Abstract
Background
Gene expression is an important process underpinning the acute and chronic adaptive response to resistance exercise (RE) training.
Purpose
To investigate the effect of training status on vastus lateralis muscle global transcriptome at rest and following acute RE.
Methods
Muscle biopsies of nine young men (age: 26(2) years; body mass: 69(9) kg; height 172(6) cm) who undertook RE training for 10 weeks were collected pre and 24 h post-RE in the untrained (W1) and trained (W10) states and analysed using microarray. Tests of differential expression were conducted for rested and after RE contrasts in both training states. To control for false discovery rate (FDR), multiple testing correction was performed at a cut-off of FDR < 0.05.
Results
Unaccustomed RE (at W1) upregulated muscle gene transcripts related to stress (e.g., heat shock proteins), damage and inflammation, structural remodelling, protein turnover and increased translational capacity. Trained muscles (at W10) showed changes in the transcriptome signature regarding the regulation of energy metabolism, favouring a more oxidative one, upregulated antioxidant- and immune-related genes/terms, and gene transcripts related to the cytoskeleton and extracellular matrix, muscle contraction, development and growth.
Conclusions
These results highlight that chronic repetition of RE changes muscle transcriptome response towards a more refined response to RE-induced stress.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The transcriptional changes that occur acutely and chronically with resistance exercise (RE) training have a substantial bearing on skeletal muscle adaptations (Gordon et al. 2012; Mackey et al. 2011; Mahoney et al. 2008). Specifically, muscle transcriptome affects some of the physiological processes, as increases in myofibrillar protein synthesis and muscle components remodelling, which may affect RE training-induced phenotypic adaptations (Drummond et al. 2009; Gordon et al. 2012; Lamas et al. 2010; Phillips et al. 2013; Thalacker-Mercer et al. 2013).
Recently, we evaluated RE-induced changes in muscle damage, repair, myofibrillar protein synthesis, and muscle hypertrophy throughout 10 weeks of RE training (Damas et al. 2016b). We demonstrated that the RE-induced increase in myofibrillar protein synthesis was attenuated as training progressed (Damas et al. 2016b). Further, we detected significant strong correlations between the protein synthetic response and muscle hypertrophy but only after muscle damage progressively subsided suggesting that the protein synthetic response can be directed towards muscle damage repair or muscle hypertrophy, depending on training status (Damas et al. 2016b). As the aforementioned intrinsic muscle processes respond differently according to training status (Damas et al. 2016b), assessing baseline and acute changes in global muscle transcriptional signature would provide information on clusters of genes responsible for muscle tissue adaptation when chronically exposed to RE loads. Global gene expression in response to 8–12 weeks RE training had been previously investigated (Gordon et al. 2012; Liu et al. 2010; Raue et al. 2012; Salanova et al. 2015). However, there is paucity of data regarding time points longer than 4 h into post-RE recovery that are important periods for muscle adaptation processes (e.g., repair due to damage, inflammation, remodelling). Additionally, to the best of our knowledge, no study has conducted a longitudinal investigation into global gene expression at rest and after acute RE in different training states (untrained and trained states) using a uniform population of young men only under normal living conditions [i.e., not using disuse models such as ‘bed rest’ applied in Salanova et al. (2015)]. A longitudinal study of this nature is important as sex and tissue heterogeneity are sources of muscle transcriptome variability both at rest and following an acute RE (Liu et al. 2010; Raue et al. 2012; Roth et al. 2002; Welle et al. 2008).
In the present study, we analysed the global transcriptome signature in different training states (i.e., untrained; and at the end of a 10-week lower limb RE training: trained) at a rested condition and a later time point into recovery, i.e., 24 h after RE completion in vastus lateralis muscle biopsies of young men.
Methods
Participants
Ten subjects performed all protocols; however, due to poor sample quality one subject was removed from the analysis. Thus, a total of nine healthy men, with at least 6 months without partaking on regular RE but with previous experience in RE training, were included in the present investigation ([mean (SD)] age: 26(2) years; body mass: 69(9) kg; height 172(6) cm). As inclusionary criteria, participants could not use supplements or anti-inflammatory drugs. All participants were instructed to refrain from exercising (such as RE and/or endurance exercise) outside of the study for 72 h before the beginning and throughout the study and to maintain their normal daily activities and nutritional habits. The present study was approved by the local Ethic Committee and all participants provided written, informed consent prior to participation. All procedures in the present study were conducted in accordance with the latest version of the Declaration of Helsinki.
Experimental design
The present study is a part of a larger study previously published (Damas et al. 2016b). In short, RE was performed twice-weekly through 10 weeks and involved lower limb exercises [six sets (three of 45° leg-press and three of leg extension exercises) of 9–12 maximum (to concentric failure) repetitions with 90 s rest intervals] (Damas et al. 2016b). Thus, at evaluation weeks (first and last weeks) the RE was performed at the same relative load. Immediately after every RE session, participants ingested 25 g of isolated whey-protein to maximally stimulate muscle protein synthesis response post-RE (Witard et al. 2014). This procedure was adopted to standardize (i.e., maximize) muscle protein synthesis response after each RE session. A standardized diet (22% protein, 41% carbohydrates and 37% fat) was provided to the subjects on the days muscle biopsies were obtained. Vastus lateralis percutaneous muscle biopsy were performed using Bergstrom needles and manual suction. Biopsies were obtained at rest (Pre) and 24 h-post (24 h) RE in the first (untrained state—W1) and 10th weeks (trained state—W10) of RE training. All biopsies were performed under 1% xylocaine local anaesthesia, and approximately 20–30 mg of muscle was collected, cleared from blood and visible connective tissue, immediately frozen in liquid nitrogen, and stored at − 80 °C. Pre-RE biopsies at W1 and W10 were obtained from the same leg, while 24 h post-RE biopsies were taken from the contralateral leg, to reduce variability between evaluation weeks. Legs were assigned to biopsy at a random fashion to dilute any effect of leg dominance.
RNA isolation, microarray procedure, data processing and statistical analyses
RNA was extracted from the muscle biopsy samples using the TRIzol/RNeasy (Qiagen Sciences, Valencia, CA, USA) method. Tissues were homogenized with 1 mL TRIzol (Life Technologies, Burlington, ON, Canada) using Lysing Matrix D tubes (MP Biomedicals, Solon, OH, USA), with FastPrep-24 Tissue and Cell Homogenizer (MP Biomedicals) for 40 s at 6 m/s. 200 µL of chloroform (Sigma–Aldrich) was added to the homogenate, the tubes vigorously agitated for 15 s and rested for 5 min at room temperature. Samples were then centrifuged at 12,000g, 4 °C for 10 min and the aqueous phase containing RNA was collected and transferred to a RNeasy minispin column. The RNA was purified according to the Total RNA Kit 1 (Omega Bio-Tek, Norcross, GA, USA) manufacturer instructions. Concentration and purity of RNA was quantified using the Nano-Drop 1000 spectrophotometer (Thermo Fisher Scientific, Rockville, MD, USA). Total RNA per muscle sample at Pre W1 vs Pre W10 was compared using a t test. P < 0.05 was set as significance level.
The microarray procedure was conducted by the Genetic and Molecular Epidemiology Laboratory (Hamilton, ON, Canada). Briefly, the amount of (normalized) RNA was determined by RiboGreen quantitation (Quant-iT RiboGreen RNA Assay Kit, Thermo Fisher Scientific, Rockville, MD, USA) before input into the Ambion Illumina Total Prep Kit (Thermo Fisher Scientific, Rockville, MD, USA) for amplification, cRNA synthesis and biotinylation. The cRNA generated was quantified and normalized by the RiboGreen fluorescence method before proceeding with the direct hybridization assay for whole genome expression (Human HT-12 v4 Expression BeadChip, Illumina, CA, USA). Samples were processed randomly to avoid systematic bias.
The Human HT-12 v4 Expression BeadChip arrays were scanned on an iScan System (Illumina, Inc., San Diego, CA, USA) with iScan Control Software 3.3.29 and saved as IDAT files, an encrypted Illumina proprietary format. The arrays raw intensity values and their corresponding manifest file were imported into R/Bioconductor (Huber et al. 2015) using the read.idat function from the limma package version 3.30.6 (Ritchie et al. 2015), which in turn uses the illuminaio package version 0.16.0 (Smith et al. 2013) to read the files. The Illumina BeadChip arrays used in this study include 770 negative probes. These negative probes were used to estimate the intensity distribution of background noise, and then background correct probe intensities with the normexp method. Following background correction, arrays were quantile normalized to ensure that the signal intensities have the same distribution across arrays. Finally, probes were filtered out if less than half of the arrays did not significantly differ in intensity from the background noise at a P < 0.05 cut-off, which increases the precision of the differential expression analysis (Shi et al. 2010a, b). All these steps were performed with the function neqc from the limma package.
Differential gene expression was assessed by adjusting linear models on normalized expression intensities using limma (Ritchie et al. 2015). To account for the repeated measurements on the same individuals, the correlation between same subject measurements was estimated with the function duplicateCorrelation, and this correlation was input into the linear model. Tests of differential expression were conducted for the following contrasts:
-
1.
Pre W10 − Pre W1;
-
2.
24 h − Pre at W1 and W10;
-
3.
[(24 h W10 – Pre W10) − (24 h W1 − Pre W1)].
To control for false discovery rate (FDR), multiple testing correction was performed (Benjamini and Hochberg 1995), at a cut-off of FDR < 0.05.
The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (Ogata et al. 1999) and Gene Ontology (GO) terms (Ashburner et al., 2000) were tested for enrichment using the CAMERA method (Wu and Smyth 2012) implemented in limma, which performs competitive tests that take into account dependence between genes in the linear model and uses a directional test to distinguish between up and downregulated gene groups. Enriched terms were defined as those having a FDR < 0.05. Significantly enriched KEGG pathways were visualized with the package pathview version 1.14.0 (Luo et al. 2009). Microarray data were submitted to GEO (http://www.ncbi.nlm.nih.gov/geo/) number GSE106865.
Results
Global gene expression and total RNA at rest in different training states
Figure 1 depicts a volcano plot and KEGG pathways for the comparison between different training states at rest (for full gene tables and GO terms please refer to Supplementary Material, Table S1). There were a number of enriched pathways when comparing trained vs untrained states at rest: (1) oxidative-related metabolic pathways; (2) immune and pathogenic responses; (3) antioxidant response; (4) extracellular matrix and cytoskeleton organization; and (5) protein metabolism and growth.
a Volcano plot of gene expression [highlighted dots represent genes with FDR < 0.05 (black horizontal dashed line) in the rested (Pre) trained (W10)—positive log2 fold change—and untrained (W1)—negative log2 fold change—states]; and b Kyoto Encyclopedia of Genes and Genomes (KEGG) enriched [false discovery rate (FDR) < 0.05, black vertical dashed line] pathways for the comparison between different training states at rest. For full gene tables and gene ontology (GO) terms please refer to Supplementary Material (Table S1)
It should also be noted that basal total RNA increased with training (P = 0.046, Fig. 2), i.e., after 10 weeks of RE training, subjects presented greater muscle total RNA compared to before training, possibly indicating heightened ribosomal density after RE training.
Changes in global gene expression Pre to 24 h after an acute RE in different training states
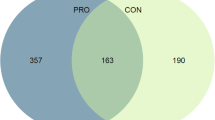
Figure 3 depicts volcano plots (Fig. 3a, b), a Venn diagram (Fig. 3c) and main enriched pathways (Fig. 3d) for the acute transcriptome response to RE in untrained and trained states separately. To perform a more thorough comparison on the impact of RE on muscle transcriptome in the untrained and trained states we contrasted the acute changes in muscle transcriptome following RE [i.e., (24 h W10 − Pre W10) − (24 h W1 − Pre W1)]. Figure 4 shows a volcano plot and the main enriched pathways regarding this comparison (for full gene tables and GO terms please refer to Supplementary Material, Table S1).
Volcano plots of gene expression [a untrained—W1; b trained—W10, highlighted dots represent genes that were up or downregulated (FDR < 0.05, black horizontal dashed line) with acute resistance exercise (RE)], c Venn diagram, and d Kyoto Encyclopedia of Genes and Genomes (KEGG) enriched [false discovery rate (FDR) < 0.05, black vertical dashed line] pathways post-RE at different training states analysed separately. For full gene tables and gene ontology (GO) terms please refer to Supplementary Material (Table S1)
a Volcano plot of gene expression [highlighted dots represent genes with FDR < 0.05 (black horizontal dashed line) for the delta in the trained (W10)—positive log2 fold change—and untrained (W1)—negative log2 fold change—states]; and b Kyoto Encyclopedia of Genes and Genomes (KEGG) enriched [false discovery rate (FDR) < 0.05, black vertical dashed line] pathways for the contrast between acute exercise deltas (i.e., 24 h post-resistance exercise − Pre) in different training states. For full gene tables and gene ontology (GO) terms please refer to Supplementary Material (Table S1)
The analyses showed that overall impact on muscle transcriptome is higher in the untrained compared with the trained state (Fig. 3a, b), with 1414 co-regulated and 5 contra-regulated genes (Fig. 3c). Results showed that RE downregulates gene transcripts related to the oxidative metabolism, but in a greater extent in the untrained vs the trained state. In addition, RE stimulates genes and terms related to ribosome biogenesis in both training states, however, to a greater extent in the untrained state. Conversely, RE performed in the trained state showed larger changes in the transcriptome signature regarding muscle structure, immune responses and protein metabolism compared to the untrained state (Fig. 3d). Specifically, RE in the untrained state promoted enriched genes/pathways in comparison to the trained state relating to: (1) glycolysis; (2) stress, damage, inflammatory response and structures remodelling; (3) protein breakdown; (4) protein processing and synthesis; and (5) cell cycle. In contrast, the RE in the trained state showed enriched pathways in comparison to the untrained state relating to: (1) oxidative metabolism; (2) muscle structural changes involving the extracellular matrix and cytoskeleton, and extra-intracellular communication; (3) muscle contraction; (4) increased antioxidant and immune response to stress; and (5) protein synthesis, muscle growth, and development.
Inter-subject muscle transcriptome dispersion with and without RE in distinct training states
We constructed a multidimensional scaling graph (Fig. 5) that takes into account the 500 genes with the greatest change in their expressions per subject and time point. The multidimensional scaling graph reduces and reorders the distances to provide insight into the relationships between the samples in a smaller number of dimensions. As depicted in Fig. 5, the analyses showed that in the rested condition (Pre W1 and Pre W10) the 500 most expressed genes have less inter-subject dispersion. With acute RE stimulation (24 h W1 and 24 h W10), the dispersion between subjects is considerably increased.
Discussion
Understanding how RE training modulates skeletal muscle global gene expression can increase our knowledge on the mechanisms regulating muscle adaptation (Zierath and Wallberg-Henriksson 2015). In the current study, we showed that the RE training status influenced both rested (Pre) and after acute-RE (24 h) changes in the muscle transcriptome signature regarding metabolic and contractile adaptations, antioxidant and immune responses, cytoskeleton and extracellular matrix organization, and cell growth/development associated with myofibrillar hypertrophy in young men.
Previous reports have demonstrated that the first RE session provokes a large disturbance in muscle homeostasis, resulting in muscle damage, inflammation, potent overall increase in protein synthesis and muscle remodelling (Damas et al. 2016b; Hyldahl et al. 2015; Mitchell et al. 2014; Peake et al. 2005). As such, we report robust changes in the global muscle transcriptome after the first RE (Fig. 3). In fact, the high stress imposed by the first RE upregulated gene expression of heat shock proteins (HSP) and inflammatory response after RE (Fig. 4). Consistently, previous investigations have shown that a damaging bout of RE promotes an increase in HSP gene expression and protein content and inflammatory transcripts 1–48 h post-RE (Dickinson et al. 2018; Gordon et al. 2012; Mackey et al. 2011; Thalacker-Mercer et al. 2010; Thompson et al. 2003). Thus, increased gene expression of HSPs and inflammation 24 h post the first RE compared to the session in the trained state is in accordance with the greater stress/muscle damage observed initially in a RE training program (Damas et al. 2016b). Unaccustomed RE triggers a remodelling process, which we show resulted in an up-regulation of transcripts related to structural proteins reorganization/remodelling in the untrained vs trained state (Fig. 4). Protein remodelling involves breakdown and synthesis processes, which are both more intensely stimulated post-RE in an untrained state (Damas et al. 2015; Phillips et al. 1999; Tang et al. 2008). The up-regulation of genes related to proteolytic processes suggests a higher protein catabolism activity. Indeed, we observed enriched genes and pathways related to the remodelling process, specifically the ubiquitin–proteasome and calpain systems (Fig. 4). In accordance with our previously reported increase in protein synthesis 24 h post-RE in W1 vs W10 (Damas et al. 2016b), we observed increased gene expression related to protein processing in the endoplasmic reticulum, RNA synthesis and processing, and several ribosome biogenesis pathways (Fig. 4). Past studies have shown that an unaccustomed RE promotes a potent increase in muscle protein synthesis, which is higher in overall magnitude than the one promoted in a trained state (Damas et al. 2015, 2016b; Tang et al. 2008). However, recently ourselves (Damas et al. 2016b; Mitchell et al. 2014) and others (Mayhew et al. 2009) showed that even though the increase in muscle protein synthesis is potent in an untrained state, it does not correlate with eventual muscle hypertrophy. Interestingly, although lower in magnitude compared to the untrained state, the increase in muscle protein synthesis after RE in a more ‘trained’ state was shown to be correlated with muscle hypertrophy (Damas et al. 2016b). These past results indicate that the potent initial increase in muscle protein synthesis is somewhat non-specific to RE training-induced hypertrophy, but chronic RE results in a progressive adaptation with the increase in protein synthesis contributing more specifically to muscle hypertrophy (Damas et al. 2018a). Our current results appear congruent with a greater overall increase in muscle protein synthesis observed in an untrained state and a more refined response in the trained state (see discussion below regarding the trained state). In addition, our data indicate that protein translation capacity is acutely increased after RE at a gene level in the untrained and trained states, but more potently in the former. Specifically, gene/pathways of total RNA content and processing, as well as transcripts related to ribosome biogenesis were higher in the untrained compared with the trained state (Figs. 1, 3). Increased expression of the 45S pre-rRNA and the 40S ribosomal subunits protein components in young subjects was previously shown 24 h after a single RE session (Stec et al. 2015), and we expand these findings indicating that even after 10 weeks of RE training, RE still upregulated ribosomal biogenesis gene expression though in a lower magnitude. In addition, we report here that the trained state showed higher total RNA content than the untrained state (Fig. 2). The largest part of total RNA is composed by rRNA, thus an increase in total RNA in the trained state might indicate an increase in rRNA and, consequently, translational capacity due to increased ribosome density (Chaillou et al. 2014). Accordingly, it was shown that an 8-week RE training resulted in a basal increase of ribosomal subunits (18S, 5.8S, and 28S rRNA, and 45S pre-rRNA) (Figueiredo et al. 2015). Altogether, an increase in ribosome density (indicated by greater muscle total RNA levels), but reduced ribosomal biogenesis transcripts with training are in line with a recent study showing increased muscle total RNA, but a down-regulation of the 45S pre-rRNA expression levels with RE training (Mobley et al. 2018). Thus, we propose an increased transcriptional capacity due to enhanced ribosome density in the rested trained state, but a reduced up-regulation of ribosomal biogenesis after RE compared with the untrained state. Unfortunately, we could not directly assess specific ribosomal subunits expression and protein levels, such as 18S, 5.8S and 28S rRNA or even the 45S pre-rRNA, which would allow inferring, for example, if a downregulation of rDNA transcription could be responsible for the difference between training states as proposed by Mobley et al. (2018). These suggestions require further scrutinizing. Finally, the RE at W1 vs W10 has also upregulated genes associated with cell cycle (Fig. 4), which is in line with the increased satellite cell activity and commitment in an untrained state in response to a novel RE-induced stress (Bellamy et al. 2014; Damas et al. 2018b; Hyldahl et al. 2015).
Chronic repetition of RE stimuli results in positive muscle adaptations to stress, including reducing the magnitude of muscle damage, successfully remodelling/strengthening tissues, and attenuating and specializing muscle protein synthetic response towards muscle hypertrophy (Chen et al. 2009; Damas et al. 2016b; Tang et al. 2008). Changes in the muscle transcriptome at both rested and post-RE seem to reflect those stress adaptations (Figs. 1, 3, 4). Indeed, the analyses of muscle transcriptome at rest showed that the sole increased gene expression (Log2FoldChange > |1|) in the untrained vs trained muscles was MYH1 (Fig. 1), which transcribes the myosin heavy chain IIx. Similar results were found by Gordon et al. (2012). This result is in line with a change from myosin heavy chain IIx to IIa with RE training, a more oxidative form of type II fibres (Adams et al. 1993). Accordingly, the resting transcriptome of trained muscles suggests: (1) regulation of energy metabolism, favouring a more oxidative metabolism (with several genes and enriched terms involving oxidation pathways and mitochondria); (2) antioxidant and immune responses; and (3) cytoskeleton and extracellular matrix organization (Fig. 1 and Table S1). All of the above are in line with the trained muscle being less susceptible to stress and damage (Damas et al. 2016b). Finally, we found enriched terms related to protein metabolism and growth for the comparison between rested trained and untrained states. Specifically, enriched PI3K-Akt signalling pathway, which involves the mechanistic target of rapamycin (mTOR)–p70 ribosomal S6 kinase (p70S6K) down-stream [one of the main regulators of muscle growth (Bodine 2006)], suggest that the trained muscle is more prepared to increase muscle proteins translation to contribute to muscle hypertrophy. This finding is in line with data from Wilkinson et al. (2008), which demonstrated that proteins in PI3K-Akt pathway (e.g., Akt and the eukaryotic translation initiation factor 4E, eIF4E) showed greater phosphorylation at rest for the resistance-trained compared with the untrained state.
Similarly to rest, muscle transcriptome after RE in different training states indicates changes in the regulation of energy metabolism with RE training development. Specifically, the untrained vs the trained state showed enriched genes/terms related to glycolysis (Fig. 4). In addition, regardless of training state, oxidative metabolism pathway is downregulated after RE (Fig. 3), although it is worth noting that this acute down-regulation seems to be attenuated in the trained state, probably due to the basal improvement after prolonged period of RE training. Previously, it was found that RE could counteract the disuse-induced (i.e., 60 days of bed rest) downregulation of genes associated with the oxidative metabolism (e.g., tricarboxylic acid cycle-, key enzymes of fatty acid oxidation- and mitochondria-related genes) (Salanova et al. 2015). Together, these results indicate that trained muscles show transcriptome changes towards greater energetic efficiency also in response to acute RE. Additionally, the RE session upregulated genes and enriched pathways/terms in the trained vs untrained state related to structural changes of the extracellular matrix and muscle cytoskeleton, extra-intra cellular signalling transmission and muscle contraction (Fig. 4). Accordingly, Hyldahl et al. (2015) showed increased transcripts of integrin [a transmembrane protein that transmits mechanical forces among muscle fibres, extracellular matrix and tendon (Grounds et al. 2005)] 48 h into recovery after a RE session, and expression of extracellular matrix collagens later on (27 days) recovery after the same RE session. In addition, Gordon et al. (2012) demonstrated that RE training practice resulted in up-regulation of several transcripts related to extracellular matrix, muscle cytoskeleton and contractile machinery post-RE. These studies corroborate with our results suggesting a continuous adaptation on muscle mechanical structures/properties throughout RE training.
Our results also showed that in response to RE, trained muscles have greater up-regulation of genes related to antioxidant and immune response to stress (Fig. 4). Accordingly, Gordon et al. (2012) showed that RE training downregulated the expression of genes related to unfolded proteins, HSP, inflammation and apoptosis 4 h following RE. The analysis in a time-point further into recovery (i.e., 24 h) post-RE allowed discovering other muscle physiological processes occurring at the transcriptome level, showing that the trained vs untrained state upregulated genes related to: (1) cellular growth and muscle development; (2) WNT pathway (e.g., gene: SFRP4), suggested to be involved in mechanical overload, muscle hypertrophy and satellite cell proliferation (Leal et al. 2011; Steelman et al. 2006); (3) aminoacids biosynthesis and protein absorption; and (4) increased myofibrillar protein synthesis (Fig. 4 and Table S1), which we previously showed to be directed to promote muscle hypertrophy in a more-trained phase (Damas et al. 2016b). Finally, it is interesting to note that the between-subject variability in the transcriptome response is increased with RE compared to rest regardless of training state (Fig. 5). This is in line with a large inter-subject variability in response to the stress induced by an unaccustomed RE as well as the hypertrophic adaptations resulted from repeated RE sessions. Specifically, we (Damas et al. 2016a) and others (Chen 2006; Gulbin and Gaffney 2002; Paulsen et al. 2010) have previously shown a large inter-subject variability to muscle damage and inflammation in response to RE in an untrained state. In addition, other research have shown a high variability between subjects in muscle hypertrophic response after RE training period (Ahtiainen et al. 2016; Churchward-Venne et al. 2015).
The analysis and results presented herein on the regulation of specific genes and enriched terms provide information on the changes in global muscle transcriptional with acute and chronic RE training practice, thus does not necessarily result in changes in the respective physiological functions. However, a strength of our manuscript is the ability to relate the transcriptional changes to previous research evaluating the adaptations in physiological function. Therefore, our data of rested and 24 h post-RE global muscle transcriptome analyses in different training states contribute to our understanding of skeletal muscle adaptation throughout RE training.
Conclusion
The modulation of gene transcripts reflected the high stress that was promoted by an unaccustomed RE, relating to muscle damage and inflammation, muscle scaffold structure (extracellular matrix and cytoskeleton) remodelling, high protein processing and turnover and increased translational capacity. As RE training progresses, our results show changes in the metabolic response, a refined response to stress (increasing antioxidant- and immune-related genes/terms), and upregulation of gene transcripts related to muscle structure and contractile function. In addition, changes in the transcriptome of trained muscles relate to muscle growth and development that contribute to muscle hypertrophy.
Abbreviations
- FDR:
-
False discovery rate
- GO:
-
Gene ontology
- HSP:
-
Heat shock proteins
- mTOR:
-
Mechanistic target of rapamycin
- p70S6K:
-
p70 Ribosomal S6 kinase
- RE:
-
Resistance exercise
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- W1:
-
Untrained state
- W10:
-
Trained state
References
Adams GR, Hather BM, Baldwin KM, Dudley GA (1993) Skeletal muscle myosin heavy chain composition and resistance training. J Appl Physiol 74:911–915. https://doi.org/10.1152/jappl.1993.74.2.911
Ahtiainen JP et al (2016) Heterogeneity in resistance training-induced muscle strength and mass responses in men and women of different ages. Age 38:10. https://doi.org/10.1007/s11357-015-9870-1
Bellamy LM et al (2014) The acute satellite cell response and skeletal muscle hypertrophy following resistance training. PLoS One 9:e109739. https://doi.org/10.1371/journal.pone.0109739
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Methodol 57:289–300
Bodine SC (2006) mTOR signaling and the molecular adaptation to resistance exercise. Med Sci Sports Exerc 38:1950–1957. https://doi.org/10.1249/01.mss.0000233797.24035.35
Chaillou T, Kirby TJ, McCarthy JJ (2014) Ribosome biogenesis: emerging evidence for a central role in the regulation of skeletal muscle mass. J Cell Physiol 229:1584–1594. https://doi.org/10.1002/jcp.24604
Chen TC (2006) Variability in muscle damage after eccentric exercise and the repeated bout effect Res. Q Exerc Sport 77:362–371. https://doi.org/10.1080/02701367.2006.10599370
Chen TC, Chen HL, Lin MJ, Wu CJ, Nosaka K (2009) Muscle damage responses of the elbow flexors to four maximal eccentric exercise bouts performed every 4 weeks. Eur J Appl Physiol 106:267–275. https://doi.org/10.1007/s00421-009-1016-7
Churchward-Venne TA, Tieland M, Verdijk LB, Leenders M, Dirks ML, de Groot LC, van Loon LJ (2015) There are no nonresponders to resistance-type exercise training in older men and women. J Am Med Dir Assoc 16:400–411. https://doi.org/10.1016/j.jamda.2015.01.071
Damas F, Phillips S, Vechin FC, Ugrinowitsch C (2015) A review of resistance training-induced changes in skeletal muscle protein synthesis and their contribution to hypertrophy. Sports Med 45:801–807. https://doi.org/10.1007/s40279-015-0320-0
Damas F, Nosaka K, Libardi CA, Chen TC, Ugrinowitsch C (2016a) Susceptibility to exercise-induced muscle damage: a cluster analysis with a large sample. Int J Sports Med 37:633–640. https://doi.org/10.1055/s-0042-100281
Damas F et al (2016b) Resistance training-induced changes in integrated myofibrillar protein synthesis are related to hypertrophy only after attenuation of muscle damage. J Physiol 594:5209–5222. https://doi.org/10.1113/JP272472
Damas F, Libardi CA, Ugrinowitsch C (2018a) The development of skeletal muscle hypertrophy through resistance training: the role of muscle damage and muscle protein synthesis. Eur J Appl Physiol 118:485–500. https://doi.org/10.1007/s00421-017-3792-9
Damas F et al (2018b) Early- and later-phases satellite cell responses and myonuclear content with resistance training in young men. PLoS One 13:e0191039. https://doi.org/10.1371/journal.pone.0191039
Dickinson JM et al (2018) Transcriptome response of human skeletal muscle to divergent exercise stimuli. J Appl Physiol 124:1529–1540. https://doi.org/10.1152/japplphysiol.00014.2018
Drummond MJ et al (2009) Expression of growth-related genes in young and older human skeletal muscle following an acute stimulation of protein synthesis. J Appl Physiol 106:1403–1411. https://doi.org/10.1152/japplphysiol.90842.2008
Figueiredo VC, Caldow MK, Massie V, Markworth JF, Cameron-Smith D, Blazevich AJ (2015) Ribosome biogenesis adaptation in resistance training-induced human skeletal muscle hypertrophy. Am J Physiol Endocrinol Metab 309:E72–E83. https://doi.org/10.1152/ajpendo.00050.2015
Gordon PM et al (2012) Resistance exercise training influences skeletal muscle immune activation: a microarray analysis. J Appl Physiol 112:443–453. https://doi.org/10.1152/japplphysiol.00860.2011
Grounds MD, Sorokin L, White J (2005) Strength at the extracellular matrix-muscle interface. Scand J Med Sci Sports 15:381–391. https://doi.org/10.1111/j.1600-0838.2005.00467.x
Gulbin JP, Gaffney PT (2002) Identical twins are discordant for markers of eccentric exercise-induced muscle damage. Int J Sports Med 23:471–476. https://doi.org/10.1055/s-2002-35076
Huber W et al (2015) Orchestrating high-throughput genomic analysis with. Bioconductor Nat Methods 12:115–121. https://doi.org/10.1038/nmeth.3252
Hyldahl RD et al (2015) Extracellular matrix remodeling and its contribution to protective adaptation following lengthening contractions in human muscle. FASEB J 29:2894–2904. https://doi.org/10.1096/fj.14-266668
Lamas L, Aoki MS, Ugrinowitsch C, Campos GE, Regazzini M, Moriscot AS, Tricoli V (2010) Expression of genes related to muscle plasticity after strength and power training regimens. Scand J Med Sci Sports 20:216–225. https://doi.org/10.1111/j.1600-0838.2009.00905.x
Leal ML, Lamas L, Aoki MS, Ugrinowitsch C, Ramos MS, Tricoli V, Moriscot AS (2011) Effect of different resistance-training regimens on the WNT-signaling pathway. Eur J Appl Physiol 111:2535–2545. https://doi.org/10.1007/s00421-011-1874-7
Liu D et al (2010) Skeletal muscle gene expression in response to resistance exercise: sex specific regulation. BMC Genom 11:659. https://doi.org/10.1186/1471-2164-11-659
Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ (2009) GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinform 10:161. https://doi.org/10.1186/1471-2105-10-161
Mackey AL et al (2011) Sequenced response of extracellular matrix deadhesion and fibrotic regulators after muscle damage is involved in protection against future injury in human skeletal muscle. FASEB J 25:1943–1959. https://doi.org/10.1096/fj.10-176487
Mahoney DJ et al (2008) Gene expression profiling in human skeletal muscle during recovery from eccentric exercise. Am J Physiol Regul Integr Comp Physiol 294:R1901–R1910. https://doi.org/10.1152/ajpregu.00847.2007
Mayhew DL, Kim JS, Cross JM, Ferrando AA, Bamman MM (2009) Translational signaling responses preceding resistance training-mediated myofiber hypertrophy in young and old humans. J Appl Physiol 107:1655–1662. https://doi.org/10.1152/japplphysiol.91234.2008
Mitchell CJ et al (2014) Acute post-exercise myofibrillar protein synthesis is not correlated with resistance training-induced muscle hypertrophy in young men. PLoS One 9:e89431. https://doi.org/10.1371/journal.pone.0089431
Mobley CB et al (2018) Biomarkers associated with low, moderate, and high vastus lateralis muscle hypertrophy following 12 weeks of resistance training. PLoS One 13:e0195203. https://doi.org/10.1371/journal.pone.0195203
Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M (1999) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 27:29–34
Paulsen G, Crameri R, Benestad HB, Fjeld JG, Morkrid L, Hallen J, Raastad T (2010) Time course of leukocyte accumulation in human muscle after eccentric exercise. Med Sci Sports Exerc 42:75–85. https://doi.org/10.1249/MSS.0b013e3181ac7adb
Peake J, Nosaka K, Suzuki K (2005) Characterization of inflammatory responses to eccentric exercise in humans. Exerc Immunol Rev 11:64–85
Phillips SM, Tipton KD, Ferrando AA, Wolfe RR (1999) Resistance training reduces the acute exercise-induced increase in muscle protein turnover. Am J Physiol 276:E118–E124
Phillips BE et al (2013) Molecular networks of human muscle adaptation to exercise and age. PLoS Genet 9:e1003389. https://doi.org/10.1371/journal.pgen.1003389
Raue U, Trappe TA, Estrem ST, Qian HR, Helvering LM, Smith RC, Trappe S (2012) Transcriptome signature of resistance exercise adaptations: mixed muscle and fiber type specific profiles in young and old adults. J Appl Physiol 112:1625–1636. https://doi.org/10.1152/japplphysiol.00435.2011
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. https://doi.org/10.1093/nar/gkv007
Roth SM, Ferrell RE, Peters DG, Metter EJ, Hurley BF, Rogers MA (2002) Influence of age, sex, and strength training on human muscle gene expression determined by microarray. Physiol Genom 10:181–190. https://doi.org/10.1152/physiolgenomics.00028.2002
Salanova M et al (2015) Vibration mechanosignals superimposed to resistive exercise result in baseline skeletal muscle transcriptome profiles following chronic disuse in bed rest. Sci Rep 5:17027. https://doi.org/10.1038/srep17027
Shi W et al (2010a) Estimating the proportion of microarray probes expressed in an RNA sample. Nucleic Acids Res 38:2168–2176. https://doi.org/10.1093/nar/gkp1204
Shi W, Oshlack A, Smyth GK (2010b) Optimizing the noise versus bias trade-off for Illumina whole genome expression BeadChips. Nucleic Acids Res 38:e204. https://doi.org/10.1093/nar/gkq871
Smith ML, Baggerly KA, Bengtsson H, Ritchie ME, Hansen KD (2013) illuminaio: an open source IDAT parsing tool for Illumina microarrays F1000Res 2:264 https://doi.org/10.12688/f1000research.2-264.v1
Stec MJ, Mayhew DL, Bamman MM (2015) The effects of age and resistance loading on skeletal muscle ribosome biogenesis. J Appl Physiol 119:851–857. https://doi.org/10.1152/japplphysiol.00489.2015
Steelman CA, Recknor JC, Nettleton D, Reecy JM (2006) Transcriptional profiling of myostatin-knockout mice implicates Wnt signaling in postnatal skeletal muscle growth and hypertrophy. FASEB J 20:580–582. https://doi.org/10.1096/fj.05-5125fje
Tang JE, Perco JG, Moore DR, Wilkinson SB, Phillips SM (2008) Resistance training alters the response of fed state mixed muscle protein synthesis in young men. Am J Physiol Regul Integr Comp Physiol 294:R172–R178. https://doi.org/10.1152/ajpregu.00636.2007
Thalacker-Mercer AE, Dell’Italia LJ, Cui X, Cross JM, Bamman MM (2010) Differential genomic responses in old vs. young humans despite similar levels of modest muscle damage after resistance loading. Physiol Genomics 40:141–149. https://doi.org/10.1152/physiolgenomics.00151.2009
Thalacker-Mercer A, Stec M, Cui X, Cross J, Windham S, Bamman M (2013) Cluster analysis reveals differential transcript profiles associated with resistance training-induced human skeletal muscle hypertrophy. Physiol Genomics 45:499–507. https://doi.org/10.1152/physiolgenomics.00167.2012
Thompson HS, Maynard EB, Morales ER, Scordilis SP (2003) Exercise-induced HSP27, HSP70 and MAPK responses in human skeletal muscle. Acta Physiol Scand 178:61–72. https://doi.org/10.1046/j.1365-201X.2003.01112.x
Welle S, Tawil R, Thornton CA (2008) Sex-related differences in gene expression in human skeletal muscle. PLoS One 3:e1385. https://doi.org/10.1371/journal.pone.0001385
Wilkinson SB, Phillips SM, Atherton PJ, Patel R, Yarasheski KE, Tarnopolsky MA, Rennie MJ (2008) Differential effects of resistance and endurance exercise in the fed state on signalling molecule phosphorylation and protein synthesis in human muscle. J Physiol 586:3701–3717. https://doi.org/10.1113/jphysiol.2008.153916
Witard OC, Jackman SR, Breen L, Smith K, Selby A, Tipton KD (2014) Myofibrillar muscle protein synthesis rates subsequent to a meal in response to increasing doses of whey protein at rest and after resistance exercise Am. J Clin Nutr 99:86–95. https://doi.org/10.3945/ajcn.112.055517
Wu D, Smyth GK (2012) Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res 40:e133. https://doi.org/10.1093/nar/gks461
Zierath JR, Wallberg-Henriksson H (2015) Looking ahead perspective: where will the future of exercise biology take us? Cell Metab 22:25–30. https://doi.org/10.1016/j.cmet.2015.06.015
Acknowledgements
We acknowledge all the volunteers that participated in this study. This study was supported by the São Paulo Research Foundation (FAPESP) Grants (#2013/21218-4 and #2017/04299-1 to CAL), CAPES-PROEX and Natural Science and Engineering Research Council (NSERC) of Canada (RGPIN-2015-04613 to SMP). SMP also acknowledges support from the Canada Research Chairs program. FD was supported by FAPESP Grants (#2012/24499-1, #2014/19594-0, #2016/24259-1 and #2018/13064-0). CU, HR and VT are supported by the National Council for Scientific and Technological Development (CNPq) Grant (CU: #303085/2015-0 and #448387/2014-0; HR: #307023/2014-1; VT: #310823/2013-7).
Author information
Authors and Affiliations
Contributions
FD had original idea of the study and the final study design was developed by FD, CU, CAL, MEL, FCV, VT, HR and SMP. Participants were recruited and trained at the School of Physical Education and Sport, University of São Paulo, by FD, MEL, FCV and PRJ. AJH and CM conducted the RNA preparation to the microarray procedure. HM performed the biostatistical procedures and analyses. All authors participated in the interpretation of the data. FD, HM and PRJ designed the figures and supplementary tables. FD wrote the first version of the manuscript. All authors contributed to the manuscript, reviewed it, and approved the content of the final version.
Corresponding author
Ethics declarations
Conflict of interest
The authors do not have any conflicts of interest financial or otherwise to declare.
Ethical standard
All procedures performed herein were in accordance with the ethical standards of the institutional and national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Additional information
Communicated by Phillip D. Chilibeck.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Damas, F., Ugrinowitsch, C., Libardi, C.A. et al. Resistance training in young men induces muscle transcriptome-wide changes associated with muscle structure and metabolism refining the response to exercise-induced stress. Eur J Appl Physiol 118, 2607–2616 (2018). https://doi.org/10.1007/s00421-018-3984-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00421-018-3984-y