai). Mol Gen Genet 261(1):42–49
Jaillon O, Aury J-M, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449(7161):463–467
Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28(1):27–30
Lara I, Miró R, Fuentes T, Sayez G, Graell J, López M (2003) Biosynthesis of volatile aroma compounds in pear fruit stored under long-term controlled-atmosphere conditions. Postharvest Biol Technol 29(1):29–39
Lee S-H, Choi J-H, Kim W-S, Han T-H, Park Y-S, Gemma H (2006) Effect of soil water stress on the development of stone cells in pear (Pyrus pyrifolia cv. ‘Niitaka’) flesh. Sci Hortic 110(3):247–253
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative pcr and the 2−ΔΔCT method. Methods 25(4):402–408
Lombard PB, Westwood MN (1987) Pear rootstocks. In: Rom RC, Carlson RF (eds) Rootstocks for fruit crops. Wiley, New York, pp 145–183
Lu X-P, Liu Y-Z, An J-C, Hu H-J, Peng S-A (2011) Isolation of a cinnamoyl CoA reductase gene involved in formation of stone cells in pear (Pyrus pyrifolia). Acta Physiologiae Plantarum 33(2):585–591
Lücker J, Laszczak M, Smith D, Lund ST (2009) Generation of a predicted protein database from EST data and application to iTRAQ analyses in grape (Vitis vinifera cv. Cabernet Sauvignon) berries at ripening initiation. BMC Genom 10(1):50
Maier T, Schmidt A, Güell M, Kühner S, Gavin AC, Aebersold R, Serrano L (2011) Quantification of mRNA and protein and integration with protein turnover in a bacterium. Mol Syst Biol 7(1):511
Marsh E, Alvarez S, Hicks LM, Barbazuk WB, Qiu W, Kovacs L, Schachtman D (2010) Changes in protein abundance during powdery mildew infection of leaf tissues of Cabernet Sauvignon grapevine (Vitis vinifera L.). Proteomics 10(10):2057–2064
Martínez-Esteso MJ, Vilella-Antón MT, Pedreño MÁ, Valero ML, Bru-Martínez R (2013) iTRAQ-based protein profiling provides insights into the central metabolism changes driving grape berry development and ripening. BMC Plant Biol 13(1):167
Ming R, Hou S, Feng Y, Yu Q, Dionne-Laporte A, Saw JH, Senin P, Wang W, Ly BV, Lewis KL (2008) The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452(7190):991–996
Morales AL, Duque C (2002) Free and glycosidically bound volatiles in the mammee apple (Mammea americana) fruit. Eur Food Res Technol 215(3):221–226
Mounet F, Moing A, Garcia V, Petit J, Maucourt M, Deborde C, Bernillon S, Le Gall G, Colquhoun I, Defernez M (2009) Gene and metabolite regulatory network analysis of early developing fruit tissues highlights new candidate genes for the control of tomato fruit composition and development. Plant Physiol 149(3):1505–1528
Nogueira SB, Labate CA, Gozzo FC, Pilau EJ, Lajolo FM, Oliveira do Nascimento JR (2012) Proteomic analysis of papaya fruit ripening using 2DE-DIGE. Journal of proteomics 75(4):1428–1439
Osorio S, Alba R, Damasceno CM, Lopez-Casado G, Lohse M, Zanor MI, Tohge T, Usadel B, Rose JK, Fei Z (2011) Systems biology of tomato fruit development: combined transcript, protein, and metabolite analysis of tomato transcription factor (nor, rin) and ethylene receptor (Nr) mutants reveals novel regulatory interactions. Plant Physiol 157(1):405–425
Palma JM, Corpas FJ, del Río LA (2011) Proteomics as an approach to the understanding of the molecular physiology of fruit development and ripening. J Proteomics 74(8):1230–1243
Pan Z, Zeng Y, An J, Ye J, Xu Q, Deng X (2012) An integrative analysis of transcriptome and proteome provides new insights into carotenoid biosynthesis and regulation in sweet orange fruits. J Proteomics 75(9):2670–2684
Pan X, Zhu B, Zhu H, Chen Y, Tian H, Luo Y, Fu D (2014) iTRAQ protein profile analysis of tomato green-ripe mutant reveals new aspects critical for fruit ripening. J Proteome Res 13(4):1979–1993
Pao SS, Paulsen IT, Saier MH (1998) Major facilitator superfamily. Microbiol Mol Biol Rev 62(1):1–34
Pedreschi R, Hertog M, Robben J, Noben J-P, Nicolaï B (2008) Physiological implications of controlled atmosphere storage of ‘Conference’pears (Pyrus communis L.): a proteomic approach. Postharvest Biol Technol 50(2):110–116
Pesis E (2005) The role of the anaerobic metabolites, acetaldehyde and ethanol, in fruit ripening, enhancement of fruit quality and fruit deterioration. Postharvest Biol Technol 37(1):1–19
Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics 3(12):1154–1169
Sauer N, Tanner W (1989) The hexose carrier from Chlorella: cDNA cloning of a eucaryotic H+-cotransporter. FEBS Lett 259(1):43–46
Schwab W, Davidovich-Rikanati R, Lewinsohn E (2008) Biosynthesis of plant-derived flavor compounds. Plant J 54(4):712–732
Seymour GB, Taylor JE, Tucker GA (1993) Biochemistry of fruit ripening. Chapman & Hall, London
Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, Delcher AL, Jaiswal P, Mockaitis K, Liston A, Mane SP (2011) The genome of woodland strawberry (Fragaria vesca). Nat Genet 43(2):109–116
Soglio V, Costa F, Molthoff J, Weemen-Hendriks W, Schouten H, Gianfranceschi L (2009) Transcription analysis of apple fruit development using cDNA microarrays. Tree Gene Geno 5(4):685–698
Suwanagul A, Richardson DG (1997) Identification of headspace volatile compounds from different pear (Pyrus communis L.) varieties. In: VII International Symposium on Pear Growing 475 605–624
Tao S, Khanizadeh S, Zhang H, Zhang S (2009) Anatomy, ultrastructure and lignin distribution of stone cells in two Pyrus species. Plant Sci 176(3):413–419
Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT, Rao BS, Kiryutin B, Galperin MY, Fedorova ND, Koonin EV (2001) The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res 29(1):22–28
Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D (2010) The genome of the domesticated apple (Malus domestica Borkh.). Nat Genet 42(10):833–839
Verde I, Abbott AG, Scalabrin S, Jung S, Shu S, Marroni F, Zhebentyayeva T, Dettori MT, Grimwood J, Cattonaro F (2013) The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat Genet 45(5):487–494
Washburn MP, Koller A, Oshiro G, Ulaszek RR, Plouffe D, Deciu C, Winzeler E, Yates JR (2003) Protein pathway and complex clustering of correlated mRNA and protein expression analyses in Saccharomyces cerevisiae. Proc Natl Acad Sci 100(6):3107–3112
Whetten RW, MacKay JJ, Sederoff RR (1998) Recent advances in understanding lignin biosynthesis. Annu Rev Plant Biol 49(1):585–609
Wilkins MR, Sanchez J-C, Gooley AA, Appel RD, Humphery-Smith I, Hochstrasser DF, Williams KL (1996) Progress with proteome projects: why all proteins expressed by a genome should be identified and how to do it. Biotechnol Genet Eng Rev 13(1):19–50
Wu J, Wang Z, Shi Z, Zhang S, Ming R, Zhu S, Khan MA, Tao S, Korban SS, Wang H, Chen NJ, Nishio T, Xu X, Cong L, Qi K, Huang X, Wang Y, Zhao X, Wu J, Deng C, Gou C, Zhou W, Yin H, Qin G, Sha Y, Tao Y, Chen H, Yang Y, Song Y, Zhan D, Wang J, Li L, Dai M, Gu C, Wang Y, Shi D, Wang X, Zhang H, Zeng L, Zheng D, Wang C, Chen M, Wang G, Xie L, Sovero V, Sha S, Huang W, Zhang S, Zhang M, Sun J, Xu L, Li Y, Liu X, Li Q, Shen J, Wang J, Paull RE, Bennetzen JL, Wang J, Zhang S (2013) The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res 23(2):396–408
Wu H-X, Jia H-M, Ma X-w, Wang S-B, Yao Q-S, Xu W-t, Zhou Y-G, Gao Z-S, Zhan R-L (2014a) Transcriptome and proteomic analysis of mango (Mangifera indica Linn) fruits. J Proteomics 105:19–30
Wu J, Xu Z, Zhang Y, Chai L, Yi H, Deng X (2014b) An integrative analysis of the transcriptome and proteome of the pulp of a spontaneous late-ripening sweet orange mutant and its wild type improves our understanding of fruit ripening in citrus. J Exp Bot 65(6):1651–1671
Xie M, Huang Y, Zhang Y, Wang X, Yang H, Yu O, Dai W, Fang C (2013) Transcriptome profiling of fruit development and maturation in Chinese white pear (Pyrus bretschneideri Rehd). BMC Genom 14(1):823
Xu Q, Chen L-L, Ruan X, Chen D, Zhu A, Chen C, Bertrand D, Jiao W-B, Hao B-H, Lyon MP (2013) The draft genome of sweet orange (Citrus sinensis). Nat Genet 45(1):59–66
Yamada K, Kojima T, Bantog N, Shimoda T, Mori H, Shiratake K, Yamaki S (2007) Cloning of two isoforms of soluble acid invertase of Japanese pear and their expression during fruit development. J Plant Physiol 164(6):746–755
Yang Y-N, Zhao G, Yue W-Q, Zhang S-L, Gu C, Wu J (2013) Molecular cloning and gene expression differences of the anthocyanin biosynthesis-related genes in the red/green skin color mutant of pear (Pyrus communis L.). Tree Genet Genomes 9(5):1351–1360
Yu K, Xu Q, Da X, Guo F, Ding Y, Deng X (2012) Transcriptome changes during fruit development and ripening of sweet orange (Citrus sinensis). BMC Genom 13(1):10
Zhou YS, Lamrani M, Chan-Park MB, Leong SSJ, Wook MC, Chen WN (2010) iTRAQ-coupled two-dimensional liquid chromatography/tandem mass spectrometric analysis of protein profile in Escherichia coli incubated with human neutrophil peptide 1-potential in antimicrobial strategy. Rapid Commun Mass Spectrom 24(18):2787–2790
Zhu M, Simons B, Zhu N, Oppenheimer DG, Chen S (2010) Analysis of abscisic acid responsive proteins in Brassica napus guard cells by multiplexed isobaric tagging. J Proteomics 73(4):790–805
Acknowledgments
The work was financially supported by the National Science Foundation of China (31171928), Ministry of Education Program for New Century Excellent Talents in University (NCET-13-0864), and the Earmarked Fund for China Agriculture Research System (CARS-29).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, J.M., Huang, X.S., Li, L.T. et al. Proteome analysis of pear reveals key genes associated with fruit development and quality. Planta 241, 1363–1379 (2015). https://doi.org/10.1007/s00425-015-2263-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-015-2263-y